Abstract
The decay rate of an mRNA and the efficiency with which it is translated are key determinants of eukaryotic gene expression. Although it was once thought that mRNA stability and translational efficiency were directly linked, the interrelationships between the two processes are considerably more complex. The decay of individual mRNAs can be triggered or antagonized by translational impairment, and alterations in the half-life of certain mRNAs can even alter translational fidelity. In this review, we consider whether mRNA translation and turnover are distinct or overlapping phases of an mRNA life cycle, and then address some of the many ways in which the two processes influence each other in eukaryotic cells.
Keywords: mRNA degradation, translational repression, quality control
Multiple modes of translation:decay interaction
After their synthesis, processing, and export to the cytoplasm, mRNA molecules are largely engaged in two activities: they serve as templates for the synthesis of specific polypeptides or as substrates for cellular degradative pathways. Translation and mRNA decay have been studied for decades and mechanistic details of both processes have been elaborated [1, 2]. Although early notions of translation:decay interactions were simplistic, e.g., that mRNAs undergoing translation are protected from decay [2], the interrelationships between mRNA stability and translation have turned out to be much more complex. Some mRNAs are highly stable although they remain untranslated, and some particularly unstable mRNAs are translated efficiently. Further, individual mRNAs can exist in an active translation state, a translationally silent state, or a state targeted for decay, with non-linear and closely intertwined transitions between these states. In this review, we first consider whether the processes of translation and mRNA decay are distinct or overlapping, and then address some of the many ways in which translation and mRNA decay influence each other in eukaryotic cells. Five different modes of translation:decay interactions will be considered, including: a) accelerated mRNA decay as a response to aberrant translation, b) modulation of mRNA decay as a response to the inhibition of translation initiation, elongation, or termination, c) promotion of mRNA decay by efficient translation, d) translation repression as a prerequisite for mRNA decay, and e) alterations in mRNA decay that promote changes in translation efficiency. The possibility that unknown alterations in one process may lead to erroneous conclusions about the other will also be considered.
mRNA decay and translation: concurrent or distinct processes?
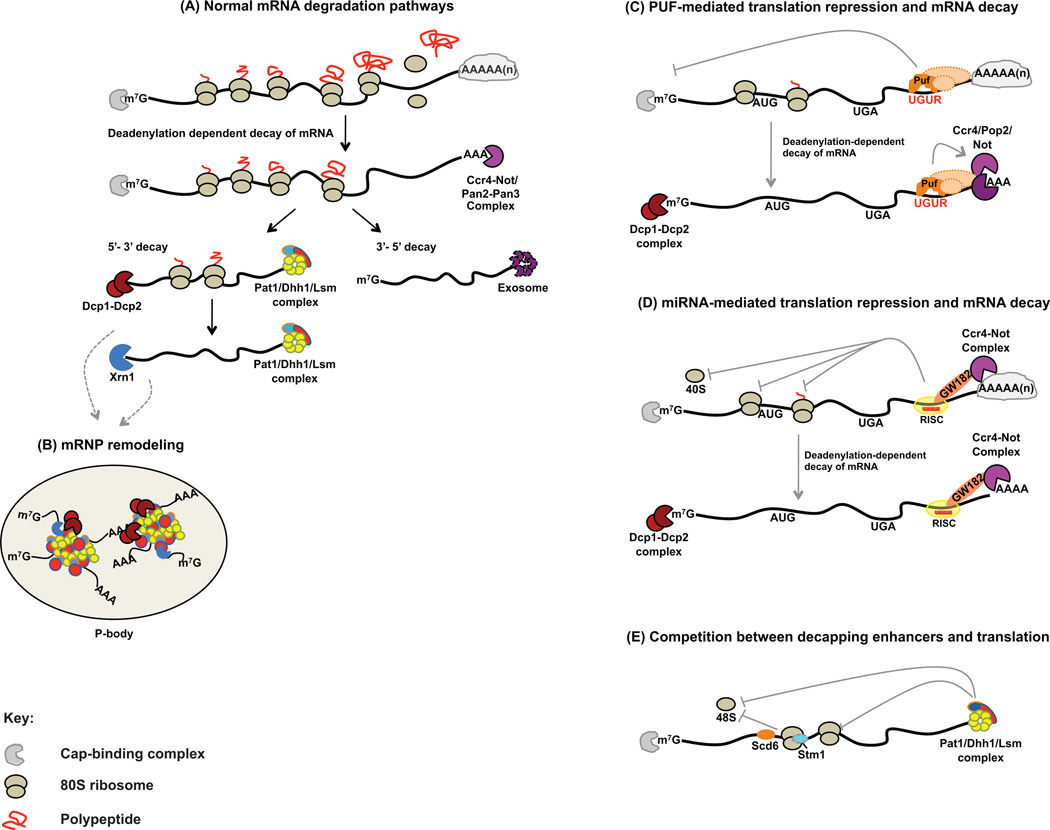
The interdependence of mRNA translation and decay is immediately evident from the functional roles of the 5’-cap and 3’-poly(A) tail, post-transcriptionally added appendages associated with most eukaryotic mRNAs. As a consequence of their respective association with specific binding proteins, both structures play a critical role in establishing the closed-loop mRNP (see Glossary) that promotes translational initiation and antagonizes mRNA decay [1–3]. This state is transient, however, and key rate-limiting events in the initiation of mRNA decay include the shortening of the poly(A) tail and removal of the cap [2] (Figure 1A). Progressive deadenylation (by the Ccr4-Not or Pan2/Pan3 complexes in yeast or PARN and PAN2/PAN3 in metazoans) leads to loss of associated poly(A)-binding protein (PABP) [1, 2] and subsequently to complete exonucleolytic digestion that proceeds either 5’ to 3’, and is decapping dependent, or 3’ to 5’, and is decapping independent [1, 2]. Decapping requires the prior activity of several accessory factors, including the Lsm 1–7 proteins, Dhh1, and Pat1, all of which are considered decapping activators [2]. These factors function post-deadenylation and loss of any one leads to accumulation of capped, deadenylated mRNAs [2]. mRNA decapping is also enhanced by Edc1, 2, and 3, factors thought to interact directly with the Dcp1/Dcp2 decapping complex [2]. Post-deadenylation mRNA degradation by the 3’ to 5’ pathway is mediated by the multisubunit exosome complex [2].
Figure 1. Decay pathways for normal mRNAs and mRNAs undergoing translational control.
mRNAs enter the cytoplasm where they get translated after associating with ribosomes and are subjected to multiple modes of post-transcriptional regulation, including: (A) mRNA degradation initiated by poly(A) shortening (catalyzed by the Ccr4–Not and poly(A)-specific deadenylases). Deadenylation is followed by 5´ −3´ decapping-dependent, Xrn1-mediated exonucleolytic decay, or, exosome-mediated 3´ -5´ exonucleolytic decay. Association of the Lsm1–7 complex with the 3’ end of the mRNA stimulates Dcp1-Dcp2-mediated decapping. (B) Untranslated transcripts are assembled into RNA-protein cytoplasmic granules called P-bodies together with the mRNA-decapping enzyme complex (Dcp1-Dcp2), Xrn1, Pat1, Dhh1, and Lsm1–Lsm7. (C) PUF proteins mediate translation repression and mRNA decay by directly binding to PUF binding elements in the mRNA or other protein partners (such as Nanos, Brat, or CPEB; not shown) in a transcript-specific manner. Recruitment of the Ccr4-Not deadenylase complex can trigger deadenylation-dependent mRNA decay. (D) Binding of the RISC complex triggers inhibition of translation initiation by interfering with cap recognition, 40S recruitment, or with 60S subunit joining. Interaction of RISC with the Ccr4-Not deadenylase complex also triggers deadenylation-dependent mRNA degradation. (E) Decapping activators stimulate mRNA decapping and inhibit translation during 48S formation (Pat1, Dhh1, Scd6) or during 80S formation (Stm1).
mRNA in the midst of interacting with these factors, i.e., in the process of being degraded, has been observed in two very different translational states. In one, mRNA is thought to first exit the translation pathway and localize to distinct cytoplasmic foci called processing bodies (P-bodies) [4], accompanied at those sites by components of the 5’-decay pathway [4] (Figure 1B). The nature of P-bodies was initially deduced from observations that their number and sizes increased with impaired translation initiation or 5’ to 3’ mRNA decay [4], conditions that would result in an increased pool of mRNAs to be degraded. Likewise, conditions that reduced the same pool, such as blocks to transcription or translation elongation, resulted in the loss of P-bodies [4]. The requirement for both P-body formation and the accumulation of 5’ to 3’ decay intermediates in P-bodies support the hypothesis that these entities are a site of decay. Consistent with this notion, mRNAs with shortened poly(A) tails only appear to leave polysomes and become targeted to P-bodies when Dhh1, Pat1, and other factors, repress the translation of those mRNAs (see below) and remodel the respective mRNPs [5].
The aforementioned results notwithstanding other experiments suggest that departure from the translation pathway and P-body localization are not obligatory steps for mRNA decay. Recent work [6] assessed the consequences of deleting the genes for Edc3 and Lsm4, two key P-body components in yeast. Although these deletions eliminated detectable P-bodies, they had no significant effect on the rates of decay of individual mRNAs. Similar experiments in metazoan cells yielded comparable results [4]. Direct evidence that polysome associated mRNAs could be substrates for decay came from experiments in yeast which showed that the polysome fraction harbored deadenylated, decapped, and partially exonucleolytically degraded mRNAs [7, 8]. Polysome targeting of mRNAs by the decay apparatus is also consistent with the observation that decay factors co-fractionate with ribosomes [9–13].
These observations raise important questions regarding the mechanism by which an mRNA can be simultaneously accessible to factors promoting both mRNA translation and decay. If poly(A) shortening minimizes or eliminates interaction between PABP and eIF4G, destabilizing the translation initiation complex [1, 2], then it is straightforward to visualize the onset of decapping and 5’ to 3’ decay while an mRNA is still associated with elongating ribosomes translating the coding region. Here, the decay apparatus could simply follow the ribosomes down the mRNA. However, 3’ to 5’ decay on the same deadenylated transcript would lead to a collision of the exosome with elongating ribosomes; therefore, some mechanism for withdrawal from the translation pathway must precede decay.
Accelerated mRNA decay as a response to aberrant translation
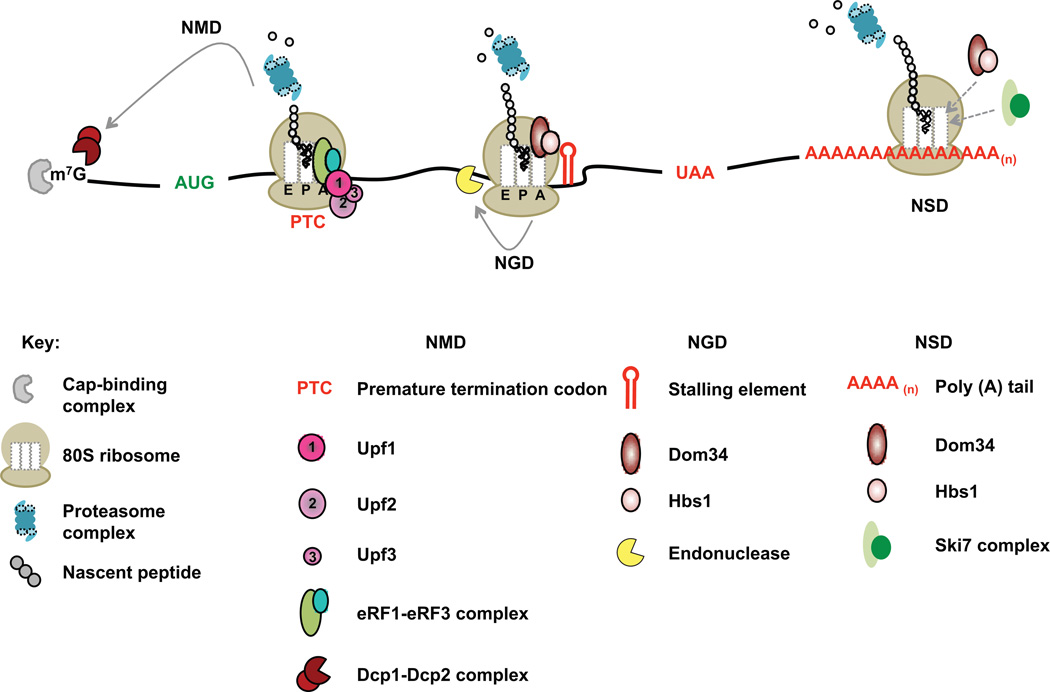
Interactions between mRNA translation and decay are emphatically evident in three quality control pathways that target defective cytoplasmic mRNAs. Nonsense mediated decay (NMD), nonstop decay (NSD), and no-go decay (NGD) [10, 14–16] respectively target mRNAs containing a premature termination codon (PTC), mRNAs lacking a termination codon, and mRNAs containing translational stall-inducing sequences. In all three pathways an aberrant translational event triggers accelerated mRNA decay (Figure 2). Additionally, it appears that NMD, NSD, and NGD must also act to dissociate and recycle paused or terminating ribosomes and tRNAs, as well as promote degradation of nascent polypeptides.
Figure 2. Abnormal translational events leading to accelerated mRNA decay.
The three major mRNA surveillance mechanisms are represented on the same mRNA. Initiation of NMD, NGD, and NSD involve recognition of abnormal translation events – a premature termination event (NMD), an elongation stall (NGD), and poly(A) translation (NSD), respectively. Recognition of the abnormal ribosomal complexes is followed by mRNA decay and proteasome-mediated degradation of the nascent peptide in all three pathways. In NMD, following recognition of a premature termination event by the Upf factors, the mRNA is subjected to accelerated decapping involving the Dcp1-Dcp2 complex. Recognition of a stalled ribosomal complex in NGD is followed by endonucleolytic cleavage of the mRNA upstream of the stalled ribosome in a Dom34-Hbs1-dependent manner. Translation of the poly(A) tail in NSD leads to recruitment of Ski7 and the exosome to the stalled ribosomal complex. Subsequent degradation of the mRNA body (in all three pathways) involves canonical 5’-3’ degradation by Xrn1 and 3’–5’ degradation by the exosome complex.
NMD targets the transcripts of nonsense alleles, as well as a wide range of endogenous substrates in which translation termination is either premature or in a sequence context characteristic of premature termination [10, 15–17]. Substrates of NMD are shunted into conventional decay pathways such that they are subject to accelerated deadenylation, deadenylation independent decapping, or increased 3’ to 5’ decay [10]. NMD depends on nonsense codon recognition by the ribosome and involves a set of conserved Upf factors that help discriminate a normal termination event from one that is premature while also promoting rapid mRNA decay, translational repression, and nascent polypeptide degradation [10]. Although the mechanisms of PTC recognition and Upf factor recruitment are still not understood well, an enhancing effect of the exon junction complex (EJC) and the atypical nature of the 3’-UTR generated by a PTC appear to be important for Upf recognition and NMD activation [10]. In most models, Upf1 (the principal NMD regulator) first associates with the premature termination complex by interacting with the ribosome-associated release factors eRF1 and eRF3 [10, 15]. NMD activation requires subsequent Upf1 interaction with a Upf2:Upf3 heterodimer. In metazoans, Upf1 and its regulator Smg-1 first form the SURF complex (Smg-1-Upf1-release factors) with Smg-8 and Smg-9 [10, 15] and then interact with Upf2 and Upf3 bound to a downstream EJC that has escaped being jettisoned by elongating ribosomes due to the upstream termination event [10]. This interaction leads to Upf1 phosphorylation by Smg-1 and activation of the ATPase and helicase activities of Upf1. Phosphorylation of Upf1 is thought to promote dissociation of the release factors and lead to activation of mRNA decay by Smg-5–7 whereas the helicase activity is thought to promote dissociation and recycling of an otherwise poorly dissociable termination complex [15]. Because intronless pre-mRNAs also give rise to mRNAs subject to NMD [10], enhancement of NMD by an EJC cannot explain all NMD events. Significantly, NMD also appears to be influenced by the markedly different efficiencies of premature and normal termination. The former are thought to lack key interactions between a terminating ribosome and specific factors localized 3’ to a normal stop codon, thereby permitting association of the Upf factors [10]. Emphasizing the crosstalk between translation and mRNA decay, Upf1 in yeast (the quintessential intronless system) is directly associated with the 40S ribosomal subunit and modulates the fate of a post-termination ribosome [9, 18].
NGD is activated by a wide range of translation elongation stalling events, including those resulting from strong stem-loops, contiguous rare codons, poly-Lys or poly-Arg tracts, or frameshift sites [14, 16, 19, 20]. NGD leads to endonucleolytic cleavage of mRNA, for which the endonuclease is yet to be identified, and to subsequent exonucleolytic digestion of the respective 5’ and 3’ cleavage products. NGD is regulated by two factors, Dom34 and Hbs1, which are structurally similar to the canonical translation release factors eRF1 and eRF3, respectively [19, 21, 22]. The endonucleolytic cleavage event in NGD is stimulated by the presence of Dom34 itself [19]. In both yeast and mammalian systems, Dom34 and Hbs1 bind the A site of the ribosome to promote recycling of stalled ribosomes and tRNAs [16], a step that may be dispensable with some specific strong translational pauses [22–24]. Recognition of an NGD substrate is dependent on the presence of a short stretch of mRNA on the 3’ end of the stalled ribosomal complex. This mRNA overhang appears to be monitored by Hbs1, which is positioned near the mRNA entry channel [25]. Endonucleolytic cleavage of NGD substrates generates 5’ and 3’ mRNA fragments that are then subjected to Xrn1 and exosome-mediated degradation. Cleavage of the mRNA is followed by release of the stalled ribosome by a mechanism similar to ribosome recycling at a bona fide translation termination codon [16, 20] while the translationally-stalled polypeptide is degraded in a proteasome-dependent process. The latter step requires the RING finger domain of Not4, a component of the Ccr4-Not complex, further illustrating yet another link between components of the mRNA decay and translation pathways [20].
NSD is triggered when ribosomes fail to encounter a stop codon during translation of an mRNA, continuing instead to the mRNA’s 3’ end and creating an unoccupied A site [16, 20]. A major class of mRNAs comprising NSD substrates is thought to arise as a consequence of premature polyadenylation within a transcript’s open reading frame [26–28]. In yeast, NSD triggers 3’ to 5’ decay and requires the exosome and the Ski7, Ski2, Ski3, and Ski8 proteins [20]. Ski7, a protein similar to Hbs1 in structure [26], is thought to recognize a ribosome stalled at the 3’ end of an mRNA and to recruit Ski2, Ski3, Ski8, and the exosome to commence 3 ’ to 5’ degradation of the mRNA using both the exo- and endonucleolytic activities of Rrp44 [29]. Very little is understood about how the stalled ribosome is removed from the mRNA and how Ski7 dissociates from the aberrant complex, although it has been suggested that the early steps of NSD may convert an mRNA into a substrate for the NGD pathway [16]. In the absence of Ski7 (or the exosome), nonstop mRNAs undergo accelerated 5’ to 3’ degradation and decapping [30], suggesting that failure to remove the NSD-stalled complex could affect the efficiency of translation initiation.
Modulation of mRNA decay as a consequence of reductions in the rates of translation initiation, elongation, or termination
As noted above, the roles of the mRNA 5’ cap and 3’ poly(A) tail in both translation initiation and mRNA stability illustrate the interrelationships of the two processes. Further manifestations of related phenomena include demonstrations that inhibition of translation initiation by mutations in the genes encoding eIF4E, eIF4G, or a component of the eIF3 complex, lead to accelerated mRNA deadenylation and subsequent decapping [2, 31], Additionally, as do reductions in translation initiation caused by a stem-loop in the mRNA 5’-UTR [2]. These consequences of initiation blockade suggest that diminished stability of the closed-loop mRNP enhances accessibility of the mRNA ends to their respective decay factors.
The mRNA decay effects associated with the inhibition of translational elongation are two-sided. On the one hand, inhibiting ribosomal translocation with the antibiotic cycloheximide [2, 32] or a mutation in the gene encoding eIF5A [33, 34] promotes mRNA stabilization, whereas the triggering of any of the three mRNA quality control pathways (NMD, NGD, or NSD) by premature termination, mRNA secondary structure, or the lack of a termination codon respectively lead to accelerated decay [16]. This dichotomy remains to be fully explained, but it may well depend on whether or not the ribosome’s A site is occupied by an aminoacyl-tRNA. Only in those instances in which elongation blockage is accompanied by an occupied A site is the mRNA stabilized.
Although the experimental evidence is limited to premature termination, it appears so far that all modes of inhibiting termination lead to mRNA stabilization. For example, overexpression of suppressor tRNAs has been shown in multiple instances to antagonize NMD [35]. Similarly, the RSE, a sequence element within the unspliced Rous sarcoma virus (RSV) RNA, is thought to promote stability of that RNA by antagonizing the utilization of a termination codon separating the gag and pol open reading frames, thereby preventing the activation of NMD [36–39]. A seemingly related type of mRNA-stabilizing sequence element has been detected between upstream open reading frames and the main open reading frame of two yeast mRNAs. These cis-acting stabilizer elements (STEs) in the 5' leader regions of the yeast GCN4 and YAP1 mRNAs inactivate NMD of the respective transcripts when positioned downstream of a uORF termination codon [40]. STEs appear to bind the Pub1 protein [40] which, in turn, prevents activation of the NMD apparatus. These phenomena may well be related, i.e., both the yeast STEs and the RSV RSE could function by mimicking the RNP context of a normal 3'-UTR, making the termination codon appear to be “normal” rather than premature.
Promotion of mRNA decay by enhanced translation
In principle, the efficiency with which a cis-acting instability element exerts its mRNA destabilizing activity ought to be directly related to the extent to which it is recognized by some factor involved in triggering mRNA decay. If it is the ribosome that recognizes the element then the degree to which decay is triggered ought to be directly related to the translational efficiency of the mRNA in question. This notion is nicely illustrated by NMD in both yeast and metazoans.
Translation of upstream open reading frames (uORFs) in mRNA 5′ leaders can reduce translation of downstream ORFs and also decrease overall mRNA stability by triggering NMD. One of the best examples of uORF translation and its effect on mRNA stability has come from the study of yeast CPA1, a gene that encodes the small subunit of arginine-specific carbamoyl phosphate synthase [41]. The CPA1 uORF encodes the arginine attenuator peptide (AAP), whose translation is critical for arginine-specific negative regulation. The AAP stalls elongating ribosomes at the uORF termination codon in response to arginine, thereby blocking access of scanning ribosomes to the downstream CPA1 initiation codon. Arg-regulated ribosome stalling by the AAP is thought to stabilize a conformation of the nascent peptide that interferes with peptidyltransferase function [42]. Ribosome stalling by the AAP also triggers NMD of the CPA1 mRNA [43], an event that is dependent on the extent of ribosome occupancy of the uORF termination codon. This relationship between the extent of termination codon occupancy by the ribosome and the degree to which NMD is triggered is supported by two additional experiments. First, a mutation in the uORF sequence (D13N) that nullifies the ribosome stalling effect of the AAP was shown to diminish NMD activation [43]. Second, improving the initiation codon context of the D13N uORF, i.e., increasing the number of ribosomes translating the uORF, led to enhanced NMD [43]. In three related scenarios, the C. elegans GLD1 protein inhibits translation and subsequent NMD of the uORF-containing gna-2 mRNA, as well as that of other target mRNAs that have acquired premature translational termination codons [44], and the yeast ASH1 mRNA remains translationally silenced and refractory to NMD while associated with Puf6 and Khd1 as it is transported to the cell’s bud tip [45]. Although ASH1 mRNA is insensitive to NMD when translation is repressed during transport, it becomes susceptible to NMD once repression is relieved [45]. In another example, the expression of Robo3.2, a receptor for axonal guidance cues, is regulated by its localized translation coupled to NMD [46]. However, how NMD is regulated concurrently with localized translation is not clear.
These observations have interesting implications for the role of the EJC in metazoan NMD. Pre-mRNA splicing deposits multiprotein EJCs 20–24 nt upstream of splice sites and these complexes serve as binding platforms for factors essential to other steps in posttranscriptional control, including the Upf2 and Upf3 factors required for NMD [15, 47]. As EJC factors have also been shown to have a positive influence on the translatability of mRNPs with which they are associated [48, 49], and recent studies have demonstrated that the EJC core component MLN51 interacts with eIF3 to activate translation [50], EJCs may not only deliver key NMD factors, but may also promote sufficient mRNA translation to ensure that the nonsense codon is recognized and NMD is actually activated. Two significant corollaries of this concept are that NMD may not be so efficient as to be triggered by a single interaction between an elongating ribosome and a premature termination codon and that some putative inhibitors of NMD may actually work indirectly by inhibiting translation.
Translational repression as a prerequisite for mRNA decay
Consistent with the notion that mRNA translation and decay can be separate phenomena, there are several well-characterized examples of translational silencing preceding the initiation of mRNA decay. Several trans-acting mediators of decay-promoting translational repression are discussed below. In addition, Box 1 illustrates that cellular mechanisms of translational repression can also target viral mRNAs and Box 2 demonstrates that specific cis-acting sequences can determine a transcript’s susceptibility to repression.
Box 1: Host-mediated viral mRNA translation repression and decay Zinc-finger antiviral protein (ZAP).
Zinc-finger antiviral protein (ZAP) is an inducible host factor that inhibits replication of alphaviruses, filoviruses, and retroviruses by restricting viral mRNA accumulation [73]. ZAP binds directly to ZAP-responsive elements (ZREs) of specific viral mRNAs and recruits the host PARN deadenylase complex to initiate viral mRNA degradation [73, 74]. Apart from its function in 3’–5’ mRNA decay via the exosome, ZAP interaction with the DEAD-box helicase p72 recruits the decapping complex and activates 5’-3’ decay [73]. Binding of ZAP to target mRNA also represses translation of the target mRNA. Translation repression is mediated by an interaction of ZAP with eIF4A that disrupts eIF4A’s ability to interact with eIF4G [73]. Downregulation of PARN and Dcp2 affects mRNA degradation with little effect on translation repression, suggesting that translation repression precedes mRNA decay [73, 74]. The mechanism by which ZAP represses the translation and/or promotes the degradation of its target mRNA is still unclear. Furthermore, whether or not ZAP affects cellular mRNA is yet to be elucidated.
Box 2: Translation repression and mRNA decay mediated by intronic elements of YRA1 pre-mRNA.
Intron-containing pre-mRNAs are normally retained and processed in the nucleus. However, there are some instances when they are exported to the cytoplasm and degraded by NMD because they contain intronic premature translation termination codons [10]. An exception to this rule is the yeast YRA1 pre-mRNA, an exported intron-containing transcript that evades NMD and is instead targeted by a specific decapping-dependent, 5’ to 3’ cytoplasmic decay pathway mediated by the decapping activator, Edc3 [75, 76]. YRA1 pre-mRNA decay is independent of translation and requires five structurally distinct but functionally interdependent modular elements in the YRA1 intron [75]. Two of these elements target the pre-mRNA as a substrate for Edc3, and the other three mediate transcript-specific translational repression [75]. Translational repression of YRA1 pre-mRNA also requires the heterodimeric Mex67/Mtr2 mRNA export receptor, but not Edc3 [75]. Interestingly, elimination of translational repression, e.g., by deletions within specific intron modules, converts the YRA1 pre-mRNA to an NMD substrate [75], suggesting that translational repression of YRA1 pre-mRNA enhances Edc3 substrate specificity by inhibiting the susceptibility of this pre-mRNA to NMD.
PUF proteins and deadenylases
Puf proteins are mRNA regulatory factors that play significant roles in development, differentiation, neural regulation, and organelle function [51, 52]. These proteins are sequence-specific RNA-binding proteins whose association with a targeted mRNA alters that transcript’s translation and/or decay rates. The primary characteristic of Puf proteins is a highly conserved Pumilio homology domain, often referred to as the Puf repeat domain, that consists of eight consecutive Puf repeats, each about 40 amino acids long [52]. Even though a wide array of protein interactions is involved, repression of mRNA function by Pufs is promoted by disruption of ongoing translation and/or recruitment of components of the decay machinery. In higher eukaryotes, interaction of Puf proteins with cofactors such as Brat and other RNA-binding proteins such as Nanos, CPEB, DAZ, DAZL, and BOL, are required for repression of mRNA targets [52]. Repression of the hunchback mRNA in Drosophila requires formation of a quaternary complex composed of Brat/Nanos/Pumilio/mRNA that recruit the Ccr4–Pop2/Caf1–Not or Pan2/Pan3 deadenylases [52]. Likewise, yeast Puf proteins recruit and directly bind Pop2, which, in turn, bridges interactions between the Puf and the Ccr4-Not complex [52]. Recruitment of Ccr4, the catalytic subunit of the deadenylase complex, by Puf3, Puf4, or Puf5 results in deadenylation of the mRNA target [52], a step that is a prelude to decapping and exonucleolytic degradation (Figure 1C). Surprisingly, in some cases recruitment of the Ccr4-Not deadenylase complex is sufficient to repress mRNA expression without concomitant deadenylation [53]. Analysis of the mechanism underlying this phenomenon in Xenopus demonstrated that Caf1/Pop2 is inherently capable of repressing translation independent of its deadenylation activity [53]. This observation provided considerable insight into the mechanism by which miRNAs down-regulate gene expression (see next section).
miRNAs
miRNAs mediate mRNA repression by base-pairing with partially complementary sequences in the 3′-UTRs of target mRNAs, an event that leads to recruitment of the miRNA-induced silencing complex (miRISC) containing a miRNA-loaded Argonaute protein and a glycine-tryptophan repeat–containing protein, GW182 [54]. miRISC appears to inhibit capdependent translation at multiple steps and to concurrently promote deadenylation and subsequent decapping and decay of its target mRNAs [54–56]. The latter events are initiated by miRNA-mediated deadenylation, requiring the Ccr4–Not and the Pan2/Pan3 deadenylation complexes, with GW182 acting as a binding platform [55] (Figure 1D). However, several studies have also suggested that miRNAs can repress target mRNA translation with no impact on mRNA levels [54]. In zebrafish embryos and D. melanogaster cell-free extracts, addition of an extra 10–40 non-A nucleotides to the poly(A) tail blocked miRNA-mediated deadenylation, but had no effect on translation repression [54, 55, 57]. Further, miRNAs could still repress translation of nonadenylated mRNAs in Drosophila S2 cells [55] Also, tethering of CAF1-CNOT7 subunit of the CCR4–NOT deadenylase complex to reporter mRNAs could repress cap-dependent translation independent of deadenylation in Xenopus laevis oocytes [58]. Repression of non-adenylated mRNAs has been observed to be impaired under Ccr4-Not knockdown conditions [59, 60]. Hence, much like Puf-mediated regulation, GW182-mediated recruitment of the Ccr4-Not complex can promote translational repression independently of deadenylation.
Decapping activators
Some decapping activators, a set of proteins that include Dhh1, Pat1, Edc1–3, Lsm1–7, Scd6, and Stm1, appear to promote decapping by translational repression, a step that may render the cap more accessible to decapping enzymes [2] (Figure 1E). For example, Dhh1 (DDX6/p54 RNA helicase) overexpression inhibits translation and leads to P-body accumulation. In yeast, tethering of Dhh1 to actively translating mRNAs promoted their transition to a translationally repressed state [61]. These effects have been attributed to Dhh1 inhibition of the interaction between the translation initiation complex and the mRNA cap, possibly by altering the competition between eIF4E and Dcp2 for binding to the mRNA cap [2]. Consistent with this idea, Xenopus p54 was found to form a complex with the eIF4E inhibitor, eIF4E-T [62]. Furthermore, Dhh1 has been shown to inhibit 48S complex formation in vitro, but had little effect on mRNAs undergoing IRES-dependent translation [2, 58]. Although these observations suggest that Dhh1 might affect cap-mediated translation initiation, they are to be contrasted with experiments showing that mRNA decay can be a co-translational process [7, 8], i.e., decapping does not require the mRNA to be completely devoid of ribosomes. More recent studies in yeast have shown that Dhh1’s effect on translation is independent of the initiation factors eIF3b and eIF4E [63] and that tethering of Dhh1 to mRNA led to the accumulation of ribosomes on the mRNA [61, 63]. Indeed, elongation-impaired ribosomes displayed enhanced Dhh1-dependent decapping [63]. Other evidence supports the notion of inhibition of decapping by alterations in translation elongation. First, decapping rates have been found to decrease after treatment with translation elongation inhibitor cycloheximide [32]. Furthermore, it has been argued that the length of the ORF, a direct measurement of the number of elongating ribosomes, affects the decapping rates of both normal as well as aberrant mRNAs [64]. Hence, the rate of translation elongation may affect the functions of decapping activators. Similar to Dhh1, other general mRNA decapping activators also repress translation both in vivo and in vitro [65]. Pat1, a scaffolding protein stimulating Dcp2, is also associated with the translating mRNP and interacts with eIF4E, eIF4G and PABP1 [66]. Overexpression of Pat1 leads to translation repression and increased P-body formation [2]. Pat1 deletion together with Dhh1, resulted in loss of translation repression in response to glucose deprivation as observed by polysome analysis [5]. Scd6, another decapping activator, is an RNA binding protein that interacts with Dhh1, Dcp2, and Pat1 [2]. Pat1 and Scd6 repress translation during 48S initiation complex formation and subsequently affect decapping [2, 65, 67]. Direct binding of Scd6 to eIF4G has been found to block 43S complex formation [67]. Another decapping activator, Stm1, is a ribosome binding protein and affects decapping of a subset of yeast mRNAs [68]. Stm1 inhibits translation after the 80S complex formation and is perceived to be mediated via its interaction with the ribosome [68, 69]. How translation inhibition leads to mRNA decapping, is still unresolved.
Getting fooled: alterations in mRNA decay that promote changes in translation efficiency
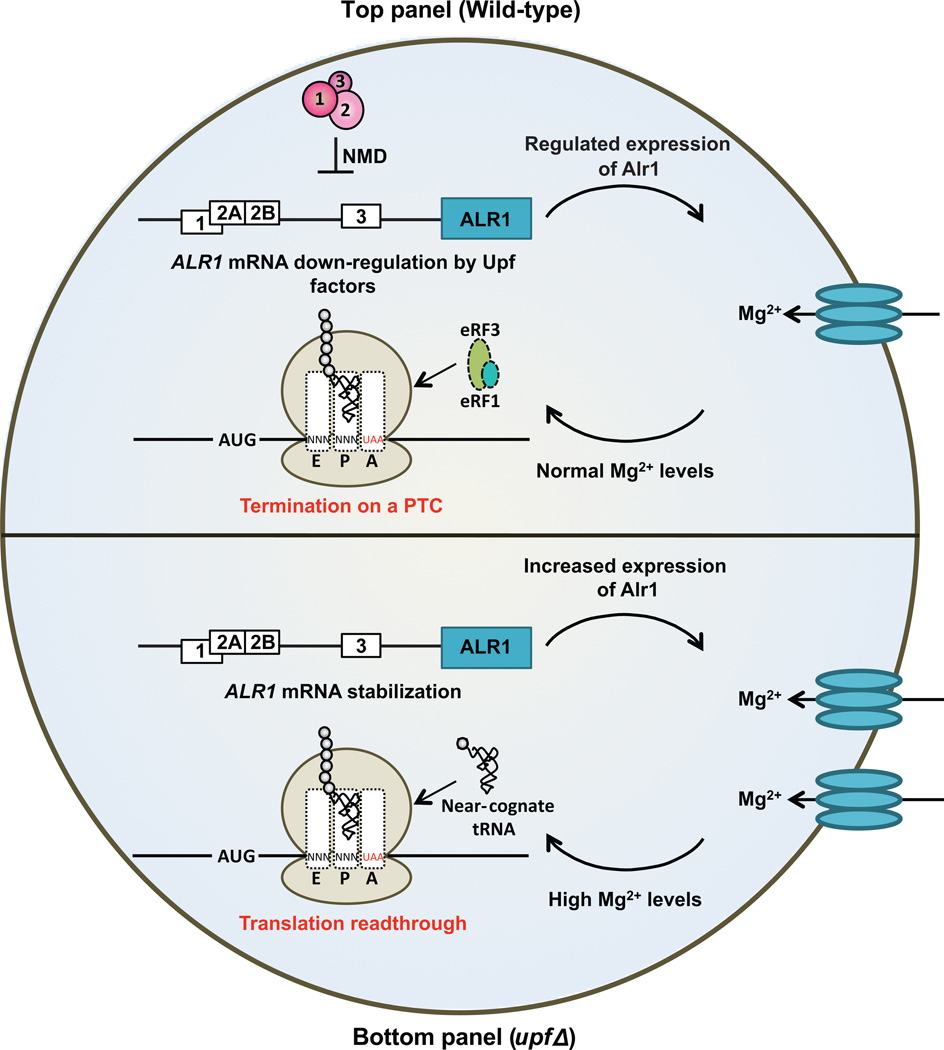
When encountered in-frame by an elongating ribosome, the nonsense codons UAA, UAG, and UGA usually lead to the termination of protein synthesis. However, when the fidelity of translation termination is compromised, e.g., by mutations in the genes encoding translation factors or the presence of specific ribosome-binding small molecules, termination efficiency can be reduced, thereby allowing a near cognate tRNA to compete effectively with eRF1 and to base-pair with a nonsense codon [10, 70]. The resulting insertion of an amino acid at a termination codon is called nonsense suppression or nonsense codon readthrough. Mutations in the yeast UPF genes that regulate NMD have been known to promote nonsense suppression and that effect was thought to reflect interactions between the Upfs and the release factors [10]. An alternative explanation arose from experiments seeking anti-suppressor mutations that could reverse the readthrough phenotype of upf1 mutants. The largest complementation group in that screen was comprised of mutations in ALR1, a gene encoding yeast’s principal Mg2+ transporter [71]. The mutant alr1 alleles did not affect the accumulation of nonsense-containing mRNAs in upf1 cells, indicating that the anti-suppression phenotype was not caused by decreased mRNA levels. Moreover, the alr1 alleles also prevented nonsense suppression in upf2Δ and upf3Δ cells, demonstrating that they counteracted the effect of NMD inactivation and were not specific to the absence of Upf1. An explanation for these phenotypes followed from the observation that the ALR1 mRNA contains several uORFs in its 5’ leader that render it an endogenous substrate of the NMD pathway. This mRNA shows an increased abundance and extended half-life in NMD-deficient cells, and these changes correlate with an accumulation of the Alr1 transporter and with increased levels of intracellular Mg2+ (Figure 3, lower panel). Thus, nonsense suppression caused by upf mutations in yeast can be explained, at least in part, by the indirect effects of NMD inactivation: stabilization of the ALR1 mRNA leading to increased levels of Alr1 protein and increased intracellular Mg2+ levels which, in turn, have significant effects on translational fidelity [71] (Figure 3).
Figure 3. Nonsense suppression as a result of altered mRNA stability.
Upper panel - Upf proteins regulate the stability of the mRNA encoding the magnesium transporter, Alr1. The uORFs in the 5’ leader of the ALR1 mRNA confer NMD sensitivity and regulate expression of the Alr1 transporter on the cell surface. This leads to normal magnesium levels in the cell, which in turn regulates the fidelity of the termination event at a premature termination codon.
Lower panel - In the absence of Upf proteins, ALR1 mRNA is stabilized due to loss of NMD. Stabilization of the mRNA leads to increased expression of Alr1 protein and increased uptake of magnesium. Elevated intracellular magnesium levels affect the fidelity of translation resulting in incorporation of a near cognate tRNA at PTCs, leading to nonsense suppression.
Concluding remarks
Translation and degradation, two fundamental gene expression processes in which all mRNAs are engaged, are temporally separable for some mRNAs and intimately integrated for others. When separated, translational repression by diverse mechanisms is sometimes a prerequisite for the onset of decay. When the two processes are integrated, the translation status of an mRNA can have varying effects. Aberrant translation almost always triggers accelerated decay. However, reductions in the rate of translation initiation frequently promote enhanced mRNA degradation, whereas inhibition of translation elongation or termination generally lead to mRNA stabilization. That said, the only reliable rule is that translation and mRNA decay are sufficiently interrelated that both processes must be understood in detail when modeling the expression of any gene. Complicating any follow through on that rule, and providing fodder for further analyses, are observations that factors influencing translation and mRNA decay can enter the gene expression pathway as early as the synthesis of the primary pre-mRNA transcript [2, 72].
Highlights.
Translation and decay have complex effects on mRNA expression.
Translational repression is a prerequisite for the onset of decay of some mRNAs.
Aberrant translation almost always triggers accelerated mRNA decay.
Reductions in translation initiation frequently promote enhanced mRNA degradation.
Inhibition of translation elongation or termination often lead to mRNA stabilization.
Acknowledgments
This work was supported by grants to A. J. from the US National Institutes of Health, the Human Frontier Science Program, and the US-Israel Binational Science Foundation.
Glossary
- Anti-suppressor
A molecule that prevents nonsense suppression
- Capped mRNA
mRNAs with a 7-methyl guanosine group at the 5’ end of eukaryotic mRNAs
- Closed-loop mRNP
Interaction between the 5’ and the 3’ end of the mRNA mediated by protein factors resulting in a closed configuration
- Deadenylation/ poly(A) shortening
Removal of the adenylate groups from the 3’ end of the mRNA with the aid of deadenylases
- Endonucleolytic cleavage
Enzyme-dependent cleavage of a phosphodiester bond within (endo-) a nucleotide chain resulting in a 5’ and a 3’ fragment
- Exonucleolytic digestion
Enzyme-dependent hydrolysis of nucleotides from the 5’- or 3’- end of a nucleotide chain
- mRNA decapping
Hydrolysis of the 7-methyl guanosine group at the 5’ end of eukaryotic mRNAs by decapping enzymes
- mRNA stability
Equilibrium between the rate of mRNA synthesis and degradation
- P-bodies
Cytosolic foci consisting of mRNA targeted for mRNA decay and proteins required for decay
- Polysome–associated mRNAs
mRNA molecules associated with multiple ribosomes engaged in translation
- Ribosome A site
Ribosomal site occupied by the incoming aminoacyl-tRNA
- Termination readthrough/nonsense suppression
Insertion of an amino acid at a stop codon through a near-cognate aminoacyl tRNA resulting in polypeptide chain elongation
- Translational stall
A stop in the movement of the translating ribosome in response to either the sequence features of the mRNA, secondary structures, rare codons in the coding sequence, or a sequence of the nascent peptide
- Translation repression
Inhibition of protein synthesis of all or a subset of mRNAs in response to stimuli (external or internal)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ghosh S, Jacobson A. RNA decay modulates gene expression and controls its fidelity. WIREs RNA. 2010;1:351–361. doi: 10.1002/wrna.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckmann CR, et al. Control of poly(A) tail length. Wiley Interdiscip Rev RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 4.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decker CJ, et al. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell. Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu W, et al. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu W, et al. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:244–247. doi: 10.1038/nsmb.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Min E, et al. Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit. RNA. 2013;19:1105–1115. doi: 10.1261/rna.039396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangus DA, Jacobson A. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- 12.Atkin AL, et al. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atkin AL, et al. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 14.Graille M, Seraphin B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat Rev Mol Cell Biol. 2012;13:727–735. doi: 10.1038/nrm3457. [DOI] [PubMed] [Google Scholar]
- 15.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh S, et al. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA. 2010;16:1832–1847. doi: 10.1261/rna.1987710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inada T. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim Biophys Acta. 2013;1829:634–642. doi: 10.1016/j.bbagrm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Graille M, et al. Structure of yeast Dom34: a protein related to translation termination factor Erf1 and involved in No-Go decay. J Biol Chem. 2008;283:7145–7154. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, et al. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol. 2010;17:1233–1240. doi: 10.1038/nsmb.1922. [DOI] [PubMed] [Google Scholar]
- 23.Kuroha K, et al. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 2010;11:956–961. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Passos DO, et al. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell. 2009;20:3025–3032. doi: 10.1091/mbc.E09-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker T, et al. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 26.Frischmeyer PA, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 27.van Hoof A, et al. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 28.Klauer AA, van Hoof A. Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip Rev RNA. 2012;3:649–660. doi: 10.1002/wrna.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc Natl Acad Sci U S A. 2011;108:2366–2371. doi: 10.1073/pnas.1013180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3'-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in. Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beelman CA, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 33.Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saini P, et al. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gozalbo D, Hohmann S. Nonsense suppressors partially revert the decrease of the mRNA level of a nonsense mutant allele in yeast. Curr. Genet. 1990;17:77–79. doi: 10.1007/BF00313252. [DOI] [PubMed] [Google Scholar]
- 36.Weil JE, Beemon KL. A 3' UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA. 2006;12:102–110. doi: 10.1261/rna.2129806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weil JE, et al. Structural characterization of the Rous sarcoma virus RNA stability element. J Virol. 2009;83:2119–2129. doi: 10.1128/JVI.02113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Withers JB, Beemon KL. Structural features in the Rous sarcoma virus RNA stability element are necessary for sensing the correct termination codon. Retrovirology. 2010;7:65. doi: 10.1186/1742-4690-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Withers JB, Beemon KL. The structure and function of the rous sarcoma virus RNA stability element. J Cell Biochem. 2011;112:3085–3092. doi: 10.1002/jcb.23272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz-Echevarria MJ, Peltz SW. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–751. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 41.Gaba A, et al. Ribosome occupancy of the yeast. CPA. 1. upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 42.Wei J, et al. The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol Cell Biol. 2012;32:2396–2406. doi: 10.1128/MCB.00136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amrani N, et al. Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell. Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 44.Lee MH, Schedl T. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C elegans. Genes Dev. 2004;18:1047–1059. doi: 10.1101/gad.1188404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heym RG, Niessing D. Principles of mRNA transport in yeast. Cell Mol Life Sci. 2012;69:1843–1853. doi: 10.1007/s00018-011-0902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colak D, et al. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153:1252–1265. doi: 10.1016/j.cell.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh G, et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isken O, et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma XM, et al. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 50.Chazal PE, et al. EJC core component MLN51 interacts with eIF3 and activates translation. Proc Natl Acad Sci U S A. 2013;110:5903–5908. doi: 10.1073/pnas.1218732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wharton RP, Aggarwal AK. mRNA regulation by Puf domain proteins. Sci STKE. 2006;2006:pe37. doi: 10.1126/stke.3542006pe37. [DOI] [PubMed] [Google Scholar]
- 52.Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA. 2011;2:471–492. doi: 10.1002/wrna.69. [DOI] [PubMed] [Google Scholar]
- 53.Goldstrohm AC, et al. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- 54.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 55.Djuranovic S, et al. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fabian MR, et al. Understanding how miRNAs post-transcriptionally regulate gene expression. Prog Mol Subcell Biol. 2010;50:1–20. doi: 10.1007/978-3-642-03103-8_1. [DOI] [PubMed] [Google Scholar]
- 57.Mishima Y, et al. Translational inhibition by deadenylation-independent mechanisms is central to microRNA-mediated silencing in zebrafish. Proc Natl Acad Sci U S A. 2012;109:1104–1109. doi: 10.1073/pnas.1113350109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cooke A, et al. Translational repression by deadenylases. J Biol Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chekulaeva M, et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braun JE, et al. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 61.Carroll JS, et al. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol. 2011;194:527–537. doi: 10.1083/jcb.201007151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minshall N, et al. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- 63.Sweet T, et al. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- 65.Nissan T, et al. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marnef A, Standart N. Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem Soc Trans. 2010;38:1602–1607. doi: 10.1042/BST0381602. [DOI] [PubMed] [Google Scholar]
- 67.Rajyaguru P, et al. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol Cell. 2012;45:244–254. doi: 10.1016/j.molcel.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balagopal V, Parker R. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA. 2011;17:835–842. doi: 10.1261/rna.2677311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balagopal V, Parker R. Stm1 modulates mRNA decay and Dhh1 function in Saccharomyces cerevisiae. Genetics. 2009;181:93–103. doi: 10.1534/genetics.108.092601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peltz SW, et al. Ataluren as an agent for therapeutic nonsense suppression. Annu Rev Med. 2013;64:407–425. doi: 10.1146/annurev-med-120611-144851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johansson MJ, Jacobson A. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 2010;24:1491–1495. doi: 10.1101/gad.1930710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lotan R, et al. The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J Cell Biol. 2007;178:1133–1143. doi: 10.1083/jcb.200701165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu Y, et al. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J. 2012;31:4236–4246. doi: 10.1038/emboj.2012.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu Y, et al. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci U S A. 2011;108:15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dong S, et al. Degradation of YRA1 Pre-mRNA in the cytoplasm requires translational repression, multiple modular intronic elements, Edc3p, and Mex67p. PLoS Biol. 2010;8:e1000360. doi: 10.1371/journal.pbio.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dong S, et al. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]