Abstract
Microsatellite markers were used to genetically characterize 19 Culex pipiens complex populations from California. Two populations showed characteristics of earlier genetic bottlenecks. The overall FST value and a neighbor-joining tree suggested moderate amounts of genetic differentiation. Analyses using Structure indicated K = 4 genetic clusters: Cx. pipiens form pipiens L., Cx. quinquefasciatus Say, Cx. pipiens form molestus Forskäl, and a group of genetically similar individuals of hybrid origin. A Discriminant Analysis of Principal Components indicated that the latter group is a mixture of the other three taxa, with form pipiens and form molestus contributing somewhat more ancestry than Cx. quinquefasciatus. Characterization of 56 morphologically autogenous individuals classified most as Cx. pipiens form molestus, and none as Cx. pipiens form pipiens or Cx. quinquefasciatus. Comparison of California microsatellite data with those of Cx. pipiens pallens Coquillett from Japan indicated the latter does not contribute significantly to genotypes in California.
Introduction
The Culex pipiens complex is an important group of mosquito vectors that transmit several encephalitic viruses, including West Nile and St. Louis encephalitis viruses.1,2 Members are found in temperate and tropical regions throughout the world, and the complex is notable for the variety of ecological, behavioral, and physiological adaptations present within such a closely related group of organisms. In the United States, members include Cx. pipiens form pipiens L., Cx. quinquefasciatus Say, their hybrids, and the autogenous form of Cx. pipiens known as form molestus Forskäl. Adaptations among taxa in the complex include epidemiologically relevant characters such as the presence or absence of seasonal reproductive diapause, which can influence the degree and nature of virus transmission,3 and whether females are autogenous (capable of producing a first batch of eggs without a blood meal). Variation in vector competence has been observed in Cx. pipiens complex populations in the state of California, and it has been suggested that such variation has a genetic component.4–6
A characteristic that potentially effects the distribution of adaptations within the Cx. pipiens complex is that Cx. pipiens and Cx. quinquefasciatus are fully interfertile, as are the resultant hybrids. This feature has resulted in a large stable hybrid zone across the United States, the limits of which Barr7 set at 36°N and 39°N based on measurements of the dorsal and ventral arms of the male genitalia (DV/D ratio). Subsequent work using microsatellites8–11 has indicated that the hybrid zone extends farther north and south than suggested by Barr.7 Whether this difference is caused by hybrid zone expansion over time or to the improved power of microsatellites over morphology to detect admixed mosquitoes is unknown, and both scenarios may be true. Regardless, there are areas in the United States where Cx. pipiens complex populations consist largely of genetically admixed mosquitoes. Interspecific hybridization produces new genotypes through genetic recombination, and thus provides natural selection with more variation on which to act. Such genetic variation may enable populations to adapt to local conditions, and can lead to genetic differentiation. Recent work has examined the possible effect of genetic introgression on host choice among Cx. pipiens complex mosquitoes, and a small number of studies have suggested the possibility that genes that confer a preference for biting humans could be transferred from form molestus to form pipiens mosquitoes, which could potentially increase the transmission of arboviruses to humans.12–15
The interaction of these two features of the complex, interfertility and a variety of life-history strategies makes the taxonomic designation of a particular Cx. pipiens complex population occasionally ambiguous. Characterizing the Cx. pipiens complex in California is made more challenging because its distribution is also influenced by geographic (and associated temperature) regimens that are more complex than simple clines. Barr7 examined DV/D ratios from individuals in several populations in California when describing the distribution of species and hybrids in North America, and noted the existence of admixed populations. A dissertation by Iltis (Iltis WG, unpublished dissertation) concluded that Cx. quinquefasciatus was found in expected locations in the southern part of the state, but also north of Sacramento, whereas Cx. pipiens was found in Sacramento and again in the northern part of the state. The work of Iltis also characterized hybridization within the complex, noting several stable zones of hybridization. Tabachnick and Powell16 sought to confirm the results of the study of Iltis, and in addition to DV/D ratios, examined several allozyme loci. Although morphologic data from both studies agreed with respect to the distribution of species (then called subspecies) around Sacramento, variation in allozyme frequencies did not correlate well with that of DV/D ratios. Tabachnik and Powell16 also made the distinction between hybrid populations made up of largely F1 individuals and the stable freely interbreeding populations they observed, and noted that taxonomic classification of populations in California as Cx. pipiens or Cx. quinquefasciatus for epidemiologic reasons could be misleading.
Urbanelli and others 17 used DV/D ratios and a panel of six allozymes that included several new loci to assay populations in California. They noted that temperature alone was insufficient to explain the distribution of DV/D ratios and that the pattern of allozyme frequencies suggested that populations were responding to microclimatic changes over the past several decades. This response resulted in the movement of Cx. pipiens southward, which shifted the southern limit of the hybrid zone. Comparing Cx. pipiens complex populations in California and South Africa using DV/D ratios and 11 isoenzyme loci, Cornell and others18 noted differences between the two sampling locations, which suggested Cx. pipiens complex taxa were less taxonomically distinct in California than in South Africa, where populations displayed limited to no introgression between species. Using a single-gene assay (Ace.2) and DV/D ratios to identify Cx. pipiens complex mosquitoes in Fresno County, California (located within the hybrid zone) McAbee and others19 sought to correlate West Nile virus infection rate with taxonomic designation. They found both identification methods uninformative with regard to classifying Cx. pipiens complex individuals and concluded that there is “no appropriate diagnostic character” to separate Cx. pipiens from Cx. quinquefasciatus in California.
The presence of autogenous individuals in California was noted by Barr.7 Iltis also mentions autogeny in CA populations, but because the DV/D ratio is identical in form molestus and form pipiens, few morphologic studies have knowingly included form molestus mosquitoes. A recent study by Strickman and Fonseca20 used morphology and eight microsatellite loci to characterize autogenous individuals in 13 above-ground sites around San Francisco, California. They observed a variety of phenotypes among and within sites, and suggested such variety could promote adaptation to local microclimates.
As the above mentioned studies suggest, there is considerable ambiguity in the taxonomic designations of Cx. pipiens complex populations in California. Furthermore, to our knowledge, the degree and extent of genetic admixture of Cx. pipiens complex populations in the state has not been recently characterized using a north–south transect. Although it is true that no one characteristic reliably distinguishes taxa within admixed populations, a panel of microsatellite markers can be useful for this purpose. Our aim in the current study was to characterize populations that represent much of the genotypic variation in California with microsatellites. Specifically, our objectives were to 1) quantify genetic diversity and differentiation within and among populations; 2) determine the most likely number of genetically distinct clusters, and to assign individuals to those clusters; 3) evaluate the respective contributions of Cx. pipiens and Cx. quinquefasciatus to admixed clusters; 4) determine whether other Culex taxa have introgressed into Cx. pipiens complex populations in California; and 5) determine whether morphological determinations of autogeny (or anautogeny) are consistent with genetic cluster assignments.
Methods
Sampling.
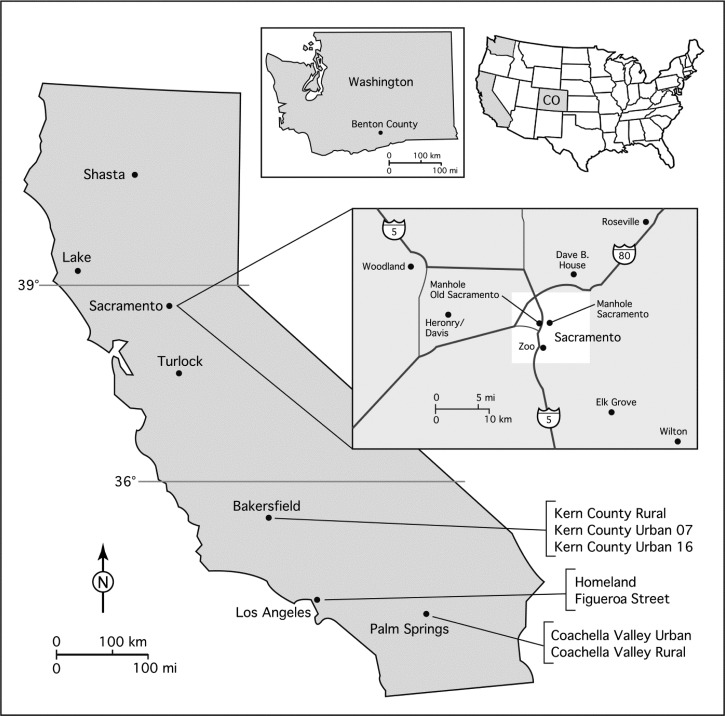
Specimens were collected from 19 sites in California and one site in Benton County, Washington (Figure 1 and Table 1). Sites were chosen that would sample the genetic diversity of Cx. pipiens complex populations. In particular, several populations were sampled around the Sacramento area because of published7,16 and observed (Nelms B, unpublished data) reports of autogenous mosquitoes and mixed autogenous and anautogenous populations being present at the same localities in the area. Specimens were collected with a variety of methods, including gravid traps,21 encephalitis vector survey suction traps (Bioquip, Rancho Domingo, CA) baited with and without CO2, CDC light traps (John W. Hock Company, Gainesville, FL), vacuum aspiration, and larval collections. In cases where gravid females were allowed to oviposit and families were reared for other experiments, only one mosquito per family was used for microsatellite analysis. Collection details varied by site and are shown in Supplemental Table 1.
Figure 1.
Map of sample sites. Shading on small United States map indicates states where specimens were collected. Map inset shows populations sampled around the city of Sacramento, California. The state of Colorado is indicated as CO on the small map of the United States.
Table 1.
Site names with abbreviations, number of specimens sampled, and location information for populations in this study, California
| Location | No. | Latitude, °N | Longitude, °W |
|---|---|---|---|
| Coachella Valley Rural (CVRu) | 13 | 33.470 | −116.090 |
| Coachella Valley Urban (CVUrb) | 13 | 33.780 | −116.430 |
| Homeland (Home) | 42 | 33.937 | −117.991 |
| Figueroa Street (FigSt) | 10 | 33.782 | −118.280 |
| Kern County Rural (KCRu) | 43 | 35.494 | −119.170 |
| Kern County Urban07 (KCU7) | 32 | 35.380 | −119.111 |
| Kern County Urban16 (KCU6) | 18 | 35.410 | −119.126 |
| Turlock (Turl) | 37 | 37.513 | −121.117 |
| Wilton (Wilt) | 42 | 38.383 | −121.219 |
| Elk Grove (ElkG) | 42 | 38.418 | −121.357 |
| Zoo | 35 | 38.540 | −121.505 |
| Manhole Sacramento (ManS) | 44 | 38.585 | −121.491 |
| Manhole Old Sac (ManOS) | 81 | 38.584 | −121.504 |
| Heronry/Davis (Heron) | 28 | 38.603 | −121.711 |
| Dave B House (DBH) | 33 | 38.661 | −121.447 |
| Woodland (Wood) | 67 | 38.673 | −121.782 |
| Roseville (Rose) | 14 | 38.748 | −121.279 |
| Lake | 34 | 38.938 | −122.667 |
| Shasta (Shas) | 46 | 40.480 | −122.347 |
| Benton County, WA (Bent) | 24 | 46.199 | −119.893 |
In addition, 193 females from the Sacramento Zoo (n = 39), Manhole Old Sacramento (n = 67), Dave B House (n = 36), Woodland (n = 39), and Heronry/Davis (n = 12) populations were reared from field-collected gravid females and examined under an SZ3060 (Olympus America Inc., Lake Success, NY) dissecting microscope to determine whether they were autogenous. Primary follicles were classified morphologically by the degree of vitellogenesis in the most mature follicles.22,23 A female was classified as autogenous when, in most follicles, the oocyte occupied between 50% (stage III) and 100% (stage V, fully formed egg) of the follicle length. Microsatellite analysis was performed on a subset of these mosquitoes (see Cluster Analyses).
Initial morphological screening.
Specimens were sorted under a dissecting microscope. Using dichotomous keys,24 those positively identified as belonging to the Cx. pipiens complex were placed in individual tubes and frozen at −80°C until they could be processed for DNA extraction. No attempt was made to morphologically distinguish Cx. pipiens from Cx. quinquefasciatus or hybrids.
Using a tissue homogenizer (Qiagen, Valencia, CA), individual mosquitoes were ground with a copper BB in 0.5 mL of BA-1 diluent.1 DNA was extracted using a liquid handling robot (Qiagen) from a 220 μl aliquot of the homogenate and the remaining sample was frozen. One hundred microliters of DNA were eluted from each sample, and two microliters of DNA were used in each subsequent PCR reaction.
Microsatellite analysis.
A panel of 17 microsatellite loci was used to generate a multilocus genotype for each individual. The panel consisted of two multiplexes of eight and nine markers each. The forward primer of each primer pair was fluorescently labeled for subsequent visualization. Primer names and concentrations per PCR are similar to those of Kothera and others25 and are shown in Supplemental Table 2. Each PCR included approximately 20 ng of genomic DNA, 1× PCR buffer containing Mg, 0.6 μM additional Mg, 200 μM each dNTP (Invitrogen, Grand Island, NY), 0.5 units of hot-start Taq polymerase (Hotstar; QIAGEN), and primers as described above. The reaction volume was 20 μL, and samples were run on a DNA Engine (Bio-Rad Laboratories, Hercules, CA) with the following program: 10 minutes at 95°C (which activates the hot-start Taq); 5 minutes at 96°C; 35 cycles for 45 seconds at 94°C, 45 seconds at 54°C, and 45 seconds at 72°C; and 10 minutes at 72°C. Labeled PCR products were visualized on a Beckman-Coulter (Fullerton, CA) CEQ8000 sequencer with a 400-basepair standard included in each sample. Multilocus genotypes were generated using the manufacturer's Fragment Analysis Module software. Approximately 10% of samples were run more than once and the results from each run were identical.
Data analysis: genetic diversity.
The program Convert26 was used to format genotypic data for use in the programs Arlequin,27 Structure,28 and FSTAT,29 and to generate a table of allele frequencies. The within-population estimates of genetic diversity, Observed (HO) and Expected (HE) heterozygosity were generated using Nei's unbiased estimate30 in Arlequin. Arlequin was also used to calculate the statistical significance of departures from Hardy-Weinberg Equilibrium (HWE) and the degree of linkage disequilibrium (LD) using Fisher's Exact tests.31 To determine how genetic variance was partitioned at various levels of organization (individual, within populations and among populations), an analysis of molecular variance (AMOVA) was performed in Arlequin. Fixation indices (F statistics) were generated as part of the AMOVA output using the method of Weir and Cockerham.32 Allelic richness was calculated using FSTAT's sample size-corrected method and averaged over loci for each population. The data were also analyzed with the program Bottleneck33 to determine whether there was a significant excess of heterozygotes, which is typical of populations growing in size after a reduction in their numbers. We used two of Bottleneck's analyses to determine if populations had the genetic signature of a bottleneck, the one-tailed Wilcoxon test for heterozygote excess, and the Mode Shift test, a graphic analysis that examines the distribution of allele frequencies.
Data analysis: genetic differentiation.
Pairwise FST values between populations were generated in Arlequin. The statistical significance of pairwise FST comparisons was determined by permutation test. A neighbor-joining tree was constructed using the following steps in Phylip.34 First, 1,000 bootstrap replicate datasets were generated by using the program SeqBoot. Next, GenDist was used to calculate a distance matrix for each replicate data set based on chord distances of Cavalli-Sforza and Edward.35 The program Neighbor was then used to make a neighbor-joining tree from each distance matrix, and Consense was used to select a consensus tree with bootstrap support values. Finally, Drawtree was used to make an unrooted phenogram of the consensus tree. In addition to the California populations, two known Cx. pipiens form pipiens populations were included in this analysis, the one collected for this study from Washington and another from Grand Junction, Colorado.25
Cluster analyses.
The program Structure was used in several ways. First, we used it to determine the most likely number of genetic groups, or clusters (K). Each run consisted of 50,000 burn-in steps and 100,000 data collecting steps using the default parameters (i.e., the Admixture Model without prior population information) and ten runs were performed for each value of K ranging from 1 to 10. Structure determines a posterior likelihood value (LnP(D)) for each run, and these values were compared across runs to determine which value of K was most likely for the data. One set of runs included only the California populations, and a second set included microsatellite data from California, as well as the two Cx. pipiens form pipiens populations from Washington and Colorado. Both the California-only run and the run with the Washington and Colorado populations were repeated with a series of longer runs (50,000 burn-ins and 1,000,000 iterations of the model) to assess the stability of assignments of mosquitoes to clusters by using the program CLUMPP.36 CLUMPP uses Structure output files as input and returns values for H (the average symmetric similarity coefficient), in which a maximum value of 1 indicates that each individual was assigned to the same cluster(s) consistently across runs. The ΔK method of Evanno and others37 was also examined. Results from Structure runs were visualized by using the program Distruct.38
Second, Structure was used to assign individuals to clusters. Specimens from the Washington and Colorado populations were included to ensure that pure Cx. pipiens form pipiens individuals were present in the analysis. Individual assignments to each cluster (i.e., q values) were examined to determine which individuals and populations were assigned to each cluster, and the number of clusters was determined by the previous Structure analyses. Given the observed taxonomic uncertainty of specimens collected in California, we set a q value ≥ 0.80 as representative of a pure individual in a cluster. Mosquitoes with q values < 0.80 are probably hybrids of one or more clusters. After individuals were assigned to clusters, another Structure run was performed with the subset of individuals (n = 472 of 739) with q values ≥ 0.80 (again with no prior population information) to determine whether further population subdivision would be evident without the presence of highly admixed individuals.
Third, we used Structure to examine a subset of the microsatellite data for females from the Zoo, Manhole Old Sacramento, Dave B House, Heronry/Davis, and Woodland populations that were examined morphologically before DNA extraction to determine whether they were autogenous. The assignments of these mosquitoes were assessed to determine how many assigned to a genetic cluster or were deemed hybrids.
Finally, we genotyped small numbers of mosquitoes from several other taxa to explore whether hybridization with them had left a discernible genetic signature on California populations. DNA from 15 Cx. pipiens pallens Coquillett individuals collected in Sakata City, Amagata Prefecture, northern Honshu, Japan, was processed and genotyped as above. In addition, three Cx. stigmatosoma Dyar and six Cx. restuans Theobald collected in Fresno County near Centerville, California, were examined morphologically using dichotomous keys24,39 and genotyped as described above.
After a most likely value of K was determined, the data from the run including the California, Washington, and Colorado populations were converted to the GENEPOP40,41 format so they could be analyzed with a Discriminant Analysis of Principal Components (DAPC) using the R package adegenet.42,43 This procedure maximizes the amount of separation between groups, which was useful for visualizing relationships among genetic clusters. Before analysis, an a-score test in adegenet was run to determine the optimal number of principal components to retain in the DAPC. The number of groups was set equal to the most likely number of clusters determined by Structure, although the data were organized by population. Unlike Structure, a DAPC assigns entire individuals to a cluster instead of assigning proportion of membership (q values) in those clusters. We therefore confirmed that assignments of proportions of populations to clusters were similar in both analyses (Supplemental Figure 1). Consequently, the DAPC was not used as a clustering method per se, but rather to discern how the clusters already determined by Structure would plot relative to each other in the multidimensional space of the DAPC analysis. After the analysis, a Loading Plot was generated using adegenet that indicated which loci contributed most to separating the groups.
Results
Genetic diversity.
Because of PCR amplification issues, two loci, Cxpq68 and Cxpq114, were excluded, leaving 15 loci for subsequent analyses. Genetic diversity measures for loci and populations in the study are shown in Table 2. The Manhole Sacramento population had the lowest genetic diversity of all sites, and the Roseville population had the highest. Overall HE ranged from 0.519 to 0.658. Several loci exhibited noticeably different degrees of diversity between groups of populations. For example, locus Cxpq78 had high diversity in Cx. quinquefasciatus populations and low diversity in the other populations. This pattern was reversed for two other loci, CxqTri4F and CxqCTG10, in which genetic diversity was low in Cx. quinquefasciatus and higher in Cx. pipiens and admixed populations. Allelic richness varied from 3.09 in the Manhole Sacramento population to 4.14 in Roseville. Three other populations had allelic richness values close to that of Roseville: Woodland, Heronry/Davis and Lake (A = 4.13). There was no statistically significant difference among allelic richness values among populations (P = 0.457, by one-way ANOVA on ranks).
Table 2.
Genetic diversity measures for loci and population in this study, California*
| Location | A | H | Cxpq5154 | Cxpq5954 | Cxpq6954 | Cxpq7854 | Cxpq7954 | Cxpq10954 | Cxpq11054 | Cxpq11754 | Cxpq11954 | CxqGT4F55 | CxqTri4F55 | CxpGT46F56 | CxpGT51F56 | CxqCTG1057 | CxqCAG10157 | Population averages | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coachella | HO | 0.692 | 0.154 | 0.909 | 0.692 | 0.800 | 0.769 | 0.615 | 0.200 | 0.308 | 0.462 | N/A | 0.538 | 0.692 | 0.077 | 0.462 | HO | 0.526 | |
| Valley Rural | 3.16 | HE | 0.711 | 0.151 | 0.788 | 0.729 | 0.742 | 0.723 | 0.732 | 0.442 | 0.271 | 0.618 | N/A | 0.711 | 0.772 | 0.077 | 0.492 | HE | 0.569 |
| Coachella | HO | 0.615 | 0.385 | 0.364 | 0.769 | 0.429 | 0.667 | 0.615 | 0.455 | 0.538 | 0.692 | N/A | 0.692 | 0.615 | 0.077 | 0.462 | HO | 0.527 | |
| Valley Urban | 3.36 | HE | 0.615 | 0.342 | 0.745 | 0.711 | 0.791 | 0.725 | 0.711 | 0.571 | 0.538 | 0.680 | N/A | 0.566 | 0.788 | 0.077 | 0.492 | HE | 0.597 |
| Homeland | HO | 0.659 | 0.500 | 0.800 | 0.762 | 0.625 | 0.667 | 0.643 | 0.357 | 0.333 | 0.667 | 0.143 | 0.512 | 0.690 | 0.024 | 0.452 | HO | 0.522 | |
| 3.41 | HE | 0.644 | 0.430 | 0.822 | 0.772 | 0.796 | 0.660 | 0.771 | 0.495 | 0.397 | 0.717 | 0.136 | 0.676 | 0.777 | 0.024 | 0.512 | HE | 0.575 | |
| Figueroa | HO | 0.700 | 0.500 | 0.778 | 0.800 | 0.625 | 0.429 | 0.700 | 0.125 | 0.600 | 0.700 | 0.300 | 0.500 | 0.500 | 0.100 | 0.600 | HO | 0.530 | |
| Street | 3.87 | HE | 0.626 | 0.563 | 0.863 | 0.811 | 0.700 | 0.604 | 0.789 | 0.508 | 0.626 | 0.758 | 0.268 | 0.767 | 0.847 | 0.100 | 0.505 | HE | 0.622 |
| Kern County | HO | 0.721 | 0.558 | 0.561 | 0.674 | 0.610 | 0.738 | 0.674 | 0.476 | 0.535 | 0.814 | 0.093 | 0.674 | 0.884 | 0.256 | 0.419 | HO | 0.579 | |
| Rural | 3.73 | HE | 0.649 | 0.501 | 0.785 | 0.717 | 0.796 | 0.703 | 0.759 | 0.655 | 0.584 | 0.728 | 0.090 | 0.746 | 0.812 | 0.270 | 0.498 | HE | 0.619 |
| Kern County | HO | 0.781 | 0.313 | 0.645 | 0.750 | 0.531 | 0.750 | 0.688 | 0.548 | 0.406 | 0.781 | 0.094 | 0.688 | 0.750 | 0.031 | 0.500 | HO | 0.550 | |
| Urban007 | 3.67 | HE | 0.629 | 0.304 | 0.762 | 0.682 | 0.733 | 0.724 | 0.775 | 0.559 | 0.523 | 0.751 | 0.092 | 0.732 | 0.801 | 0.091 | 0.496 | HE | 0.577 |
| Kern County | HO | 0.778 | 0.500 | 0.588 | 0.833 | 0.389 | 0.611 | 0.647 | 0.500 | 0.389 | 0.778 | 0.222 | 0.889 | 0.778 | 0.056 | 0.556 | HO | 0.568 | |
| Urban 016 | 3.95 | HE | 0.705 | 0.495 | 0.725 | 0.719 | 0.827 | 0.754 | 0.761 | 0.633 | 0.544 | 0.735 | 0.203 | 0.821 | 0.859 | 0.157 | 0.541 | HE | 0.632 |
| Turlock | HO | 0.703 | 0.595 | 0.343 | 0.324 | 0.469 | 0.444 | 0.857 | 0.714 | 0.514 | 0.459 | 0.676 | 0.694 | 0.622 | 0.568 | 0.649 | HO | 0.575 | |
| 3.94 | HE | 0.698 | 0.671 | 0.369 | 0.280 | 0.806 | 0.737 | 0.851 | 0.745 | 0.656 | 0.586 | 0.505 | 0.760 | 0.817 | 0.540 | 0.626 | HE | 0.643 | |
| Wilton | HO | 0.595 | 0.667 | 0.238 | 0.238 | 0.474 | 0.400 | 0.738 | 0.700 | 0.714 | 0.405 | 0.595 | 0.659 | 0.905 | 0.357 | 0.643 | HO | 0.555 | |
| 4.04 | HE | 0.672 | 0.648 | 0.340 | 0.212 | 0.775 | 0.758 | 0.816 | 0.716 | 0.717 | 0.449 | 0.542 | 0.731 | 0.904 | 0.540 | 0.582 | HE | 0.627 | |
| Elk Grove | HO | 0.643 | 0.683 | 0.250 | 0.143 | 0.622 | 0.463 | 0.833 | 0.718 | 0.762 | 0.452 | 0.429 | 0.744 | 0.929 | 0.439 | 0.512 | HO | 0.575 | |
| 4.04 | HE | 0.741 | 0.672 | 0.425 | 0.178 | 0.743 | 0.702 | 0.808 | 0.722 | 0.712 | 0.485 | 0.488 | 0.730 | 0.911 | 0.527 | 0.500 | HE | 0.623 | |
| Zoo | HO | 0.657 | 0.714 | 0.382 | 0.086 | 0.879 | 0.143 | 0.906 | 0.829 | 0.914 | 0.286 | 0.600 | 0.629 | 0.914 | 0.457 | 0.314 | HO | 0.581 | |
| 3.46 | HE | 0.660 | 0.674 | 0.324 | 0.084 | 0.746 | 0.707 | 0.795 | 0.750 | 0.760 | 0.448 | 0.583 | 0.637 | 0.783 | 0.544 | 0.316 | HE | 0.587 | |
| Manhole | HO | 0.523 | 0.500 | 0.182 | 0.318 | 0.605 | 0.488 | 1.000 | 0.886 | 0.512 | 0.227 | 0.614 | 0.318 | 0.767 | 0.558 | 0.386 | HO | 0.526 | |
| Sacramento | 3.09 | HE | 0.647 | 0.568 | 0.208 | 0.277 | 0.716 | 0.720 | 0.772 | 0.739 | 0.532 | 0.226 | 0.505 | 0.374 | 0.705 | 0.487 | 0.315 | HE | 0.519 |
| Manhole Old | HO | 0.700 | 0.617 | 0.179 | 0.013 | 0.560 | 0.324 | 0.713 | 0.577 | 0.613 | 0.383 | 0.605 | 0.538 | 0.753 | 0.395 | 0.235 | HO | 0.480 | |
| Sacramento | 3.67 | HE | 0.676 | 0.674 | 0.331 | 0.013 | 0.803 | 0.736 | 0.808 | 0.722 | 0.670 | 0.336 | 0.564 | 0.602 | 0.840 | 0.483 | 0.260 | HE | 0.568 |
| Heronry/Davis | HO | 0.607 | 0.679 | 0.333 | 0.107 | 0.478 | 0.462 | 0.808 | 0.607 | 0.786 | 0.464 | 0.536 | 0.630 | 0.893 | 0.464 | 0.500 | HO | 0.557 | |
| 4.13 | HE | 0.592 | 0.708 | 0.371 | 0.103 | 0.804 | 0.654 | 0.813 | 0.756 | 0.758 | 0.477 | 0.590 | 0.801 | 0.908 | 0.507 | 0.519 | HE | 0.624 | |
| DaveBHouse | HO | 0.667 | 0.697 | 0.074 | 0.152 | 0.483 | 0.548 | 0.758 | 0.710 | 0.758 | 0.364 | 0.606 | 0.758 | 0.727 | 0.515 | 0.364 | HO | 0.545 | |
| 3.91 | HE | 0.728 | 0.671 | 0.209 | 0.195 | 0.770 | 0.780 | 0.793 | 0.744 | 0.747 | 0.440 | 0.538 | 0.675 | 0.807 | 0.615 | 0.453 | HE | 0.611 | |
| Woodland | HO | 0.606 | 0.657 | 0.177 | 0.197 | 0.613 | 0.429 | 0.716 | 0.672 | 0.701 | 0.507 | 0.515 | 0.677 | 0.761 | 0.507 | 0.328 | HO | 0.538 | |
| 4.13 | HE | 0.688 | 0.694 | 0.367 | 0.207 | 0.766 | 0.745 | 0.853 | 0.744 | 0.747 | 0.562 | 0.546 | 0.808 | 0.831 | 0.516 | 0.477 | HE | 0.637 | |
| Roseville | HO | 0.571 | 0.714 | 0.545 | 0.154 | 0.714 | 0.364 | 0.643 | 0.538 | 0.714 | 0.786 | 0.500 | 0.643 | 0.857 | 0.643 | 0.357 | HO | 0.583 | |
| 4.14 | HE | 0.664 | 0.672 | 0.584 | 0.271 | 0.813 | 0.732 | 0.817 | 0.698 | 0.733 | 0.664 | 0.516 | 0.791 | 0.913 | 0.632 | 0.370 | HE | 0.658 | |
| Lake | HO | 0.559 | 0.559 | 0.414 | 0.375 | 0.560 | 0.452 | 0.606 | 0.515 | 0.618 | 0.618 | 0.559 | 0.788 | 0.794 | 0.265 | 0.588 | HO | 0.551 | |
| 4.13 | HE | 0.531 | 0.561 | 0.479 | 0.416 | 0.802 | 0.688 | 0.809 | 0.746 | 0.734 | 0.566 | 0.532 | 0.823 | 0.903 | 0.472 | 0.526 | HE | 0.639 | |
| Shasta | HO | 0.318 | 0.778 | 0.314 | 0.065 | 0.516 | 0.442 | 0.578 | 0.667 | 0.717 | 0.435 | 0.609 | 0.609 | 0.761 | 0.609 | 0.348 | HO | 0.518 | |
| 4.00 | HE | 0.491 | 0.701 | 0.481 | 0.064 | 0.798 | 0.619 | 0.778 | 0.716 | 0.716 | 0.380 | 0.653 | 0.771 | 0.851 | 0.592 | 0.382 | HE | 0.600 | |
| Benton | HO | 0.542 | 0.667 | 0.214 | 0.087 | 0.700 | 0.364 | 0.667 | 0.727 | 0.625 | 0.250 | 0.417 | 0.750 | 0.833 | 0.542 | 0.417 | HO | 0.520 | |
| 3.70 | HE | 0.621 | 0.710 | 0.466 | 0.128 | 0.858 | 0.444 | 0.723 | 0.733 | 0.684 | 0.231 | 0.508 | 0.698 | 0.841 | 0.542 | 0.488 | HE | 0.578 | |
A = allelic richness; H = heterozygosity; HO, = observed heterozygosity; HE, = expected heterozygosity; NA = not applicable because this locus was monomorphic. Superscript numbers in top row are references for original source of primer sequences. Values in bold indicate significant departures from Hardy-Weinberg equilibrium.
There were 21 instances of significant departures from HWE after Bonferroni correction, distributed among 10 populations (Table 2). The Manhole Sacramento and Woodland populations had the most instances (n = 4 each), followed by the Manhole Old Sacramento and Zoo populations (n = 3 each). Seven of the departures from HWE were caused by locus Cxpq79, but because its inclusion did not change the results of analyses, it was not excluded. Of the 21 departures, 18 were characterized by HO < HE and three were HE < HO (two in the Zoo population and one in the Manhole Sacramento population). Instances of significant LD (n = 54) occurred most often in populations in and around the city of Sacramento. The Manhole Sacramento population had the most instances (n = 27), followed by Zoo (n = 19), Manhole Old Sacramento (n = 5), Dave B House (n = 2), and Woodland (n = 1). The AMOVA analysis showed that most of the genetic variation in the populations resided at the level of individual mosquitoes (83%) (Table 3). At higher levels of organization, approximately half as much genetic variation was partitioned within populations (6%) as among populations (10%). The Bottleneck results suggested there was an excess of heterozygotes in 13 of the 20 populations. Populations with a mode-shift and a significant one-way Wilcoxon test result were the following: Coachella Valley Rural, Coachella Valley Urban, Homeland, Figueroa Street, Kern County Rural, Kern County Urban 007, Kern County Urban 016, Zoo, Manhole Sacramento, Heronry/Davis, Woodland, Roseville, and Shasta.
Table 3.
Analysis of molecular variance (AMOVA) results for Culex pipiens complex populations, California*
| Source of variation | df | Sum of squares | Variance components | % Variation |
|---|---|---|---|---|
| Among populations | 19 | 689.202 | 0.465 | 10.49 |
| Among mosquitoes within populations | 678 | 2875.817 | 0.273 | 6.17 |
| Within mosquitoes | 698 | 2579.000 | 3.695 | 83.34 |
| Total | 1395 | 6144.019 | 4.433 |
Corresponding fixation indices are given in the text. df = degrees of freedom.
Genetic differentiation.
The overall FST value (0.105) suggested a moderate degree of differentiation among populations. Most pairwise FST comparisons were significant by permutation test except for populations in close proximity to each other (Table 4). The highest pairwise FST values (range = 0.22–0.33) were between the Manhole populations (in Sacramento and Old Sacramento) and the Cx. quinquefasciatus populations in southern California, and between the Shasta population and Cx. quinquefasciatus populations (range = 0.17–0.27).
Table 4.
Pairwise FST values for Culex pipiens complex populations in this study, California*
| Parameter | CVRu | CVUr | Home | FigSt | KCRu | KCU7 | KCU6 | Turl | Wilt | ElkG | Zoo | ManS | ManOS | Heron | DBH | Wood | Rose | Lake |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CVUr | 0.013 | |||||||||||||||||
| Home | 0.012 | 0.015 | ||||||||||||||||
| FigSt | 0.014 | 0.022 | −0.004 | |||||||||||||||
| KCRu | 0.030 | 0.026 | 0.025 | 0.003 | ||||||||||||||
| KCU7 | 0.023 | 0.028 | 0.016 | 0.004 | 0.002 | |||||||||||||
| KCU6 | 0.067 | 0.043 | 0.051 | 0.016 | 0.008 | 0.029 | ||||||||||||
| Turl | 0.187 | 0.174 | 0.160 | 0.100 | 0.102 | 0.127 | 0.076 | |||||||||||
| Wilt | 0.248 | 0.236 | 0.216 | 0.155 | 0.164 | 0.192 | 0.143 | 0.027 | ||||||||||
| ElkG | 0.241 | 0.227 | 0.207 | 0.147 | 0.155 | 0.184 | 0.136 | 0.026 | −0.003 | |||||||||
| Zoo | 0.263 | 0.246 | 0.223 | 0.161 | 0.176 | 0.202 | 0.152 | 0.042 | 0.033 | 0.025 | ||||||||
| ManS | 0.332 | 0.318 | 0.286 | 0.241 | 0.222 | 0.251 | 0.219 | 0.086 | 0.053 | 0.054 | 0.076 | |||||||
| ManOS | 0.295 | 0.281 | 0.256 | 0.204 | 0.202 | 0.231 | 0.194 | 0.050 | 0.017 | 0.018 | 0.035 | 0.035 | ||||||
| Heron | 0.229 | 0.214 | 0.196 | 0.130 | 0.140 | 0.175 | 0.117 | 0.016 | 0.002 | 0.001 | 0.027 | 0.064 | 0.026 | |||||
| DBH | 0.267 | 0.248 | 0.230 | 0.167 | 0.171 | 0.199 | 0.146 | 0.029 | 0.003 | 0.004 | 0.023 | 0.027 | 0.008 | 0.009 | ||||
| Wood | 0.197 | 0.189 | 0.169 | 0.110 | 0.120 | 0.144 | 0.096 | 0.012 | 0.015 | 0.013 | 0.027 | 0.063 | 0.033 | 0.003 | 0.012 | |||
| Rose | 0.175 | 0.160 | 0.143 | 0.083 | 0.092 | 0.114 | 0.081 | 0.015 | 0.014 | 0.010 | 0.033 | 0.077 | 0.032 | 0.005 | 0.015 | 0.004 | ||
| Lake | 0.172 | 0.165 | 0.153 | 0.099 | 0.107 | 0.134 | 0.086 | 0.029 | 0.032 | 0.029 | 0.060 | 0.097 | 0.061 | 0.013 | 0.032 | 0.016 | 0.007 | |
| Shas | 0.268 | 0.267 | 0.236 | 0.166 | 0.193 | 0.222 | 0.174 | 0.063 | 0.034 | 0.033 | 0.048 | 0.092 | 0.052 | 0.020 | 0.039 | 0.030 | 0.048 | 0.052 |
Values in bold indicate significant pairwise FST values. Site abbreviations are shown in Table 1.
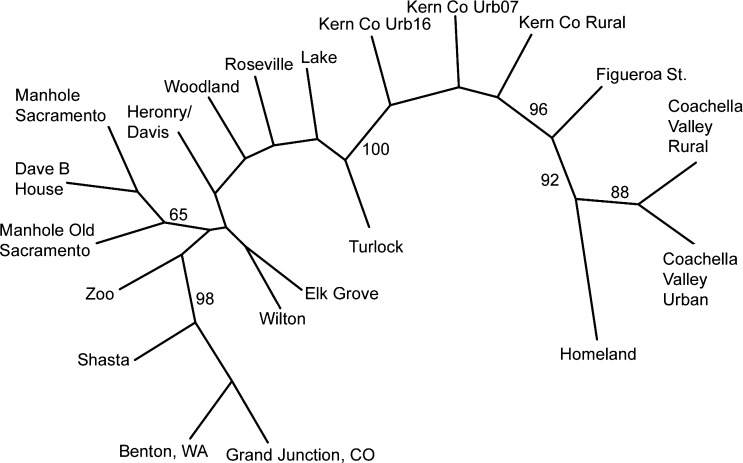
The neighbor-joining tree topography was consistent with the geographic locations of populations, as well as the degree of differentiation among them (Figure 2 ). The Cx. quinquefasciatus populations (Coachella Valley Rural through Kern County Urban 016 on Figure 1 and Table 1) formed a branch with 100% bootstrap support. The branch including the two northernmost sites along the transect, Shasta, and Benton, Washington, and a Cx. pipiens form pipiens population from Grand Junction, Colorado, had a bootstrap value of 98%. Three populations with a large proportion of autogenous individuals grouped together (Manhole Sacramento, Manhole Old Sacramento, Dave B House), but with weak bootstrap support (65%).
Figure 2.
Unrooted consensus neighbor-joining tree based on 1,000 bootstrap replicates using Cavalli-Sforza and Edwards chord distances. Numbers indicate percentage bootstrap support.
Cluster analyses.
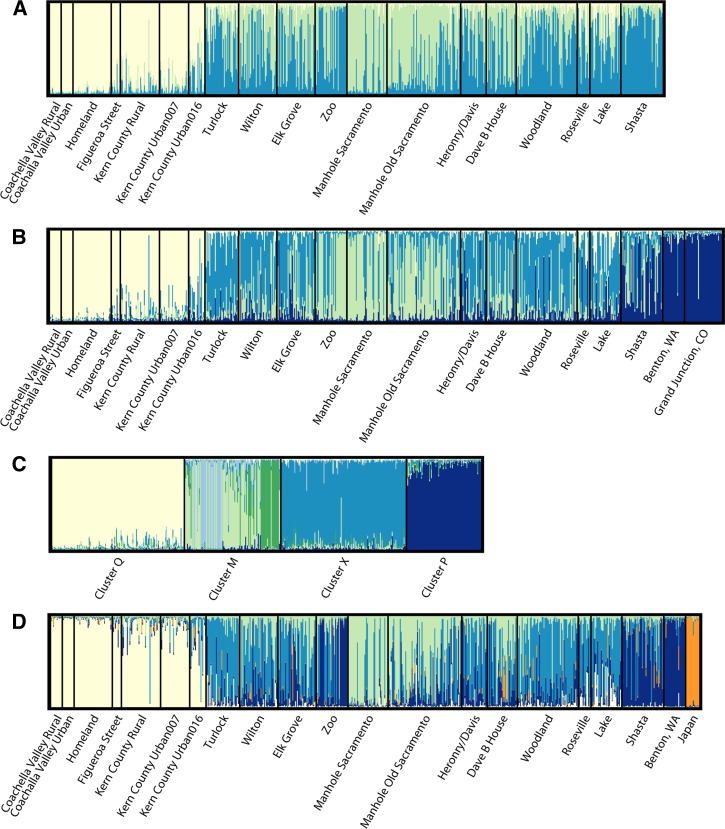
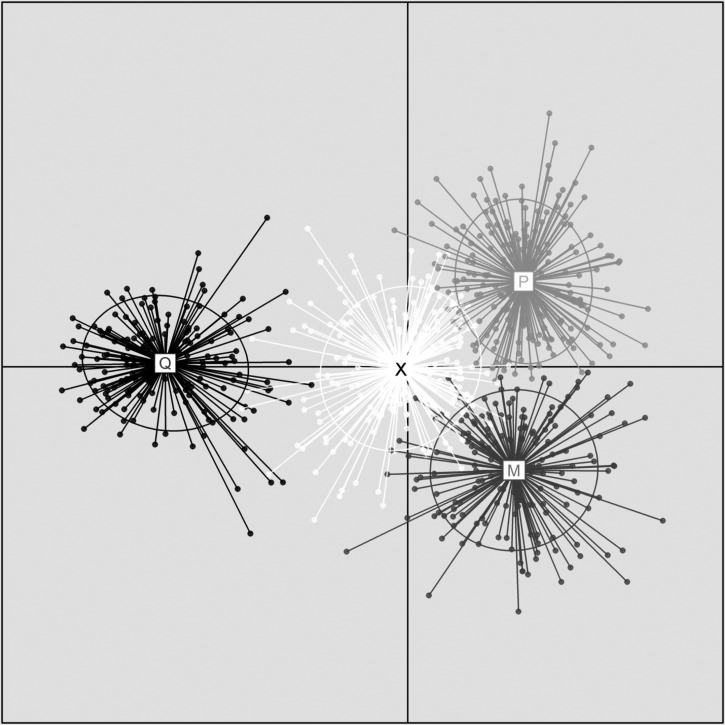
Structure results for the analysis of California populations indicated that a value of K = 3 was associated with the highest posterior probability (Figure 3A). Repeating the Structure analysis with the addition of the Washington and Colorado Cx. pipiens populations resulted in approximately equal likelihoods that K = 4, K = 5, and K = 6. Given that Structure's documentation recommends choosing the smallest value of K that captures the structure of the data, we chose K = 4 (Figure 3B). As discussed below, a comparison of Structure results with and without the Washington and Colorado populations suggests the K = 3 clusters are Cx. quinquefasciatus, Cx. pipiens form molestus (from large portions of the Manhole Sacramento, Manhole Old Sacramento, Zoo, and Dave B House populations), and a genetically similar group of mosquitoes of hybrid ancestry we call Cluster X. Members of this cluster comprise large portions of the Turlock population, as well as those around Sacramento. We make the distinction between hybrids, which we define as genetically admixed individuals with q values < 0.80, and Cluster X individuals, which have q values ≥ 0.80. Cluster X individuals are a group of genetically similar mosquitoes, yet are derived from the Cx. pipiens complex parent taxa. The fourth cluster (here called Cluster P) that resulted from the addition of the Washington and Colorado populations was comprised of Cx. pipiens form pipiens and comprised nearly 100% of those populations, as well as smaller portions of the Shasta, Heronry/Davis, Lake, and Elk Grove populations.
Figure 3.
Structure diagrams showing most likely numbers of clusters. Colors correspond to the same clusters for each panel: Cluster Q, light yellow; Cluster P, dark blue; Cluster M, light green; Cluster X, medium blue. A, California Culex pipiens complex populations, K = 3. B, populations as in A with the addition of populations from Benton, Washington and Grand Junction, Colorado, K = 4 (see text). C, When highly admixed individuals are removed from the analysis, the most likely number of clusters is K = 6, and cluster M further subdivides, with the Manhole Sacramento (light blue) and Zoo (green) populations becoming distinct. D, California and Benton, Washington populations, with the addition of 15 Cx. pipiens pallens individuals from Japan (in orange), K = 5.
When the subset of individuals with q ≥ 0.80 was analyzed, the most likely number of clusters was K = 6. Clusters Q, P, and X remained the same as when all individuals were used, but Cluster M was further subdivided into three clusters, two of which comprised most of the Manhole Sacramento and Zoo populations, respectively (Figure 3C). The CLUMPP analysis that used only the California populations indicated that mosquitoes were assigned most consistently to the same clusters when K = 2 or K = 4 (H = 0.99 and 0.93), with other values of K having lower values of H. When the Washington and Colorado populations were included, the most stable values of K were 2, 3, and 5, all with H = 0.99. The Structure runs on individuals whose q values were ≥ 0.80 were not subject to a stability analysis because the analysis only used part of the data. The ΔK method of determining the most likely number of clusters returned a value of K = 2 for both the California only data and the data set that included Washington and Colorado.
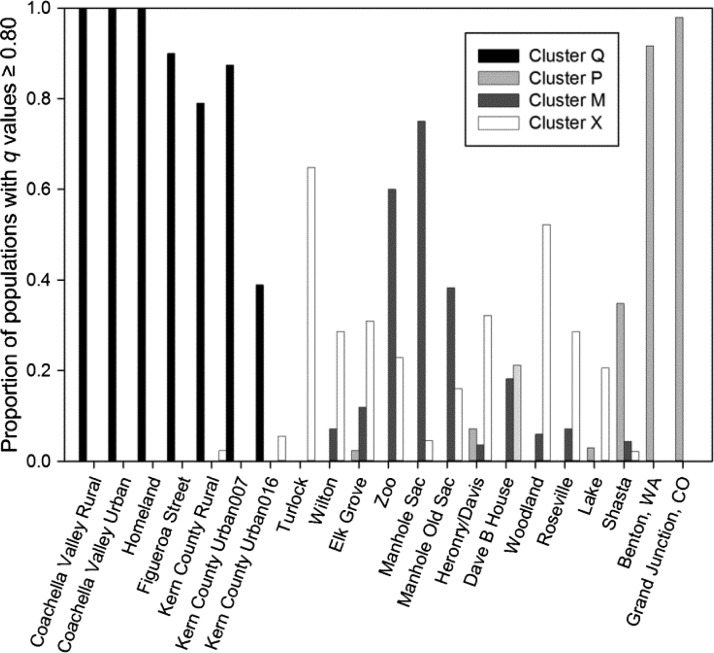
The proportion of each population that assigned with q values ≥ 0.80 to the K = 4 clusters in Structure (n = 427 of 713 mosquitoes) is shown in Figure 4. The southernmost sites assigned with a high proportion of their populations to Cluster Q. Overall, the California populations had a low proportion of individuals assigned to Cluster P. Shasta had the highest proportion of Cluster P individuals (35%). Several other populations had smaller proportions of their populations assigned to Cluster P: Heronry/Davis (7%), Lake (3%), and Elk Grove (2%). Cluster M was comprised of presumably autogenous mosquitoes from populations around Sacramento (range = 4% in Heronry/Davis and 75% in Manhole Sacramento). Turlock had the highest proportion of individuals assigned to Cluster X (65%), followed by Woodland (52%). Most of the populations around Sacramento had approximately 25% of their individuals assigned to Cluster X. Moreover, as indicated by the proportion of populations having q values less than the 0.80 threshold (i.e., the proportion of the population that is not represented in Figure 4), most of the populations around Sacramento have approximately 40% of their population made up of hybrid mosquitoes. Finally, 90% and 98%, respectively, of the Washington and Colorado populations were assigned to Cluster P, and no individuals in these populations were assigned to any other cluster.
Figure 4.
Bar graph of California Culex pipiens populations showing proportion of populations whose mosquitoes had structure q values ≥ 0.80. Mosquitoes with q values ≤ 0.80 are not shown on the graph and represent hybrids.
Of the 193 specimens morphologically classified as autogenous or anautogenous, 137 produced acceptable microsatellite genotypes (35 from Zoo, 42 from Manhole Old Sacramento, 33 from Dave B House, 0 from Heronry/Davis, and 27 from Woodland). Twenty-eight of 35 mosquitoes examined from the Zoo population were autogenous. Of those individuals, 20 (71%) assigned to Cluster M, 5 (18%) assigned to Cluster X, and 3 (11%) had q values less than the cutoff of 0.80. Of the 42 Manhole Old Sacramento mosquitoes examined, 27 were morphologically autogenous, and 12 (44%) were assigned to Cluster M, 4 (15%) were assigned to Cluster X, and 11 (41%) were not assigned to a cluster because their q values were too low. For the Dave B House population, 7 of 33 individuals were morphologically autogenous, but only 1 (14%) was assigned to Cluster M and the remaining mosquitoes had q values < 0.80 and were not assigned to a cluster. All 12 mosquitoes from the Heronry/Davis population were anautogenous, but all had PCR amplification problems and were not included in the analysis. All 27 individuals sampled from the Woodland population were anautogenous. None were assigned to Cluster M, 15 (56%) were assigned to Cluster X, and the remainder were not assigned to any cluster because of low q values.
The results from the Structure run that included Cx. pipiens pallens mosquitoes are shown in Figure 3D. All of the loci used in other analyses in this study amplified for these individuals, indicating that they are closely related taxonomically to California Cx. pipiens complex populations. However, this taxon forms its own cluster, and does not appear to be substantially admixed with any mosquitoes in the sampled California populations. Nevertheless, there are small numbers of individuals in the Shasta and Dave B House populations, as well as in the populations around Sacramento that could have a very low degree of admixture with Cx. p. pallens. Microsatellite assays on the three Cx. stigmatosoma and six Cx. restuans individuals resulted in amplification of only 2 of the 15 markers. Markers GT4F and GT51F amplified in all individuals. In addition, the allele sizes were monomorphic within taxa and not characteristic of the Cx. pipiens complex; Cx. stigmatosoma had an 88-basepair allele for GT4F and a 161-basepair allele for GT51F, and Cx. restuans had an 80-basepair allele for GT4F and a 159-basepair allele for GT51F.
The DAPC results are shown in Figure 5. Cluster Q is well defined and its confidence ellipse does not overlap those of the other clusters. The ellipses for the P and the M clusters do not overlap with each other and occupy the opposite side of the plot. Cluster X is located between clusters Q, P, and M, but aligns more closely to Cluster P and Cluster M, and is approximately equidistant from each. The Loading Plot generated to show which loci best discriminated the groups indicated that Cxpq78 was by far the most informative, followed by GT51F (Supplemental Figure 2).
Figure 5.
Discriminant analysis of principal components plot showing position of four clusters (determined by Structure). This procedure was used as a means to visualize the relative positions of populations, not as a clustering method per se. See text for details. Colors correspond to clusters in Figure 3: cluster Q, black; cluster P, light gray; cluster M, dark gray, cluster X, white.
Discussion
Overall genetic diversity was similar among populations despite several loci (CxqTri4F, CxqCTG10, Cxpq78) having lower diversity values in some populations (Table 2). Most of the significant departures from HWE were caused by an excess of homozygotes (i.e., a deficit of heterozygotes). Often, an excess of homozygotes is suggestive of the Wahlund Effect,44 in which allele frequencies are sampled after some degree of population subdivision has occurred. However, in the Manhole Sacramento and Zoo populations, there was a small number of significant HWE departures in the opposite direction (HO > HE), indicating an excess of heterozygotes (Table 2).
These findings were consistent with genetic characterizations of populations whose allele frequencies have changed because of some perturbation.45 One possibility is that the Manhole Sacramento and Zoo populations have each experienced a genetic bottleneck. Another possibility, although not mutually exclusive, is that the Manhole Sacramento and Zoo populations were receiving gene flow from genetically dissimilar populations, a condition referred to as isolate breaking,44 which is supported by the high degree of LD (27 in Manhole Sacramento and 19 in Zoo) in both populations. The Bottleneck analysis suggested both the Manhole Sacramento and Zoo populations have experienced bottlenecks, but also detected the signature of a bottleneck in several other populations that would not be expected to have them. A recent review by Peery and others46 suggests that the program Bottleneck can give spurious results when sample sizes are small, and that results can be affected by the duration and time since the bottleneck event. Conversely, some mosquito populations regularly experience reductions in numbers because of vector control efforts, which could influence allele frequencies in these natural populations even though they are in HWE. For example, Cartaxo and others47 found that genetic diversity of populations of Cx. quinquefasciatus in Brazil decreased over a three-year period because of control efforts. Our study used specimens collected by vector control programs from sites known to harbor mosquitoes and probably included in control programs. Nevertheless, the Manhole Sacramento and Zoo populations, and to a lesser degree Manhole Old Sacramento, had distinct patterns of HWE and LD.
Another commonality of the Zoo and Manhole Sacramento populations was that both had autogenous mosquitoes. The low genetic diversity found in the Manhole Sacramento population was in contrast with the Zoo population's genetic diversity values, which were towards the middle of the distribution of values. One explanation could be that the two populations have had different patterns of gene flow subsequent to the proposed genetic bottleneck, in which Manhole Sacramento has been isolated and the Zoo has been receiving gene flow. Several findings support this assertion. First, Structure assigned many Zoo specimens to Cluster M when the Washington and Colorado Cx. pipiens populations were included, but these individuals were assigned to Cluster P when they were excluded (compare panels A and B in Figure 3). Also, despite the presence of large proportions of autogenous mosquitoes at the Zoo site, the neighbor-joining tree placed this population away from the other autogenous populations (Figure 2). Finally, of the 35 specimens examined morphologically and determined to be autogenous in the Zoo population, most (71%) assigned to Cluster M, suggesting at least some individuals are genetically distinct enough to be classified as form molestus. Taken together, it appears that the allele frequencies in the Zoo population were less fixed than in the Manhole Sacramento population. This finding might have been caused by characteristics of the Zoo collection site, which made it more accessible to colonizing mosquitoes. The Zoo site was a catch basin containing water 1–1.5 meters below street level and thus was an enclosed underground site, although not as isolated as the Manhole Sacramento site (Nelms B, unpublished data).
The AMOVA results indicated that although most diversity resides at the level of the individual, almost twice as much genetic variation was partitioned among populations versus among individuals within populations. This distribution of variance was similar to that in another study48 we conducted that involved form molestus mosquitoes in Chicago, Illinois, and New York, New York, but contrasted with our work with Cx. quinquefasciatus, Cx. pipiens, and their hybrids along a transect from New Orleans, Louisiana, to Chicago, Illinois, in which variation was distributed equally at the two upper levels of organization in the AMOVA.8 Inclusion of a group with low genetic diversity such as form molestus might result in a greater proportion of variance at the among-population level.
The overall FST value reflects differences between many of the pairs of populations in this study, as well as among the four genetically distinct groups discerned by the Structure analyses. With regard to comparisons among pairs of populations, the largest degree of genetic differentiation was between the Cx. quinquefasciatus populations and the autogenous populations, followed by that between Cx. quinquefasciatus populations and Cx. pipiens populations (Table 4). Nonsignificant pairwise FST values resulted from comparisons between pairs of populations in the areas surrounding Sacramento, suggesting that these populations had a similar mixture of genotypes. The neighbor-joining tree was consistent with these findings, because the populations around Sacramento had low bootstrap support but were located together between the Cx. quinquefasciatus populations, the Cx. pipiens populations, and the form molestus populations. The low bootstrap support might have been caused by the populations being similar mixtures of the other three larger genetic groupings, such that their consensus order on the branch is equivalent to several other possible orders.
If Cx. pipiens form pipiens in the United States can be defined as genetically similar to populations in Washington and Colorado, the Structure results suggest there is a low frequency of Cx. pipiens genotypes in California. When only California populations were included in the analysis, K = 3 was the most likely number of clusters. The addition of Washington and Colorado Cx. pipiens populations resulted in a fourth genetic cluster being derived from the data. The comparison of panels A and B in Figure 3 shows that Cluster X when K = 4 is still present when K = 3. When K = 4, Cx. pipiens individuals represented mostly by the Washington and Colorado populations, form a distinct group. This finding might be caused by the small number of pure Cx. pipiens form pipiens in California, and those individuals although similar enough to group with Cluster X when K = 3, were more similar to the Washington and Colorado Cx. pipiens when those data were included in the analysis. It is clear that mosquitoes in Cluster X are Cx. pipiens complex mosquitoes because the microsatellite loci used in the study amplified in these populations similarly to the other populations and had no unique alleles.
The CLUMPP analyses of the Structure runs with and without the Washington and Colorado populations yielded interesting results that likely reflect the ability of this software to detect genetic structure at more than one level of biological organization. When only California populations were considered, individuals were most consistently assigned to the same cluster when K = 2 or K = 4. When K = 2, genotypes are organized rather broadly into those that belong to Cluster Q and those that do not, and the populations from Turlock northwards appear as one group. The results of the ΔK analysis may have detected only this broad pattern in the data. When K = 4, the analysis appears to detect the small number of Cx. pipiens form pipiens in the northern CA populations, as well as the large number of Cluster X mosquitoes. When the Washington and Colorado populations are included, several stable configurations of cluster assignments were evident: K = 2, K = 3 and K = 5. At K = 3, mosquitoes are still partitioned into logical groups: Cluster Q, Cluster P, and a group comprised of Cluster X and Cluster M. When K = 5, Cluster Q, Cluster P, and Cluster X are present. However, many individuals that assigned to Cluster M when K = 4 were deemed hybrids (i.e., individual specimens did not assign to any cluster with a q value ≥ 0.80). The inability of the program to assign such a large proportion of mosquitoes to a cluster indicated that K = 4 (Clusters Q, P, X, and M) was realistic, despite not having as strong support in the CLUMPP analysis.
Interestingly, removing hybrids (when K = 4) from the subsequent Structure analysis suggests additional population subdivision within Cluster M that was otherwise obscured. In particular, the Manhole Sacramento and Zoo populations are genetically distinct from each other and from the other California populations studied. This finding is consistent with those of previous studies,48,49 which suggested form molestus populations in the United States are genetically divergent from each other and from local above-ground form pipiens populations.
The choice of a q cutoff value of 0.80 may have overestimated the number of pure Cx. pipiens individuals which is somewhat surprising given the low proportions of such mosquitoes in California. Populations that had Cluster P individuals were found as far south as Elk Grove (2%), which is relatively close to the 39°N latitude distribution boundary of Cx. pipiens in the United States reported by Barr.7 However, the two populations above that latitude, Lake and Shasta, had only 3% and 35% of individuals respectively, assigned to Cluster P and < 10% of individuals from the remaining California populations assigned to this cluster. Most genotypes found at the Lake and Shasta sites were not assigned to any cluster because their q values were divided among two or more clusters, indicating that these populations have a high proportion of hybrid individuals. Our findings that Cx. pipiens form pipiens is rare in northern California is consistent with those of a related study on overwintering Cx. pipiens complex females from the Shasta population, where an unexpectedly high proportion of females did not enter reproductive diapause under midwinter conditions.50 Benton, Washington, also had a small proportion (2%) of hybrids, but consisted mostly (98%) of Cx. pipiens form pipiens mosquitoes.
From its position on the DAPC plot (Figure 5), Cluster X appears to be made up of contributions from Cx. pipiens form pipiens, Cx. pipiens form molestus, and Cx. quinquefasciatus. It appears from the location of Cluster X on the plot that its ancestry contains somewhat more form pipiens and form molestus than Cx. quinquefasciatus. If Cluster X was a hybrid only of Cx. pipiens form pipiens and Cx. quinquefasciatus, it would be expected to be located directly between those clouds of points on the DAPC plot. Instead, Cluster X was positioned in the middle of the plot, but clearly was closer to the P and M clusters. In addition, our analyses indicate autogenous mosquitoes in California largely belong to Cluster M, and were more closely related to Cx. pipiens and Cluster X mosquitoes than to Cx. quinquefasciatus. This finding is in contrast to a recent study of Cx. pipiens complex mosquitoes in the San Francisco area by Strickman and Fonseca, which concluded that autogenous mosquitoes there had hybridized with Cx. quinquefasciatus.20 The results of the Loading Plot analysis showed that data from one marker in the microsatellite panel, Cxpq78, was best at discriminating among the groups in the DAPC analysis. For this reason, this marker could be useful in future examinations of introgression in the Cx. pipiens complex.
Several studies have noted the presence of what have been referred to as stable hybrid populations in and around the central valley of California, which historically were presumed to consist of Cx. pipiens–Cx. quinquefasciatus hybrids.16,19 Unlike some areas subjected to temperature extremes that shift the balance of taxa over the course of a year, the existence of areas with entirely admixed populations has been known for some time.16 Our study detected a distinct genetic group (Cluster X) that is closely related to other members of the Cx. pipiens complex, and comprised of Cx. pipiens form pipiens, Cx. pipiens form molestus, and Cx. quinquefasciatus. That the Structure analyses found Cluster X to be a discernible group suggests that it does not consist of F1 hybrids, which would be the case if there was a high degree of gene flow from the parental taxa. Instead, Cluster X appears to consist of advanced-generation hybrids. An intriguing possibility is that this group may be undergoing speciation as a result of genetic differentiation from the other taxa over time.
Evaluating temporal and spatial patterns of hybridization in California Cx. pipiens complex populations is important to ongoing disease modeling and vector control efforts because the high degree of interfertility within the complex means that advantageous traits can spread via gene flow. For example, McAbee and others19 noted that the same molecular pathways for two kinds of insecticide resistance were present in Cx. pipiens and Cx. quinquefasciatus populations from California. Although the two mechanisms might have evolved in each taxon independently, the authors emphasize a more likely scenario in which these advantageous mutations arose in one taxa and spread to the other via interspecific hybridization. Another example, based on more recent work, involves a shift in host preference in Cx. pipiens form pipiens. Several studies12–15 have suggested gene flow to form pipiens from form molestus is a contributing factor to comparatively high rates of feeding on humans in populations of Cx. pipiens in the midwestern and eastern United States. Although form molestus is believed to have evolved as an urban mosquito in close proximity to humans, form pipiens is believed to prefer feeding on birds. Interestingly, recent blood meal analysis studies in southern51 and central52,53 California indicate that few Culex pipiens complex females feed on humans, even in urban Los Angeles or Sacramento. If traits such as insecticide resistance and host preference can transfer among closely related forms, other traits that are genetically controlled could also transfer, such as those related to vector competence, autogeny, and seasonal diapause (or the absence of it). The current study is a snapshot of gene flow within the Cx. pipiens complex in California. Subsequent sampling and genetic analysis of these populations would enable inferences to be made as to how the taxonomic composition of populations is changing over time.
Given the high degree of admixture in this system, it was logical to assess whether related species of Culex have contributed genes to the California Cx. pipiens complex populations in the current study. The Structure analysis that included Cx. pipiens pallens mosquitoes from Japan clearly showed that taxon to be distinct from the California populations. The two other Culex taxa whose DNA was assessed with the panel of microsatellites, Cx. stigmatosoma and Cx. restuans, proved not to be similar genetically to the Cx. pipiens complex and most of the microsatellite loci did not amplify.
Comparing the data from morphologic assessments of autogeny with those from the cluster assignments in Structure indicate a fairly good ability of this panel of microsatellites to detect autogenous mosquitoes. In the Zoo population, most (71%) individuals that were morphologically autogenous and assayed with the panel of microsatellites were correctly assigned to Cluster M. The percentage of correct determinations was lower for the other two populations examined, with Manhole Old Sacramento having 44% of autogenous individuals assigned to Cluster M and Dave B House, where one of eight individuals was assigned to Cluster M. The remaining morphologically autogenous individuals in these populations were either assigned to Cluster X or were deemed hybrids because of low q values. None of the morphologically autogenous specimens was assigned to the P or Q clusters. The variety of genotypes present in morphologically autogenous mosquitoes suggests specimens with diverse genetic backgrounds may express autogeny. Given the high proportion of hybrid individuals found especially in populations around Sacramento, it is unknown whether a similar diversity of genotypes will persist over time, or whether subsequent sampling would find ever larger proportions of morphologically autogenous mosquitoes that would be assigned to Cluster M.
The diversity of epidemiologically important life history traits that vary within the Cx. pipiens complex combined with extensive hybridization and adaptation to a variety of local conditions make it an important group of vector species to study. Our study measured genetic diversity and differentiation among California Cx. pipiens complex populations, and characterized the degree and extent of hybridization within the state. Our data will add to future work on Cx. pipiens complex populations in California and elucidate whether the observed patterns of genetic variation continue to evolve, or represent a stable configuration of genotypes.
Supplementary Material
ACKNOWLEDGMENTS
We thank personnel from multiple mosquito control agencies in California for collecting, storing, and shipping specimens to us for this study. They include Paula Macedo, Sacramento-Yolo Mosquito and Vector Control District (MVCD); Bonnie Ryan, Lake County VCD; Hugh Lothrop, Center for Vectorborne Diseases at the Coachella Valley MVCD; Richard Takahashi, Kern VCD; John Albright, Shasta MVCD; Susanne Kluh, Greater Los Angeles County VCD; Karen Mellor, Antelope Valley MVCD; Jerry Davis, Turlock Mosquito Abatement District; and Matt Hoefer and Kevin Shoemaker, Benton County Mosquito Control, Washington State. We thank Dr. Motoyoshi Mogi for kindly providing specimens of Cx. pipiens pallens from Japan and Tara Thiemann and Sarah Wheeler (Center for Vectorborne Diseases, University of California, Davis) for providing specimens from the Heronry/Davis and Roseville populations, respectively. Harry M. Savage thanks Charlie W. Smith (Consolidated Mosquito Abatement District, Selma) for assistance with field collection of Cx. stigmatosoma and Cx. restuans egg rafts, and Martin R. Williams (Centers for Disease Control and Prevention) for assistance with rearing. We also thank Mark Delorey (Centers for Disease Control and Prevention) and Chris Richards (National Center for Genetic Resources Preservation) for assisting with statistical analyses.
Footnotes
Financial support: This study was supported in part by a research grant from the Sacramento-Yolo MVCD and grant RO1 AI55607 from the National Institutes of Allergy and Infectious Diseases, National Institutes of Health (NIH). Brittany M. Nelms was supported in part by an NIH fellowship from the Training Program in Biology of Disease Vectors, grant number T32-AI074550. William K. Reisen was supported by the Research and Policy for Infectious Disease Dynamics Program of the Science and Technology Directorate, Department of Homeland Security and the Fogarty International Center, NIH. Linda Kothera and Harry M. Savage are supported by the Centers for Disease Control and Prevention in Fort Collins, Colorado.
Authors' addresses: Linda Kothera and Harry M. Savage, Division of Vector-Borne Diseases, Centers for Disease Control and Prevention, Fort Collins, CO, E-mails: lkothera@cdc.gov and hms1@cdc.gov. Brittany M. Nelms and William K. Reisen, Center for Vectorborne Diseases, School of Veterinary Medicine, University of California, Davis, CA, E-mails: bnelms@wesphotos.com and wkreisen@ucdavis.edu.
References
- 1.Savage HM, Aggarwal D, Apperson CS, Katholi CR, Gordon E, Hassan HK, Anderson M, Charnetzky D, McMillen L, Unnasch EA, Unnasch TR. Host choice and West Nile virus infection rates in blood-fed mosquitoes, including members of the Culex pipiens complex, from Memphis and Shelby County, Tennessee, 2002–2003. Vector Borne Zoonotic Dis. 2007;7:365–386. doi: 10.1089/vbz.2006.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell CJ, Francy DB, Monath TP. Arthropod vectors. In: Monath TP, editor. St. Louis Encephalitis. Washington, DC: American Public Health Association; 1980. pp. 313–379. [Google Scholar]
- 3.Nasci RS, Savage HM, White DJ, Miller JR, Cropp BC, Godsey MS, Kerst AJ, Bennet P, Gottfried K, Lanciotti RS. West Nile virus in overwintering Culex mosquitoes, New York City, 2000. Emerg Infect Dis. 2000;7:742–744. doi: 10.3201/eid0704.010426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McAbee RD, Kang K-D, Stanich MA, Christiansen JA, Wheelock CE, Inman AD, Hammock BD, Cornel AJ. Pyrethroid tolerance in Culex pipiens pipiens var molestus from Marin County, California. Pest Manag Sci. 2004;60:359–368. doi: 10.1002/ps.799. [DOI] [PubMed] [Google Scholar]
- 5.Reisen WK, Barker CM, Fang Y, Martinez VM. Does variation in Culex (Diptera: Culicidae) vector competence enable outbreaks of West Nile virus in California? J Med Entomol. 2008;45:1126–1138. doi: 10.1603/0022-2585(2008)45[1126:dvicdc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Vaidyanathan R, Scott TW. Geographic variation in vector competence for West Nile virus in the Culex pipiens (Diptera: Culicidae) complex in California. Vector Borne Zoonotic Dis. 2007;7:193–198. doi: 10.1089/vbz.2006.0589. [DOI] [PubMed] [Google Scholar]
- 7.Barr AR. The distribution of Culex p. pipiens and C. p. quinquefasciatus in North America. Am J Trop Med Hyg. 1957;6:153–165. doi: 10.4269/ajtmh.1957.6.153. [DOI] [PubMed] [Google Scholar]
- 8.Kothera L, Zimmerman EM, Richards CM, Savage HM. Microsatellite characterization of subspecies and their hybrids in Culex pipiens complex (Diptera: Culicidae) mosquitoes along a north-south transect in the central United States. J Med Entomol. 2009;46:236–248. doi: 10.1603/033.046.0208. [DOI] [PubMed] [Google Scholar]
- 9.Edillo F, Kiszewski A, Manjourides J, Pagano M, Hutchinson M, Kyle A, Arias J, Gaines D, Lampman R, Novak R, Foppa I, Lubelcyzk C, Smith R, Moncayo A, Spielman A. The Culex pipiens Working Group Effects of latitude and longitude on the population structure of Culex pipiens s.l., vectors of West Nile virus in North America. Am J Trop Med Hyg. 2009;81:842–848. doi: 10.4269/ajtmh.2009.08-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S, Molaei G, Andreadis TG. Genetic insights into the population structure of Culex pipiens (Diptera: Culicidae) in the northeastern United States by using microsatellite analysis. Am J Trop Med Hyg. 2008;79:518–527. [PubMed] [Google Scholar]
- 11.Huang S, Molaei M, Andreadis TG. Reexamination of Culex pipiens hybridization zone in the eastern United States by ribosomal DNA-based single nucleotide polymorphism markers. Am J Trop Med Hyg. 2011;85:434–441. doi: 10.4269/ajtmh.2011.10-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilpatrick AM, Kramer LD, Jones MJ, Marra PP, Daszak P, Fonseca DM. Genetic influences on mosquito feeding behavior and the emergence of zoonotic pathogens. Am J Trop Med Hyg. 2007;77:667–671. [PubMed] [Google Scholar]
- 13.Hamer GL, Kitron UD, Brawn JD, Loss SR, Ruiz MO, Goldberg TL, Walker ED. Culex pipiens (Diptera: Culicidae): a bridge vector of West Nile virus to humans. J Med Entomol. 2008;45:125–128. doi: 10.1603/0022-2585(2008)45[125:cpdcab]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Huang S, Hamer GL, Molaei G, Walker ED, Goldberg TL, Kitron UD, Andreadis TG. Genetic variation associated with mammalian feeding in Culex pipiens from a West Nile virus epidemic region in Chicago, Illinois. Vector Borne Zoonotic Dis. 2009;9:637–642. doi: 10.1089/vbz.2008.0146. [DOI] [PubMed] [Google Scholar]
- 15.Savage HM, Kothera L. The Culex pipiens complex in the Mississippi River Basin: identification, distribution and bloodmeal hosts. J Am Mosquito Contr. 2012;28:93–99. doi: 10.2987/8756-971X-28.4.93. [DOI] [PubMed] [Google Scholar]
- 16.Tabachnick WJ, Powell JR. Genetic analysis of Culex pipiens populations in the central valley of California. Ann Entomol Soc Am. 1983;76:715–720. [Google Scholar]
- 17.Urbanelli S, Silvestrini F, Reisen WK, De Vito E, Bullini L. Californian hybrid zone between Culex pipiens pipiens and Cx. p. quinquefasciatus revisited (Diptera: Culicidae) J Med Entomol. 1997;34:116–127. doi: 10.1093/jmedent/34.2.116. [DOI] [PubMed] [Google Scholar]
- 18.Cornel AJ, McAbee RD, Rasgon J, Stanich M, Scott TW, Coetzee M. Differences in extent of genetic introgression between sympatric Culex pipiens and Culex quinquefasciatus (Diptera: Culicidae) in California and South Africa. J Med Entomol. 2003;40:36–51. doi: 10.1603/0022-2585-40.1.36. [DOI] [PubMed] [Google Scholar]
- 19.McAbee RD, Kang K-D, Stanich MA, Christiansen JA, Wheelock CE, Inman AD, Hammock BD, Cornel AJ. Pyrethroid tolerance in Culex pipiens pipiens var molestus from Marin County, California. Pest Manag Sci. 2004;60:359–368. doi: 10.1002/ps.799. [DOI] [PubMed] [Google Scholar]
- 20.Strickman D, Fonseca DM. Autogeny in Culex pipiens complex mosquitoes from the San Francisco Bay area. Am J Trop Med Hyg. 2012;87:719–726. doi: 10.4269/ajtmh.2012.12-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiter P. A portable, battery-operated trap for collecting gravid Culex mosquitoes. Mosq News. 1983;43:496–498. [Google Scholar]
- 22.Kawai S. Studies on the follicular development and feeding activity of the females of Culex tritaeniorhynchus with special reference to those of autumn. Trop Med. 1969;11:145–169. [Google Scholar]
- 23.Clements AN, Boocock MR. Ovarian development in mosquitoes: stages of growth and arrest, and follicular resorption. Physiol Entomol. 1984;9:1–8. [Google Scholar]
- 24.Darsie RF, Jr, Ward RA. Identification and Geographical Distribution of the Mosquitoes of North America, North of Mexico. Gainesville, FL: University Press of Florida; 2005. [Google Scholar]
- 25.Kothera L, Godsey MS, Doyle MS, Savage HM. Characterization of Culex pipiens complex (Diptera: Culicidae) populations in Colorado, USA using microsatellites. PLoS ONE. 2012;7:e47602. doi: 10.1371/journal.pone.0047602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glaubitz JC. CONVERT: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes. 2004;4:309–310. [Google Scholar]
- 27.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goudet J. FSTAT (Version 1.2): a computer program to calculate F-Statistics. J Hered. 1995;86:485–486. [Google Scholar]
- 30.Nei M. Molecular Evolutionary Genetics. New York, NY: Columbia University Press; 1987. [Google Scholar]
- 31.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 32.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 33.Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered. 1999;90:502–503. [Google Scholar]
- 34.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Seattle, WA: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 35.Cavalli-Sforza LL, Edwards AW. Phylogenetic analysis: models and estimation procedures. Evolution. 1967;32:550–570. doi: 10.1111/j.1558-5646.1967.tb03411.x. [DOI] [PubMed] [Google Scholar]
- 36.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 37.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- 39.Carpenter SJ, LaCasse WJ. Mosquitoes of North America (North of Mexico) Berkeley and Los Angeles, CA: University of California Press; 1955. [Google Scholar]
- 40.Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- 41.Rousset F. Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 42.Jombart T, Devillard S, Dufour AB, Pontier D. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity. 2008;101:92–103. doi: 10.1038/hdy.2008.34. [DOI] [PubMed] [Google Scholar]
- 43.Jombart T, Devillard SD, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartl DL, Clark AG. Principles of Population Genetics. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 45.Kimmel M, Chakraborty R, King JP, Bamshad M, Watkins WS, Jorde LB. Signatures of population expansion in microsatellite repeat data. Genetics. 1998;148:1921–1930. doi: 10.1093/genetics/148.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet-Bëer E, Robinson S, Vásquez-Carrillo C, Pauli JN, Palsbøll PJ. Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol. 2012;21:3403–3418. doi: 10.1111/j.1365-294X.2012.05635.x. [DOI] [PubMed] [Google Scholar]
- 47.Cartaxo MF, Ayresa CF, Weetman D. Loss of genetic diversity in Culex quinquefasciatus targeted by a lymphatic filariasis vector control program in Recife, Brazil. Trans R Soc Trop Med Hyg. 2011;105:491–499. doi: 10.1016/j.trstmh.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Kothera L, Godsey M, Mutebi J-P, Savage HM. A comparison of aboveground and belowground populations of Culex pipiens (Diptera: Culicidae) mosquitoes in Chicago, Illinois, and New York City, New York, using microsatellites. J Med Entomol. 2010;47:805–813. doi: 10.1603/me10031. [DOI] [PubMed] [Google Scholar]
- 49.Kothera L, Godsey M, Mutebi J-P, Savage HM. A comparison of above-ground and below-ground populations of Culex pipiens pipiens in Chicago, Illinois, and New York City, New York, using 2 microsatellite assays. J Am Mosquito Contr. 2012;28:106–112. doi: 10.2987/8756-971X-28.4.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelms BM, Macedo PA, Kothera L, Savage HM, Reisen WK. Overwintering biology of Culex mosquitoes (Diptera: Culicidae) in the Sacramento Valley, California. J Med Entomol. 2013;50:773–790. doi: 10.1603/me12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molaei G, Cummings RF, Su T, Armstrong PM, Williams GA, Cheng M-L, Webb J-P, Andreadis TG. Vector-host interactions governing epidemiology of West Nile virus in southern California. Am J Trop Med Hyg. 2010;86:1269–1282. doi: 10.4269/ajtmh.2010.10-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montgomery MJ, Thiemann T, Macedo P, Brown DA, Scott TW. Blood-feeding patterns of the Culex pipiens complex in Sacramento and Yolo Counties, California. J Med Entomol. 2011;48:398–404. doi: 10.1603/me10067. [DOI] [PubMed] [Google Scholar]
- 53.Thiemann TC, Lemenager DA, Kluh S, Carroll BD, Lothrop HD, Reisen WK. Spatial variation in host feeding patterns of Culex tarsalis and the Culex pipiens Complex (Diptera: Culicidae) in California. J Med Entomol. 2012;49:903–916. doi: 10.1603/me11272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molecular Ecology Resources Primer Development Consortium Abreu AG, Albaina A, Alpermann TJ, Apkenas VE, Bankhead-Dronnet S, Bergek S, Berumen ML, Cho C-H, Clobert J, Coulon A, de Feraudy D, Estonba A, Hankeln T, Hochkirch A, Hsu T-W, Huang T-J, Irigoien X, Iriondo M, Kay KM, Kinitz T, Kothera L, le Hénanff M, Lieutier F, Lourdais O, Macrini CM, Manzano C, Martin C, Morris VR, Nanninga G, Pardo MA, Plieske J, Pointeau S, Prestegaard T, Quack M, Richard M, Savage HM, Schwarcz KD, Shade J, Simms EL, Solferini VN, Stevens VM, Veith M, Wen M-J, Wicker F, Yost JM, Zarraonaindia I. Permanent genetic resources added to the Molecular Ecology Resources Database 1 October 2011–30 November 2011. Mol Ecol Res. 2012;12:374–376. doi: 10.1111/j.1755-0998.2011.03109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith J, Keyghobadi N, Matrone MA, Escher RL, Fonseca DM. Cross-species comparison of microsatellite loci in the Culex pipiens complex and beyond. Mol Ecol Notes. 2005;5:697–700. [Google Scholar]
- 56.Keyghobadi N, Matrone MA, Ebel GD, Kramer LD, Fonseca DM. Microsatellite loci from the northern house mosquito (Culex pipiens), a principal vector of West Nile virus in North America. Mol Ecol Notes. 2004;4:20–22. [Google Scholar]
- 57.Edillo FT, McAbee RD, Foppa IM, Lanzaro GC, Cornel AJ, Spielman A. A set of broadly applicable microsatellite markers for analyzing the structure of Culex pipiens (Diptera: Culicidae) populations. J Med Entomol. 2007;44:145–149. doi: 10.1603/0022-2585(2007)44[145:asobam]2.0.co;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.