Abstract
We propose a framework for quantifying node-level community structures between groups using anatomical brain networks derived from DTI-tractography. To construct communities, we computed hierarchical binary trees by maximizing two metrics: the well-known modularity metric (Q), and a novel metric that measures the difference between inter-community and intra-community path lengths. Changes in community structures on the nodal level were assessed between generated trees and a statistical framework was developed to detect local differences between two groups of community structures. We applied this framework to a sample of 42 subjects with major depression and 47 healthy controls. Results showed that several nodes (including the bilateral precuneus, which have been linked to self-awareness) within the default mode network exhibited significant differences between groups. These findings are consistent with those reported in previous literature, suggesting a higher degree of ruminative self-reflections in depression.
1 Introduction
Modern imaging techniques have allowed us to construct anatomical (structural) and functional networks of the human brain and a new term - the human “connectome” [1] has been recently coined to describe their mathematical properties. A brain connectivity network consists of nodes which represent gray matter regions and edges connecting nodes which represent white matter fibers. Many tools and metrics have been adapted from the field of graph-theory to help characterize and analyse the human connectome [1-2]. It has been shown that brain networks are sparse (networks having the number of nodes in the order of the number of edges) [3], thus allowing the investigation of their community structure properties in which nodes may be grouped into different communities [4, 5] (in our context, “community structure” is a property for sparse networks, which allows us to extract the topological organization of a network and to partition it into a set of non-overlapping communities, sometimes also referred to as modules or clusters).
Studying the structural organization of the brain communities has lately shed light on alterations related to aging [6, 7] as well as neuropsychiatric disorders such as schizophrenia [8-12]. In this paper, we proposed a novel metric, with which we extracted community structures in the form of top-down binary trees (also known as dendrograms). This is followed by the computation of a node “consistency metric”, designed to quantify differences between trees at each node. To our best knowledge, we are the first group to propose a recipe for quantifying node-level community differences between two groups of brain networks. Lastly, statistical analyses were conducted to detect significant community alterations on the nodal level for group studies. We tested the proposed method and compared the binary trees generated from the structural brain networks of depressed versus normal control subjects. Significant changes were found on the nodal level in the community structure in depression relative to controls.
2 Methods
2.1 Image acquisition
We scanned 47 healthy subjects (20 male/27 female; age: 59.74±14.8)) and 42 subjects with major depression as defined by DSM-IV (18 male/24 female; age: 57.64±13.4). A Philips 3.0 T Achieva scanner supplied with 8-channel SENSE head-coil located at the University of Illinois Medical Center Advanced Imaging Center was used to acquire the brain MRI data. High resolution T1-weighted images were acquired with MPRAGE sequence (FOV=240 mm; TR/TE=8.4/3.9 ms; flip angle 8°; voxel size 1.1×1.1×1.1mm). Diffusion weighted (DW) images were acquired using SS-SE-EPI sequences (FOV = 240 mm; resolution 0.83×0.83 mm; TR/TE=6994/71 ms; flip angle = 90°, 32 gradient directions, b= 700 s/mm2 and one minimally DW scan: b0 image). Parallel imaging was also used with a SENSE factor of 2.5 to reduce scan time to ~ 4 min.
2.2 Brain Network Construction
Structural brain networks were generated using a pipeline which integrates multiple image analysis techniques. First, DW images were eddy current corrected using the automatic image registration (AIR) tool embedded in DtiStudio software (http://www.mristudio.org) by registering all DW images to their corresponding b0 images with 12-parameter a ne transformations. This was followed by the computation of diffusion tensors and deterministic tractography using the DtiStudio program. T1-weighted images were used to generate label maps using the Freesurfer software (http://surfer.nmr.mgh.harvard.edu). Brain networks formed by the 68 cortical regions were generated using an in-house program in Matlab by counting the number of fibers connecting each pair of nodes.
2.3 Hierarchical Trees Creation
Measuring community structure has been a scientifically important task in network science. To this end, we note that nodes belonging to the same community should exhibit stronger connections than those in different communities. A well-known metric for such task is the Q modularity proposed in [13] and is mathematically defined as:
| (1) |
where Q is a function of a graph G, m is the total number of edges, Aij = 1 if an edge links nodes i and j and 0 otherwise, δ(i, j) = 1 if nodes i and j are in the same community and 0 otherwise, and ki is the node i’s degree (its number of edges). Extracting the community structure of a network is obtained by finding the set of non-overlapping modules that maximizes Q. Here, we further propose an alternative maximization problem with respect to a novel metric. This new metric is defined as the difference between the mean inter- and the mean intra-modular path lengths (interPL/intraPL). For two communities Ci and Cj the interPL and intraPL are defined as:
| (2) |
where Ni denotes the number of nodes in a community Ci, dnm denotes the shortest path length connecting nodes n and m. In the case of two modules, for example, the alternative metric ΨPL is defined as:
| (3) |
Here, notice that can be considered the weighted characteristic path length (a measure of global network integration) defined on the sub-network formed by Ci. Thus, maximizing ΨPL is equivalent to searching for a set of communities that exhibit stronger sub-network integration. Since path length is based on the number of fibers, ΨPL may provide a more anatomical basis for defining community structure.
There exist two main types of hierarchical clustering techniques: the agglomerative and the divisive. The agglomerative method starts bottom-up. At the first level, every node is a community by itself. In each subsequent step (level), two communities that are considered closest (with respect to a similarity measure) are merged into one, until all nodes are in one community. The divisive method follows the opposite or a top-down approach. It starts with all nodes belonging to one single community, and at each step each community is further split into several sub-communities (unlike the agglomerative method, the divisive clustering usually uses a dis-similarity measure). It should be noted that one may choose to stop the clustering process at a pre-specified level (stopping criteria can be a given number of communities or the optimization of a quality function [14]). Here, to maximize Q or ΨPL, we used a top-down hierarchical clustering approach, as it has been suggested that agglomerative methods may fail to find correct communities in networks where the community structures are known a priori [13]. At the first level, brain regions were randomly assigned to one of two communities, and optimal assignment was determined by maximizing Q or ΨPL using the well-known simulated-annealing algorithm [15]. The process is repeated at each level until a 4-level dendrogram is reached (a total of 16 communities). One million random permutations were used at each optimization stage.
2.4 Community Structures Difference Assessment
In order to assess local differences between communities in different groups, we first construct the mean binary tree for the control group (by maximizing Q or ΨPL, with respect to the mean connectivity matrix for this group). All individual subjects’ trees will then be compared to the mean control tree. To this end, a local metric is needed to quantify how two trees differ. One candidate metric is via the consistency metric assessment method introduced in [16], which yields a consistency vector V of length equal to the number of nodes in the network (68 in our case). To compute V, we need to first construct a L×M similarity matrix XR that compares any module in a test tree (i.e., the individual subject’s tree in our framework) to any module in a reference tree (the mean tree for the control group). Here L and M are the number of communities in the test and reference trees; in our case L = M = 16. Mathematically, for any two communities Cp and Cq belonging to the test tree and the reference tree respectively, the (p, q)th entry XR(p, q) in XR is calculated using the following equation:
| (4) |
where Npq denotes the number of common nodes between the two communities Cp and Cq, and Np and Nq the number of nodes in community Cp and Cq. Values in XR range from 0 to 1, with 0 indicating no overlap between the two communities and 1 identical communities (numerically, XR(p, q) thus represents the similarity between community Cp in the test tree and community Cq in the reference tree). With the computation of XR, the consistency vector V now can be constructed as follows. First, we identify the maximum entry in each column of the XR matrix. For the qth column corresponding to community Cq in the reference tree, if the maximum exists in the pth row corresponding to community Cp in the test tree, we assign this value to the kth element of V, for all k ∈ {Cp ∩ Cq}. Although V provides a node-level measurement of how “consistent” the test tree’s community structure is with respect to that of the reference tree, it should be noted that with this definition, V may yield zero values. We thus proposed a modified non-zero node-level consistency measure (Vm) for each node k as follows:
| (5) |
where Nc denotes the number of common nodes between the two communities Cp and Cq, that contain this node k in the test and reference trees respectively.
2.5 Statistical Analysis
To statistically test group differences in community structures at the nodal level, all individual subjects’ trees (both depressed and control) are compared to the mean control reference tree, thus yielding 47 V or Vm vectors in the healthy group, and 42 in the depressed group. Group differences on local community structures can now be assessed using node-wise 2-sample Mann-Whitney-Wilcoxon (MWW) tests for V (due to the existence of zero values) or node-wise 2-sample T-tests for Vm followed by correcting for multiple comparisons. Alternatively, a more powerful test can be constructed via a multivariate distribution for each community in the 4th level binary tree of the mean normal group (16 total modules), by concatenating the V or Vm vectors of all nodes in this community, on which 2-sample Hotelling’s T-squared tests can be conducted.
3 Results
3.1 Comparison of modularity measures
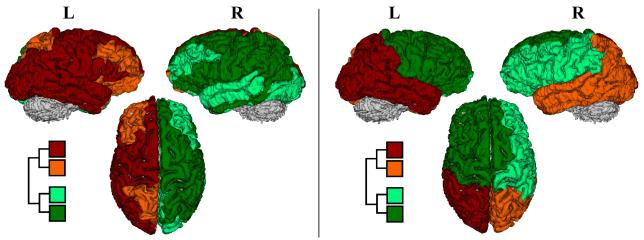
We first generated and compared the mean community structures of the normal controls in this study, using both Q and ΨPL (figure 1). Note that in the first level of the binary tree (partitioning the brain into two modules), the constructed mean tree using the Q function exhibits an exact left/right separation while the tree generated using ΨPL shows an anterior/posterior partition of the human brain (the left-right separation represents the second level partitioning using the proposed metric; the mean community structure of the studied depressed subjects is shown in right panel of figure 1). The second level partitioning using ΨPL also generated more unified communities compared to the Q function.
Fig. 1.
First two levels of modular or community structure via the Q modularity (left) and ΨPL (right). Notice that at the first level, the constructed mean tree by maximizing Q exhibits a left/right parcellation while the constructed mean tree using ΨPL exhibits an anterior/posterior partitioning along the frontal-parietal junction.
3.2 Group difference
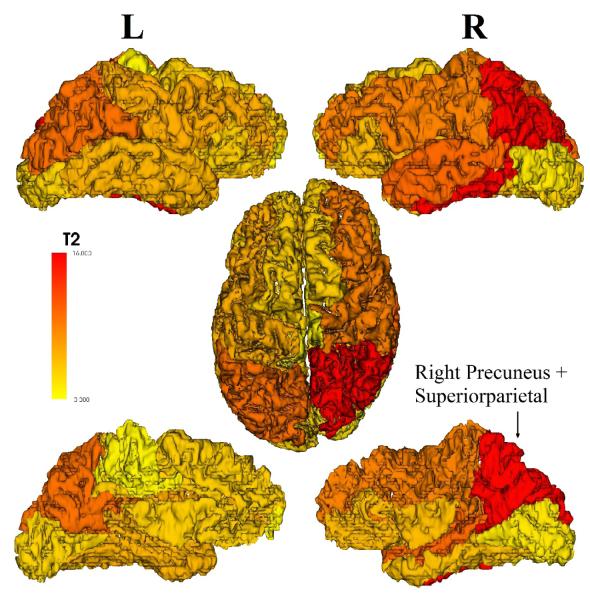
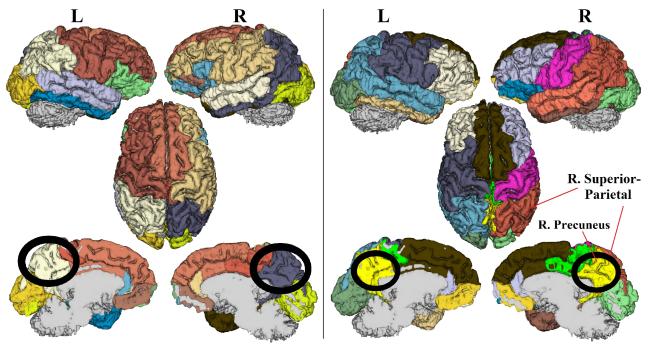
To detect community structure abnormalities in depression, we constructed the Hotelling T-squared statistics as described in the method section for all 16 communities in the reference tree. Results showed a significantly lower consistency (p = 0.0079) in depression versus control for the community containing the right precuneus, superior parietal gyrus, inferior parietal gyrus, inferior temporal gyrus, and isthmus cingulate (figures 2 and 3). On a node level, conducting 2-sample T-tests or the MWW-test on Vm or V confirmed such findings (table 1). The right precuneus and superior parietal gyrus exhibited the lowest node-level consistency (p < 0.01; uncorrected). Comparing the mean community structures between two groups, we note that the right precuenus and right superior parietal gyrus, while belonging to the same community in the healthy controls, are assigned to different communities in the depressed group. Additionally, in depression the bilateral precuneus are assigned to one community, suggesting a stronger structural integration between them (figure 3).
Fig. 2.
Hotelling’s T-Squared statistics are overlaid on the 16 communities of the reference tree, showing regional group differences in the community structure between groups. The most significant differences are in the right precuenus and superior parietal gyrus. This figure is showing a left/right lateral view (top), an axial view (middle) and a left/right mid-line view (at the bottom). The community containing the right precuneus and the right superior parietal gyrus is highlighted in red.
Fig. 3.
4-level binary trees showing mean community structures via the proposed metric ΨPL for the healthy controls (left) and the depressed subjects (right). Notice that the right precuneus and right superior parietal gyrus belong to the same community in the healthy controls but not in the depressed group. Additionally, in the depressed group the right and left precuneus are assigned to one single community (which also comprises bilateral isthmus cingulate).
Table 1.
List of nodes which showed significantly lower node-level consistency in depression relative to healthy controls at p < 0.01 (uncorrected). The table shows mean and standard deviation values using both MWW (for V) and 2-sample T-tests (for Vm), and their corresponding p values.
| Healthy | Depressed | p-value | ||
|---|---|---|---|---|
| right-precuneus | MWW-test | 0.2373 ± 0.1815 | 0.1281 ± 0.1311 | 0.0044 |
| T-test | 0.2645 ± 0.1501 | 0.1784 ± 0.0908 | 0.0019 | |
| right-superiorparietal | MWW-test | 0.2088 ± 0.1773 | 0.1007 ± 0.1376 | 0.0032 |
| T-test | 0.2374 ± 0.1475 | 0.1524 ± 0.1106 | 0.0033 | |
4 Discussion and Conclusion
In this paper we presented a new approach to construct hierarchical community structures from weighted structural brain networks. Our proposed metric took into account the path lengths within and between communities, and we used simulated annealing to construct top-down four-level binary trees. Moreover, we are the first to develop a statistical framework that allows for the quantification of node-level community structural differences between two groups of brain networks, via the consistency vector metric. This framework was validated on a sample of depressed patients. Results revealed local community differences in parts of the default mode network (especially the right precuneus), which has been previously linked to self-awareness and shown to exhibit abnormalities in depression [17, 18].
References
- 1.Sporns O, Tononi, Ktter R. The human connectome: A structural description of the human brain. PLoS Computational Biology. 2005;1:245–251. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. J of NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Bassett BS. Small World Brain Networks. The Neuroscientist. 2006;12(6):512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- 4.Meunier D, Lambiotte R, Fornito A, Ersche KD, Bullmore E. Hierarchical modularity in human brain functional networks. Front. Neuroinformatics. 2009b;3:37. doi: 10.3389/neuro.11.037.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Telesford Q, Simpson S, Burdette J, Hayasaka S, Laurienti P. The Brain as a Complex System: Using Network Science as a Tool for Understanding the Brain. Brain Connectivity. 2011;1(4):295–308. doi: 10.1089/brain.2011.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meunier D, Achard S, Morcom A, Bullmore E. Age-related changes in modular organization of human brain functional networks. J of Neuroimage. 2009a;44:715723. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- 7.Fair DA, Cohen AL, Power JD, Dosenbach NU, Church JA, Miezin FM, Schlaggar BL, Petersen SE. Functional brain networks develop from a “local to distributed” organization. PLoS Comput Biol. 2009;5(5):e1000381. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Liang M, Zhou Y, He Y, Hao Y, Song M, Yu C, Liu H, Liu Z, Jiang T. Disrupted small-world networks in schizophrenia. Brain. 2008;131:945–961. doi: 10.1093/brain/awn018. [DOI] [PubMed] [Google Scholar]
- 9.Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and Schiz-ophrenia. J of Neuroscience. 2008;28(37):9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynall ME, Bassett DS, Kerwin R, McKenna PJ, Kitzbichler M, Muller U, Bullmore E. Functional connectivity and brain networks in schizophrenia. J of Neuroscience. 2010;30(28):9477–9487. doi: 10.1523/JNEUROSCI.0333-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Den Heuvel MP, Mandl RCW, Stam CJ, Kahn RS, Hulsho Pol HE. Aber-rant frontal and temporal complex network structure in schizophrenia: A graph theoretical analysis. J of Neuroscience. 2010;30(47):15915–15926. doi: 10.1523/JNEUROSCI.2874-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander-Bloch A, Lambiotte R, Roberts B, Giedd J, Gogtay N, Bullmore E. The discovery of population differences in network community structure: New methods and applications to brain functional networks in schizophrenia. J of NeuroImage. 2012;59(4):3889–3900. doi: 10.1016/j.neuroimage.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Phys. Rev. E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 14.Fortunato S. Community detection in graphs. Physical Reports. 2010;486(3-5):75–175. [Google Scholar]
- 15.Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by Simulated Annealing. Science. 1983;220(4598):671680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 16.Steen M, Hayasaka S, Joyce K, Laurienti P. Assessing the consistency of community structure in complex networks. Physical Review E. 2011;84:016111. doi: 10.1103/PhysRevE.84.016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X, Wang X, Xiao J, Liao J, Zhong M, Wang W, Yao S. Evidence of a Dissociation Pattern in Resting-State Default Mode Network Connectivity in First-Episode, Treatment-Naive Major Depression Patients. Biol. Psychiatry. 2011 doi: 10.1016/j.biopsych.2011.10.035. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Lou HC, Luber B, Crupain M, Keenan JP, Nowak M, Kjaer TW, Sackeim HA, Lisanby SH. Parietal cortex and representation of the mental Self. Proc. of the National Academy of Sciences of the United States of America. 2004;101 doi: 10.1073/pnas.0400049101. [DOI] [PMC free article] [PubMed] [Google Scholar]