Abstract
More detailed sequence standards that keep up with revolutionary sequencing technologies will aid the research community in evaluating data.
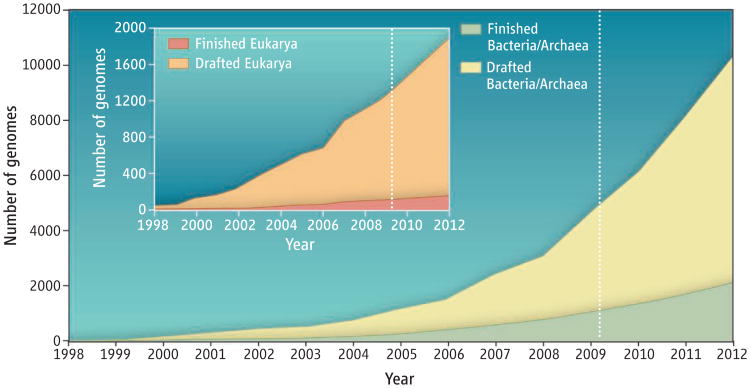
For over a decade, genome sequences have adhered to only two standards that are relied on for purposes of sequence analysis by interested third parties (1, 2). However, ongoing developments in revolutionary sequencing technologies have resulted in a redefinition of traditional whole-genome sequencing that requires reevaluation of such standards. With commercially available 454 pyrosequencing (followed by Illumina, SOLiD, and now Helicos), there has been an explosion of genomes sequenced under the moniker “draft”; however, these can be very poor quality genomes (due to inherent errors in the sequencing technologies, and the inability of assembly programs to fully address these errors). Further, one can only infer that such draft genomes may be of poor quality by navigating through the databases to find the number and type of reads deposited in sequence trace repositories (and not all genomes have this available), or to identify the number of contigs or genome fragments deposited to the database. The difficulty in assessing the quality of such deposited genomes has created some havoc for genome analysis pipelines and has contributed to many wasted hours. Exponential leaps in raw sequencing capability and greatly reduced prices have further skewed the time- and cost-ratios of draft data generation versus the painstaking process of improving and finishing a genome. The result is an ever-widening gap between drafted and finished genomes that only promises to continue (see the figure, page 236); hence, there is an urgent need to distinguish good from poor data sets.
Figure. Trends in generation of drafted and finished genomes.
A conservative estimate of future projects is shaded in light blue. Data were derived from (6, 7).
The sequencing institutes and consortia whom we represent believe that a new set of standards is required for genome sequences. The following represents community-defined categories of standards that better reflect the quality of the genome sequence, based on our understanding of the technologies, available assemblers, and efforts to improve upon drafted genomes. Due to the increasingly rapid pace of genomics, we avoided rigid numerical thresholds in our definitions to take into account products achieved by any combination of technology, chemistry, assembler, or improvement and/or finishing process.
Standard Draft
Minimally or unfiltered data, from any number of different sequencing platforms, that are assembled into contigs. This is the minimum standard for a submission to the public databases. Sequence of this quality will likely harbor many regions of poor quality and can be relatively incomplete. It may not always be possible to remove contaminating sequence data. Despite its shortcomings, Standard Draft is the least expensive to produce and still possesses useful information.
High-Quality Draft
Overall coverage representing at least 90% of the genome or target region. Efforts should be made to exclude contaminating sequences. This is still a draft assembly with little or no manual review of the product. Sequence errors and misassemblies are possible, with no implied order and orientation to contigs. This is appropriate for general assessment of gene content.
Improved High-Quality Draft
Additional work has been performed beyond the initial shotgun sequencing and High-Quality Draft assembly, by using either manual or automated methods. This should contain no discernable misassemblies and should have undergone some form of gap resolution to reduce the number of contigs and supercontigs (or scaffolds). Undetectable misassemblies are still possible, particularly in repetitive regions. Low-quality regions and potential base errors may also be present. This standard is normally adequate for comparison with other genomes.
Annotation-Directed Improvement
May overlap with the previous standards, but the term emphasizes the verification and correction of anomalies within coding regions, such as frameshifts, and stop codons. It will most often be used in cases involving complex genomes where improvement beyond this category fails to outweigh the associated costs. Gene models (gene calls, including intronexon determination for eukaryotes) and annotation of the genomic content should fully support the biology of the organism and the scientific questions being investigated. Exceptions to this gene-specific finishing standard should be noted in the submission. Repeat regions at this level are not resolved, so errors in those regions are much more likely. This standard is useful for gene comparisons, alternative splicing analysis, and pathway reconstruction.
Noncontiguous Finished
Describes high-quality assemblies that have been subject to automated and manual improvement, and where closure approaches have been successful for almost all gaps, misassemblies, and low-quality regions. Attempts have been made to resolve all gap and sequence uncertainties, and only those recalcitrant to resolution remain (with notations in the genome submission as to the nature of the uncertainty). This product is thus of “Finished” quality with the only exception being repetitive or intractable gaps, along with heterochromatic sequence for eukaryotic applications. Thus, it is appropriate for most analyses. For nearly all higher organisms, this is the grade previously called “Finished.”
Finished
Refers to the current gold standard; genome sequences with less than 1 error per 100,000 base pairs and where each replicon is assembled into a single contiguous sequence with a minimal number of possible exceptions commented in the submission record. All sequences are complete and have been reviewed and edited, all known misassemblies have been resolved, and repetitive sequences have been ordered and correctly assembled. Remaining exceptions to highly accurate sequence within the euchromatin are commented in the submission. The Finished product is appropriate for all types of detailed analyses and acts as a high-quality reference genome for comparative purposes. Some microbial genome sequences where multiple platforms have been used for the same genome have exceeded this standard, and it is believed that no bases are incorrect except for natural, low-level biological variation.
Intermediate standards often overlap, and although we do not advocate any one standard, we recommend that the target standard be based on the needs and goals of each project. There may be cases where select regions will be targeted for improvement and more than one standard may apply (such regionally improved sequences should be identified). This approach is most often used for eukaryotic whole-genome sequencing projects, where the cost of complete finishing remains prohibitive, and allows improvement to be directed at euchromatic sequence, because heterochromatic sequence remains largely recalcitrant to available approaches. Legacy eukaryotic tiling path standards will remain in use for a time.
Here, we have attempted to capture in a technology-independent fashion the types of whole-genome sequencing projects that are beginning to populate databases, and we have defined a set of standards that accommodate a growing list of alternative genome products that have been obtained via less conventional means, such as environmental (metagenomic) or single-cell sequencing. Ongoing discussions with genome database repositories have been met with enthusiasm, and the implementation of these standards as a requirement for genome submissions is expected. To aid in adoption of this classification of sequence finishing standards, we have added this classification to the Sequence Ontology (3) where it can now be used to comply with the Genomic Standards Consortium's (GSC) “Minimum Information about a Genome Sequence” standard (4) “sequencing status” descriptor. Furthermore, the efforts described here recently have been adopted under the umbrella of the GSC (5). This common currency in defining the products of genome projects enables better management of expectations and allows users of genomic data to assess the quality of the deposited available sequences and decide whether these meet their needs.
Supplementary Material
Acknowledgments
For Joint Genome Institute members, work was performed under the auspices of the U.S. Department of Energy Office of Science, Biological and Environmental Research Program; by the University of California, Lawrence Berkeley National Laboratory, contract no. DE-AC02-05CH11231; Lawrence Livermore National Laboratory contract no. DE-AC52-07NA27344; and Los Alamos National Laboratory, contract no. DE-AC02-06NA25396. Naval Medical Research Center work was supported by grant TMTI0068_07_NM_T from the Joint Science and Technology Office for Chemical and Biological Defense, Defense Threat Reduction Agency Initiative to T.D.R.
Footnotes
References and Notes
- 1.International standards for sequence fidelity, established at the Second International Strategy Meeting on Human Genome Sequencing in Bermuda in 1997 (27 February to 2 March). “Finished” quality standards, commonly known as the Bermuda standards, defined finished sequence as a contiguous sequence with less than one error per 10,000 bases. Almost everything else was “draft.”
- 2.Blakesley RW, et al. Genome Res. 2004;14:2235. doi: 10.1101/gr.2648404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eilbeck K, et al. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Field D, et al. Nat Biotechnol. 2008;26:541. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genomic Standards Consortium. http://gensc.org.
- 6.GOLD, Genomes OnLine Database. www.genomesonline.org.
- 7.Liolios K, et al. Nucleic Acids Res. 2008;36:D475. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.