Abstract
Inactivating germline mutations in DNA mismatch repair (MMR) genes are diagnostic for Lynch syndrome. However, the clinical significance of missense variants is uncertain. A threshold level of compromised MLH1 expression, correlating with greater protein instability and MMR functional defect, has been identified to help classify the pathogenicity of missense variants.
In this issue of Clinical Cancer Research, Hinrichsen and colleagues (1) report a critical threshold level of compromise in the expression of MLH1, a major DNA mismatch repair (MMR) gene, that correlated with greater protein instability and clinical data, thereby providing a means for classifying the pathogenicity of missense variants in MLH1.
Genetic testing for cancer predisposing syndromes has evolved as a powerful clinical tool. Lynch Syndrome (LS), caused by pathogenic mutations in DNA MMR genes, predisposes carriers to colorectal, endometrial/ovarian and other cancers. Detection of a pathogenic MMR mutation in the proband enables identification of other mutation carriers in the family who would benefit from risk-reduction strategies, including cancer surveillance, chemoprevention and/or prophylactic surgery. However, mutation testing can provide three categories of possible results: pathogenic mutation, variant of uncertain significance (VUS), or informative negative finding.(2) Variants, typically missense mutations, pose significant difficulties for management due to their unknown clinical impact.(3) Hinrichsen et al aimed to address this problem by providing a classifier for the pathogenic potential of MLH1 variants.
Classifying VUS involves integrating multiple lines of evidence for pathogenicity, including: 1) direct evidence, arising from clinical pedigree and phenotype data of co-segregation and co-occurrence; and 2) indirect evidence, consisting of in vitro assays of protein function, and in silico models predicting the impact of mutations on protein function based on altered splicing, protein structure, and/or evolutionary conservation.(2) Hinrichsen et al gathered 38 MLH1 missense variants from public databases, and measured the levels of MLH1 expression (by quantitative PCR) and MMR repair activity (by in vitro MMR assay), relative to the wild-type. Seven recurrent variants with proficient MMR function, termed “validating variants”, formed the basis for identifying the expression threshold. They were categorized as pathogenic vs. neutral, based on clinical data. Relative to the wild-type, the MLH1 expression levels of the putative pathogenic variants were 52% or lower, while those of the putative neutral variants were 65% or higher.(1) To further corroborate this threshold, MMR repair function was shown to be compromised when intracellular MLH1 level falls below 50%. Hinrichsen et al proposed to use MLH1 expression level of 52% (vs. wild-type) as the first cutoff for pathogenicity, and to use functional assays only to distinguish among high-expressing variants. This proposal is attractive because it is relatively simple and actionable, and its derivation took into account several lines of evidence. However, before clinical implementation, additional research should be considered. First, impaired MMR functional assay remains the gold-standard qualitative surrogate for pathogenicity, and partially eliminating it in a classification algorithm requires further validation. Second, the identified threshold was based on pooling a finite number of VUS with clinical data. Might the level change if more VUS had been investigated? Indeed, locus-specific clinicopathologic data are being accumulated in several well-annotated repositories. The collaboration merging the variant databases of international research groups including Collaborative Groups of Americas on Hereditary Colorectal Cancer (CGA), International Society of Gastrointestinal Hereditary Tumors (InSiGHT) and the Human Variome Project (http://chromium.liacs.nl/LOVD2/colon_cancer/home.php), will prove essential for validating proposed thresholds and algorithms.(4) Finally, the 2008 International Agency for Research on Cancer working group consensus standardized a variant classification system based on quantitative probability of pathogenicity. Integrating the findings of Hinrichsen et al into quantitative multifactorial likelihood models (5) will remain a challenge.
Variants can exert pathogenic effects by several different mechanisms, and Hinrichsen et al focused on defects in MLH1 protein expression level, suggesting that amino acid substitutions contributed to decreased protein stability. Low-expressing variants exhibited shorter half-life and lower de-folding temperatures; and the affected residues clustered in the core three-helix motif of the C-terminal “In” subdomain on protein structural analysis.(1) Beyond expression, other protein-level mechanisms of pathogenicity may include subcellular localization, protein-protein complex formation, and/or functional efficiency. In addition, on a RNA-level, missense mutations can affect mRNA processing, transcript levels, splicing patterns, and transcript stability. Levels of mRNA transcript (measured by RT-PCR) have been reported to allow classification of 50% of the variants.(6) Thus, comprehensive functional assessment of variants at both the RNA and the protein levels is needed to fully understand the mechanism of pathogenicity. In silico models that predict the functional impact of RNA splicing alterations (e.g. NNSplice, Spliceport) and of protein alterations (e.g. SIFT, PolyPhen-2, and MAPP-MMR) can help prioritize RNA vs. protein level analyses.(2, 5, 6)
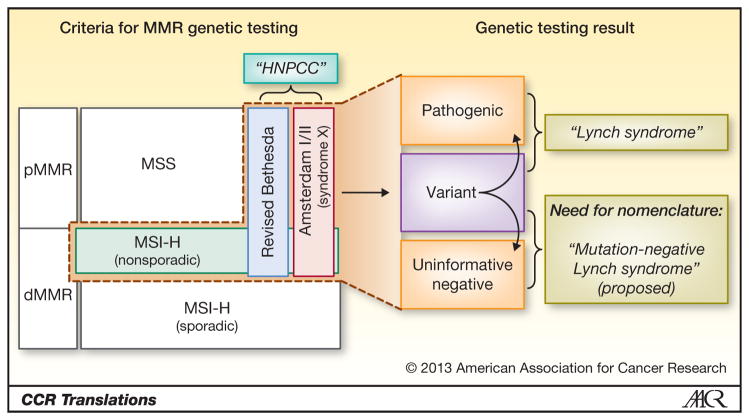
Unclassified variants can constitute up to 20–50% of all tested for MMR mutations, depending on the criteria used for testing.(2) Indeed, different nomenclature and criteria have evolved over time (Figure 1):
Figure 1.
Schematic representation of the clinical criteria for identifying patients to undergo MMR germline testing, based on familial and/or tumor-based criteria (left, within dotted line). Three categories of results from germline testing are shown, and curved arrows indicate efforts to classify variants by pathogenicity. While the term “Lynch syndrome” refers to patients with pathogenic mutations, “mutation-negative Lynch syndrome” is proposed for patients who 1) has non-sporadic MSI-high tumors (i.e. no evidence of MLH1 hypermethylation or BRAF mutation); 2) may or may not fulfill Amsterdam or Bethesda criteria; 3) have a genetic testing result that is uninformative negative or likely a non-pathogenic variant. (Figure not drawn to scale) pMMR, mismatch repair proficient; dMMR, mismatch repair deficient; MSS, microsatellite-stable; MSI-H, microsatellite-high; HNPCC, Hereditary non-polyposis colorectal cancer
Originally, “LS” described families with clustering of phenotypic cancers; but after the genetic basis of disease was discovered, “LS” has referred to patients with pathogenic MMR mutations.(7)
The term “HNPCC (hereditary nonpolyposis colorectal cancer syndrome)” was coined based on clinical and familial criteria, initially Amsterdam only and later loosely expanded to include revised Bethesda criteria, regardless of MMR mutation status.(8, 9) Although the phrase “LS also known as HNPCC” is commonplace in the literature, “LS” is actually not synonymous with “HNPCC”.
Tumor microsatellite (MSI) testing has shown that nearly all LS patients have MSI-high tumors, but many HNPCC patients have microsatellite stable tumors. This latter subgroup of “HNPCC” has been termed “Syndrome X” and does not overlap with “LS”. (10)
A growing desire to identify LS patients and availability of tumor MSI testing have led to universal tumor-based screening to identify patients with non-sporadic MSI-high tumors, who then undergo directed confirmatory genetic testing.(11)
Thus, the criteria for genetic testing for LS has expanded from the most stringent Amsterdam criteria to the revised Bethesda criteria, and now universal tumor-based screening. This movement toward increasingly sensitive and less specific criteria for genetic testing has translated to lower proportions of tested patients with pathogenic mutations (or “LS”)(9) and higher proportions of VUS and uninformative negative results. While clinical management of LS is well established, that of the latter patients is highly problematic. Specifically, these challenging patients are defined by: 1) non-sporadic MSI-high tumors (i.e. no evidence of MLH1 hypermethylation or BRAF mutation); 2) positive or negative Amsterdam/Bethesda criteria; 3) uninformative negative or likely non-pathogenic variant on directed genetic testing (Figure 1). Currently, significant knowledge gaps exist, regarding their cancer risks relative to LS patients, their needs for cancer surveillance, their familial pedigrees, and their molecular biology. In parallel, there is an apparent void in our existing nomenclature for these patients who are not described by “LS” or by “HNPCC”. While “clinical Lynch” and “Lynch-like” have appeared in the literature(12), we herein propose the term “mutation-negative Lynch Syndrome” for the patients defined above (Figure 1). This term expresses the link with LS, assumes the presence of yet unknown MMR genes, and acknowledges potential roles played by other genes yet to be discovered.
Hinrichsen et al has provided a classifier of VUS as pathogenic or not, so that patients can be managed as LS or not. Unfortunately, no consensus exists at present for the management of mutation-negative LS patients. Some clinicians have considered it safest to manage them just as LS, an approach that likely represents over-treatment. Indeed, the ability to classify missense mutations will only be truly impactful if it translates to differences in patient management, but for that, the mutation-negative Lynch Syndrome must be fully understood clinically and molecularly.
Acknowledgments
Grant support: This work was supported in part by The University of Texas Anderson Cancer Center Core Support Grant (P30 CA016672).
We thank Ester Borras, PhD for her critical comments.
This is a commentary on article Hinrichsen I, Brieger A, Trojan J, Zeuzem S, Nilbert M, Plotz G. Expression defect size among unclassified MLH1 variants determines pathogenicity in Lynch syndrome diagnosis. Clin Cancer Res. 2013;19(9):2432-41.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Hinrichsen I, Brieger A, Trojan J, Zeuzem S, Nilbert M, Plotz G. Expression defect size among unclassified MLH1 variants determines pathogenicity in Lynch Syndrome diagnosis. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-12-3299. [DOI] [PubMed] [Google Scholar]
- 2.Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Human mutation. 2008;29(11):1282–91. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Syngal S, Fox EA, Li C, Dovidio M, Eng C, Kolodner RD, et al. Interpretation of genetic test results for hereditary nonpolyposis colorectal cancer: implications for clinical predisposition testing. JAMA : the journal of the American Medical Association. 1999;282(3):247–53. doi: 10.1001/jama.282.3.247. [DOI] [PubMed] [Google Scholar]
- 4.Spurdle AB. Clinical relevance of rare germline sequence variants in cancer genes: evolution and application of classification models. Current opinion in genetics & development. 2010;20(3):315–23. doi: 10.1016/j.gde.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Thompson BA, Goldgar DE, Paterson C, Clendenning M, Walters R, Arnold S, et al. A multifactorial likelihood model for MMR gene variant classification incorporating probabilities based on sequence bioinformatics and tumor characteristics: a report from the Colon Cancer Family Registry. Human mutation. 2013;34(1):200–9. doi: 10.1002/humu.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borras E, Pineda M, Brieger A, Hinrichsen I, Gomez C, Navarro M, et al. Comprehensive functional assessment of MLH1 variants of unknown significance. Human mutation. 2012;33(11):1576–88. doi: 10.1002/humu.22142. [DOI] [PubMed] [Google Scholar]
- 7.Lynch HT, Shaw MW, Magnuson CW, Larsen AL, Krush AJ. Hereditary factors in cancer. Study of two large midwestern kindreds. Archives of internal medicine. 1966;117(2):206–12. [PubMed] [Google Scholar]
- 8.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Diseases of the colon and rectum. 1991;34(5):424–5. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 9.Colas C, Coulet F, Svrcek M, Collura A, Flejou JF, Duval A, et al. Lynch or not Lynch? Is that always a question? Advances in cancer research. 2012;113:121–66. doi: 10.1016/B978-0-12-394280-7.00004-X. [DOI] [PubMed] [Google Scholar]
- 10.Lindor NM, Rabe K, Petersen GM, Haile R, Casey G, Baron J, et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA : the journal of the American Medical Association. 2005;293(16):1979–85. doi: 10.1001/jama.293.16.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heald B, Plesec T, Liu X, Pai R, Patil D, Moline J, et al. Implementation of Universal Microsatellite Instability and Immunohistochemistry Screening for Diagnosing Lynch Syndrome in a Large Academic Medical Center. Journal of clinical oncology. 2013 doi: 10.1200/JCO.2012.45.1674. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Soler M, Perez-Carbonell L, Guarinos C, Zapater P, Castillejo A, Barbera VM, et al. Risk of Cancer in Cases of Suspected Lynch Syndrome without Germline Mutation. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.01.044. epub. [DOI] [PubMed] [Google Scholar]