Abstract
β-cells of the pancreatic islets are highly specialized and high-throughput units for the production of insulin, the key hormone for maintenance of glucose homeostasis. Elevation of extracellular glucose and/or GLP-1 levels triggers a rapid upregulation of insulin biosynthesis through the activation of post-transcriptional mechanisms. RNA-binding proteins are emerging as key factors in the regulation of these mechanisms as well as in other aspects of β-cell function and glucose homeostasis at large, and thus may be implicated in the pathogenesis of diabetes. Here we review current research in the field, with a major emphasis on RNA-binding proteins that control biosynthesis of insulin and other components of the insulin secretory granules by modulating the stability and translation of their mRNAs.
Keywords: RNA-binding proteins, β-cells, Diabetes, Insulin, mRNA stability, Translation
1. Introduction
The hallmark of the pancreatic islet β-cell is its ability to synthesize and secrete large quantities of insulin, which maintains metabolic homeostasis by lowering glycemia. Insulin is synthesized as a single-chain precursor termed preproinsulin, composed of an N-terminal signal sequence, the B chain, the connecting C-peptide, and finally the A chain, which is covalently linked to the B chain via disulfide bridges. Preproinsulin is converted into proinsulin upon removal of the signal peptide in the endoplasmic reticulum (ER). Proinsulin is sorted at the trans-Golgi network into immature secretory granules (SGs); subsequent removal of the intervening C-peptide by protein convertases during the maturation of SGs leads to the generation of insulin [1]. Each β-cell contains on average ~5×103 SGs [2], of which however only 1–2% undergo regulated exocytosis in response to elevation of glycemia [3]. Other stimuli, such as incretins, further potentiate the release of insulin induced by hyperglycemia. Each SG is an independent functional unit composed of >50 different cargo proteins, including insulin and C-peptide, other peptide hormones, convertases for peptide processing, packaging proteins such as the granins, pumps and channels, as well as ions, neurotransmitters and nucleotides [4]. Glucose-triggered SG exocytosis is coupled to a concomitant rapid increase in the biosynthesis of insulin and other components necessary for SG assembly [5]. This is especially relevant in view of the evidence that newly-synthesized SGs undergo preferential exocytosis [6–8]. Hence, proper secretory function of β-cells strictly depends on their glucose-regulated SG biogenesis [5].
Rapid induction of insulin biosynthesis by glucose involves regulation at the transcriptional, translational, and post-translational levels [9–11]. Through elevation of intracellular cAMP, GLP-1 also induces insulin expression via transcriptional and post-transcriptional mechanisms similar to those elicited by glucose [12]. Several landmark papers have shown that in the short term, glucose-induced enhancement of insulin synthesis does not require de novo transcription of preproinsulin mRNA, but is obtained by specifically increasing the efficiency of translation as well as the number of mRNA molecules available for protein synthesis [13–16]. Hence, detailed knowledge of the molecular machinery responsible for preproinsulin mRNA stability and translation is of paramount importance for understanding the physiology of β-cells and their inability to meet insulin demands in type 2 diabetes.
2. Regulatory elements in the preproinsulin mRNA
There is now ample evidence that sequence elements within the 5′- and 3′-untranslated regions (UTRs) of the preproinsulin mRNA control its translation and stability in response to stimuli such as hyperglycemia. In 2001 Rhodes and colleagues first reported that the 5′- and the 3′-UTRs of the preproinsulin mRNA act cooperatively to increase glucose-induced preproinsulin biosynthesis in rat pancreatic islets [17]. Specifically, their study identified a UUGAA motif located between the polyadenylation signal and the polyadenylation site within the 3′-UTR of rat preproinsulin 2 mRNA which is critical for its stability (Figure 1) [17]. Notably, this motif is found in all rat, human and mouse preproinsulin mRNAs. In a later study, a pyrimidine-rich sequence located just downstream of the preproinsulin 1 coding region was reported to have a similar role in the control of mRNA turnover (Figure 1) [18]. The authors further identified polypyrimidine tract-binding protein 1 (PTBP1) as a factor binding to this sequence element, and thereby regulating mRNA stability (Table 1). Interestingly, the interaction between PTBP1 and the preproinsulin mRNA was promoted by increased glucose availability and hypoxia [18,19]. Such a pyrimidine-rich tract is also present in a similar location in the other rat, human, and mouse preproinsulin transcripts.
Figure 1.
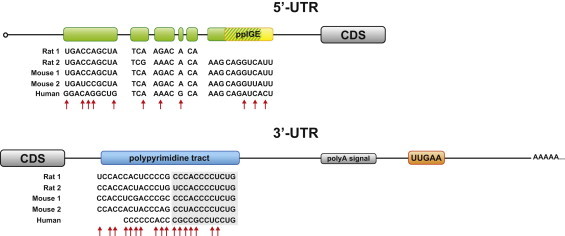
Regulatory elements in the preproinsulin mRNA UTRs. The green boxes indicate a 29 bp-sequence element, identified in the 5′-UTR of the rat preproinsulin gene 1. This element partially overlaps with the 9 bp-sequence identified in the rat proinsulin 2 mRNA as the preproinsulin glucose element (ppIGE, in yellow). The 3′-UTR of the preproinsulin transcript contains elements that control mRNA stability, including a pyrimidine-rich tract (in blue) downstream of the coding sequence (CDS), and a UUGAA motif (in orange). The UUGAA element is identical in human, rat and mouse, while other regulatory elements within preproinsulin mRNAs, though highly conserved in their location, display various degrees of sequence homology. The cross-species sequence homology of these elements among rat, mouse and human preproinsulin transcripts is shown. Red arrows point to non-conserved nucleotides. The shaded sequences are those identified as PTBP1 binding sites. A polypyrimidine tract is also found in the 5′-UTR of the human preproinsulin mRNA (not shown).
Table 1.
RNA-binding proteins with roles in β-cell function and type 2 diabetes.
|
Insulin/SG biosynthesis | ||||||
|---|---|---|---|---|---|---|
| RBP | RNA-binding domain | Target | Cis-element | Regulation of target mRNA | Model system | Ref. |
| HuD | RRM | Insulin | 5′-UTR | Translation ⇓ | Mouse, βTC6 | [61,62] |
| INS-1E | ||||||
| PDI/PABP | RRM | Insulin | 5′-UTR | Translation ⇑ | Rat, βTC6 | [60] |
| PTBP1 | RRM | Insulin | 3′-UTR | Stability ⇑ | Rat, INS-1 | [18,19] |
| ICA512 | [33,34] | |||||
| PC1/3 | ||||||
| PC2 | ||||||
| Insulin | 5′-UTR | Translation ⇑ | Human, INS-1 | [33,52] | ||
| PC2 | ||||||
| ICA512 | 3′-UTR | Translation ⇑ | INS-1 | [28,33] | ||
| PC2 | ||||||
| CGA | n.d. | Expression ⇑ | INS-1 | [28,33] | ||
| SCG2 | ||||||
| SYB2 | ||||||
| Other functions in β-cells and glucose homeostasis | ||||||
| RBP |
RNA-binding domain |
Target |
Function |
Model system |
Ref. |
|
| AUF1 | RRM | Bcl2 | β-cell death ⇑ | MIN6 | [67] | |
| Mcl1 | ||||||
| Hzf | zf-C2H2 | n.d. | Glucose metabolism ⇑ | Mouse | [72] | |
| Insulin sensitivity ⇑ | ||||||
| IGF2BP2 | RRM, KH | IGF2 | IGF2 expression ⇑ | RD | [73] | |
| Mushashi 1 | RRM | Hes1 | Insulin expression ⇓ | MIN6 | [65] | |
| β-cell proliferation ⇑ | ||||||
| β-cell death ⇓ | ||||||
| Mushashi 2 | RRM | n.d. | Insulin expression ⇓ | MIN6 | [65] | |
| PTBP1 | RRM | IR | IR expression ⇑ | HeLa | [68] | |
| RBM4 | RRM | Isl1 | Insulin expression ⇑ | Mouse | [63] | |
| Pax4 | AR42J | |||||
| IR | Insulin signaling ⇑ | AR42J | [63] | |||
| SRSF3 | RRM | Hnf1α | Glucose metabolism ⇓ | Mouse | [69] | |
| Insulin sensitivity ⇓ | ||||||
n.d.=not determined.
While the 3′-UTR of the preproinsulin mRNA contains elements critical for its stability, the 5′-UTR regulates preproinsulin translation rate. Indeed, a 2007 study reported the presence of a conserved element in the 5′-UTR of the rat preproinsulin 2 mRNA, termed the preproinsulin glucose element (ppIGE), which is required for glucose-induced preproinsulin translation (Figure 1) [20]. Luciferase reporter assays indicated that mutation or removal of this cis-element abolished glucose-induced translational activation [20]. A 29 bp-sequence element was also described in the 5′-UTR of the rat preproinsulin gene 1 by an independent study [21]. This element partially overlaps with the ppIGE. However, while the regulatory function of ppIGE lies in its primary sequence, a stem-loop secondary structure has been postulated to be critical for translational activation by this 29-bp element (Figure 1) [20,21]. On the other hand, both studies on the ppIGE and the 29-bp element proposed that induction of insulin translation occurs through glucose-dependent binding of yet unknown factors to the 5′-UTR of insulin mRNA. Intriguingly, ppIGE core sequences were also found in the mRNA 5′-UTRs of other glucose-regulated proteins of the insulin SGs, namely prohormone convertases PC1/3 and PC2, pro-islet amyloid polypeptide (proIAPP), and chromogranin A (CGA) [22].
Cis-acting sequence elements in the UTRs of mRNAs act in concert with a group of trans-acting factors to specifically and rapidly change the translational pattern of the β-cell in response to external stimuli, and thus maintain glucose homeostasis. Such trans-acting factors include microRNAs (miRNAs) and RNA-binding proteins (RBPs) [23]. RBPs compose a vast group of structurally and functionally diverse proteins which regulate virtually all aspects of RNA metabolism [24,25]. Supporting their role in mediating the adaptive response of the β-cell, mRNA-binding proteins have been identified as a major class of molecules whose expression pattern rapidly changes in response to stimulation of insulinoma cells [26]. In the following sections we review current knowledge of RBPs that specifically regulate insulin abundance and the assembly of SGs by modulating mRNA turnover and translation. Furthermore, we will briefly discuss RBPs with proposed roles in other aspects of β-cell physiology and more generally implicated in glucose homeostasis and type 2 diabetes.
3. Stability of mRNAs encoding components of the insulin SGs
Insulin mRNA stability varies according to nutrient availability. In animal studies, significantly lower insulin mRNA levels were measured in fasted rats compared to control animals [27]. In vitro studies have further shown that the half-life of insulin mRNA is markedly increased under high glucose conditions, while it decreases sharply in the absence of glucose [16,28]. Interestingly, the latter effect is reversed by elevation of intracellular cAMP levels [28].
The most thoroughly-characterized regulator of preproinsulin mRNA stability, and thereby discussed here in greater length, is the ubiquitous RNA-binding protein PTBP1 (also known as PTB or hnRNP I – heterogeneous nuclear ribonucleoprotein I), which is essential for mammalian development already at early stages of gastrulation [29,30]. Heterogeneous nuclear ribonucleoproteins (hnRNPs) represent a structurally composite family of mRNA-binding proteins. Their RNA-binding specificity resides in their inclusion of different RNA-binding domains, namely the RNA recognition motif (RRM), the K-homology (KH) domain, and the Arg–Gly–Gly (RGG) box. HnRNPs are multifunctional, as they participate in pre-mRNA processing (i.e. splicing), but are also important determinants of mRNA export, localization, translation, and stability [31]. In particular PTBP1, which contains 4 RRMs, was originally identified for its role in splicing, but is now known to function in a number of diverse cellular processes including polyadenylation, mRNA stability and translation initiation [32]. Specificity of PTBP1 function is achieved by a combination of changes in its cellular localization, and its interaction with additional proteins. As mentioned in Section 2, PTBP1 has been identified as an RBP which increases mRNA stability and protein levels of insulin through its binding to the 3′-UTR of the rat preproinsulin mRNA (Figure 2) [18]. In line with these findings, it was subsequently shown that PTBP1 knockdown by RNAi specifically decreases both intracellular and secreted insulin in rat insulinoma INS-1 cells [33]. RNA decay assays demonstrated that upon glucose stimulation cytosolic PTBP1 also binds and stabilizes the mRNA 3′-UTR of islet cell autoantigen 512 (ICA512, also known as IA-2), an integral membrane protein of the insulin SGs. Additionally, high glucose increased the stability of PC 1/3 and PC2 mRNAs, which also harbor consensus sites for PTBP1 binding in their 3′-UTRs [33]. A similar role for PTBP1 in the stabilization of preproinsulin, ICA512 and PC1/3 mRNAs was proposed in the islets of rats with insulin resistance, and thus hyperinsulinemia, secondary to pharmacologically-induced acute liver failure [34]. Hence, modulation of mRNA stability by PTBP1 represents a unifying mechanism, which is crucial for upregulation of insulin SG biosynthesis.
Figure 2.
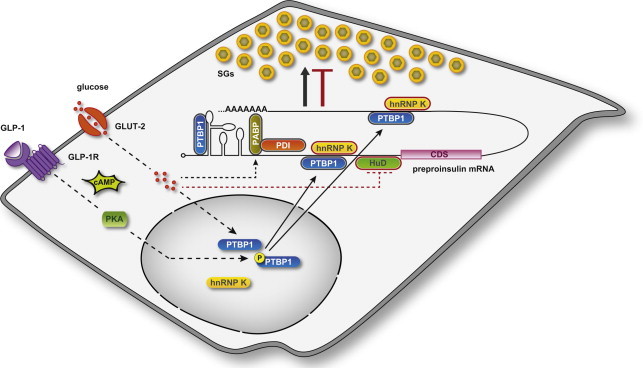
Model of RBP function in insulin expression and SG biosynthesis. Coordinated regulation by RBPs maintains insulin SG stores adequate to metabolic needs. Cytosolic RBPs with positive (PTBP1, PABP/PDI) and negative (HuD and hnRNP K) roles on insulin translation, and thereby SG biosynthesis, are outlined in black and red, respectively. Stimulation of β-cells induces cytoplasmic accumulation of PTBP1 through two distinct pathways. GLP-1 elevates intracellular cAMP levels, thereby activating PKA, which phosphorylates PTBP1 and thus prompts its redistribution into the cytoplasm. Glucose also triggers the cytosolic accumulation of PTBP1 through a phosphorylation-independent mechanism. In the cytoplasm PTBP1 binds to consensus sequences in the 5′- and 3′-UTRs of mRNAs encoding insulin and other proteins of the SGs. Glucose promotes the PABP-mediated binding of PDI to preproinsulin mRNA, whereas it inhibits that of HuD.
PTBP1 is thought to bind to the preproinsulin mRNA as part of a larger protein complex. Other candidate RBPs reported to bind in vitro to the 3′-UTR of preproinsulin mRNA include TiaR (also known as TiaL1 – Tia1 cytotoxic granule-associated RNA binding protein-like 1), hnRNP C, hnRNP E (also known as PCBP – poly(C)-binding protein), and hnRNP K [35]. Notably, binding of hnRNP E to the preproinsulin mRNA was observed using human [35], but not rat [18], islet cytosolic extracts. Unpublished work from our group indicates that hnRNP K is also implicated in controlling the stability of mRNAs encoding components of the insulin SGs (Figure 2). Specifically, decay assays showed that hnRNP K destabilizes the 3′-UTRs of preproinsulin and ICA512 mRNAs, thereby counteracting the effect of PTBP1 in insulinoma cells.
4. Translational control of insulin and SG protein expression
There is considerable evidence for the involvement of translational control in insulin biosynthesis. Indeed, in response to increased circulating glucose, insulin production is acutely regulated primarily through changes in mRNA translation [10,36]. This regulation is specific, as several independent studies have reported that high glucose induces an 8-fold or greater increase in preproinsulin translation, while overall protein synthesis only rises 2- to 3-fold compared to low glucose conditions [10,14,20,37]. The mechanisms governing glucose-induced insulin synthesis include stimulation of translation initiation and elongation, and signal recognition particle (SRP)-mediated translocation of the nascent preproinsulin into the lumen of the ER [10,36]. Indeed, through interaction with its N-terminal signal peptide, the SRP facilitates preproinsulin translocation across the ER membrane into the lumen [38]. The SRP can also block the elongation of the nascent chain and induce the “stacking” of ribosomes on the preproinsulin mRNA [39]. This brake on mRNA scanning is relieved upon interaction with the SRP receptor on the ER membrane, thus allowing active translation. In line with these findings, Herbert and colleagues showed that in mouse islets and insulinoma MIN6 cells glucose selectively stimulates the transport of the ribosome-associated preproinsulin mRNA to the ER and the recruitment of additional ribosomes for protein synthesis [40]. Unsurprisingly, the rapid translational effect exerted by glucose on insulin expression extends to the biosynthesis of other insulin SG constituents [41]. Among these are PC1/3 and PC2 [40,42–44], CGA [37], and ICA512 [45,46]. Notably, the mRNAs of human, rat and mouse CGA also contain consensus motifs for PTBP1 binding in their 5′- and 3′-UTRs.
PTBP1 not only stabilizes mRNAs encoding SG constituents, but is also a well-characterized regulator of their expression. For instance, experiments using luciferase reporter constructs have indicated that it regulates the translation of ICA512 and PC2 in INS-1 cells [28,33]. A substantial enhancement of luciferase activity was indeed observed upon the separate inclusion in the reporter constructs of the mRNA 3′-UTRs of ICA512 or PC2, or the mRNA 5′-UTR of PC2. Conversely, increments of luciferase activity were abolished upon mutation of the PTBP1-binding sites in the UTRs of PC2 mRNA [28,33].
Elucidating the mechanism by which PTBP1 enhances the translation of insulin and other SG cargoes is a major focus of current research in the field. PTBP1 is a known internal ribosomal entry site (IRES)-trans-acting factor (ITAF) which binds to elements in the 5′-UTRs of capless viral mRNAs, thereby enabling the recruitment of ribosomes and IRES-mediated translation of these transcripts [47–49]. Moreover, PTBP1 is required for function of most known cellular IRES [50,51]. Hence, it is plausible that PTBP1 may stimulate translation of insulin SG proteins by a cap-independent mechanism. Indeed, according to a recent report by the Welsh group insulin translation occurs at a low rate by cap-independent translation, which is mediated by binding of PTBP1 to the 5′-UTR of the human preproinsulin mRNA [52]. While this cap-independent translation mechanism was enhanced in conditions of cellular stress (i.e. nitrosative stress), it was seemingly unaffected by glucose stimulation. The authors therefore concluded that in human islets, glucose-stimulated insulin mRNA translation occurs exclusively via increasing cap-dependent translation. It should be noted, however, that hippuristanol, which was used in this study to abolish cap-dependent translation, by blocking the RNA helicase eIF4A, also inhibits some forms of IRES-mediated cap-independent translation [53].
Given its effect on mRNA stability and translation, PTBP1 function in β-cells is key for insulin SG biogenesis. Accordingly, knockdown of PTBP1 depletes SG stores and decreases the expression of SG components containing PTBP1 binding sites in their mRNA 3′-UTRs, such as ICA512, PC2, PC1/3, CGA, secretogranin II (SCG2/CHGC), and synaptobrevin 2 (SYB2/VAMP-2) [33]. The importance of PTBP1 in mediating the glucose response of the β-cell is also supported by observations that PTBP1 mRNA levels are significantly increased in mouse insulinoma cells following exposure to hyperglycemia for 24 h [54,55]. Short-term hyperglycemia, in contrast, does not alter PTBP1 expression [18]. On the other hand, the activity of PTBP1 is acutely regulated mainly through post-translational modifications and changes in its intracellular localization. In particular, cytoplasmic accumulation of PTBP1 is critical for its function in modulating the expression of SG proteins. Albeit predominantly nuclear, PTBP1 accumulates in the cytoplasm of β-cells upon stimulation [28,33]. This relocalization, as in the neurosecretory PC12 cell line [56,57], is triggered by PKA-dependent phosphorylation of a conserved serine residue. Accordingly, stimulation of insulinoma cells with the cAMP-elevating agent IBMX, and in part with GLP-1, induces a nucleocytoplasmic redistribution of PTBP1 and consequently upregulates the expression of insulin and other SG components containing PTBP1 binding sites in their mRNA 3′-UTRs [28]. Glucose also induces a cytosolic enrichment of PTBP1 by a still unknown mechanism that does not involve phosphorylation [28,33]. However, it is still unclear whether stimulation of β-cells leads to the cytosolic accumulation of PTBP1 by promoting its nucleocytoplasmic translocation or rather by inhibiting its nuclear import. Notably, it has been shown that prolonged culture of rat islets, which leads to reduced insulin stores and secretion, correlates with the reduced intracellular redistribution of PTBP1 [33]. Accordingly, in islets from subjects with type 2 diabetes the reduced upregulation of insulin levels in response to glucose stimulation correlates with increased nuclear retention of PTBP1 [58]. The recent report that a single nucleotide polymorphism in the PTBP1 gene is associated with reduced insulin secretion further suggests a role for PTBP1 in the pathogenesis of type 2 diabetes [59]. In this context, further study of PTBP1 function in the biogenesis of SGs in vivo using ad hoc transgenic mouse models will be of particular interest.
Though PTBP1 is the most extensively studied, other RBPs have been proposed to participate in regulation of insulin translation. Recently, polyA-binding protein (PABP) and protein-disulfide isomerase (PDI) have been reported as additional trans-acting factors that bind to the 5′-UTR of preproinsulin mRNA in pancreatic islets and mediate its glucose-stimulated translation (Figure 2) [60]. Specifically, PABP was proposed to mediate mRNA-binding of PDI, which was identified as the functional factor regulating translation. Glucose-dependent activation of insulin translation was abrogated by PDI immunodepletion, while it was restored upon transient PDI overexpression [60]. As PDI is known to reside in the lumen of the ER, its identification as a trans-acting factor in islet cytosolic extracts is however especially surprising, and requires independent validation.
Other RBPs are known to act as repressors of insulin translation. Among these is human antigen D (HuD, also known as ELAVL4 – embryonic lethal abnormal vision-like 4) (Figure 2). An elegant study by Gorospe and colleagues showed that HuD binds a 22-nucleotide segment in the 5′-UTR of the mouse preproinsulin 2 mRNA, thereby repressing its translation and decreasing insulin production [61]. However, the HuD–mRNA complex rapidly dissociated after acute exposure to glucose. In keeping with HuD suppressing translation, insulin levels were higher in the β-cells of knockout mice, whereas HuD-overexpressing mice displayed lower insulin expression, decreased plasma insulin, and impaired glucose tolerance [61]. A recent report specifically implicates HuD in the inhibition of insulin production and secretion in response to melatonin and ER stress [62]. Interestingly, unpublished evidence from our group suggests that hnRNP K may also exert a function similar to HuD. Indeed, hnRNP K binds to the mRNA 5′-UTR of rat preproinsulin 1 and other proteins of the SGs and negatively regulates their expression as well as insulin secretion. However, its direct role in repressing mRNA translation has not been tested. Similarly, it should be noted that 13 additional mRNA-binding proteins are rapidly regulated, in their expression or phosphorylation, in INS-1 cells after stimulation with glucose and IBMX [26]. Conceivably these RBPs may also have important roles in the post-transcriptional regulation of insulin SG biogenesis. However, their specific functions and mechanisms of action remain to be elucidated.
5. RBPs with other known functions in β-cells
Other RBPs have been shown to affect insulin expression and β-cell function, albeit not by directly interacting with the preproinsulin mRNA. For instance, a role in promoting insulin expression has been reported recently for the RNA-binding motif protein 4 (RBM4) [63]. RBM4-deficient mice exhibited hyperglycemia and reduced levels of serum insulin, and did not efficiently respond to a glucose challenge. RBM4 was found to promote the expression of insulin primarily at the transcriptional level, by regulating alternative splicing of transcripts encoding factors required for pancreas cell differentiation and endocrine function [63]. Interestingly, RBM4 is also known to repress the expression of PTBP1 via alternative splicing-coupled nonsense-mediated decay during muscle cell differentiation [64]. However, it has not been investigated whether it may also regulate PTBP1 expression in β-cells in response to varying glucose levels.
A transcriptional effect on insulin gene expression has also been reported for the RNA-binding proteins Mushashi-1 (MSI1) and Mushashi-2 (MSI2) [65]. Specifically, overexpression and knockdown studies demonstrated that both Mushashi isoforms downregulate insulin expression in insulinoma cells. Accordingly, conditions associated with type 2 diabetes, such as ER stress and lipotoxicity, increased the expression of the Mushashi genes. In the same study MSI1 was also found to promote the proliferation of MIN6 cells and strongly reduce apoptosis [65]. Conversely, an opposite role in regulating β-cell death has been shown for ARE/poly(U)-binding factor 1 (AUF1, also known as hnRNP D), a well-characterized RBP which binds to AU- or U-rich sequence elements located in the 3′-UTR of target mRNAs, and thereby controls their stability [66]. Recently Regazzi and colleagues have shown that exposure of insulinoma cells to inflammatory cytokines leads to the activation of AUF1 and its translocation from the nucleus to the cytoplasm [67]. Although altered AUF1 levels did not influence insulin synthesis and secretion, its overproduction significantly increased the apoptotic rate of MIN6 cells. Conversely, blockade of AUF1 production by RNAi protected MIN6 and human primary β-cells from cytotokine-induced apoptosis. The authors further clarified that AUF1 contributes to the death of insulin-producing cells by reducing the expression of anti-apoptotic proteins B cell leukaemia/lymphoma 2 (BCL2) and myeloid cell leukaemia sequence 1 (MCL1). Particularly for BCL2 this effect was directly related to changes in mRNA stability [67].
6. RBPs with proposed roles in type 2 diabetes
Several other RBPs have been implicated in glycemic control, and thus type 2 diabetes. As these proteins do not exert their function specifically in β-cells and affect components of glucose homeostasis other than insulin biosynthesis and secretion, we will only briefly review them here. Among these RBPs is RBM4, which was previously mentioned (Section 5) for its role in insulin expression and pancreas cell differentiation. RBM4, in addition, influences glucose-induced insulin receptor (IR) signaling by promoting a splicing switch of IR pre-mRNA towards an isoform that exhibits a high affinity for insulin [63]. Incidentally, PTBP1, a target of RBM4 splicing regulation, in turn regulates IR expression via IRES-mediated translation [68]. A recent study reported that serine/arginine-rich splicing factor 3 (SRSF3, previously known as SRp20) also affects glucose homeostasis. Indeed, mice carrying a liver-specific deletion of Srsf3 showed fasting hypoglycemia and increased insulin sensitivity [69]. Notably, SRSF3 regulates the splicing and expression of Hnf1α, which is responsible for maturity onset diabetes of the young type 3 [70] and is one of the type 2 diabetes susceptibility genes [71].
An impact on glucose homeostasis has also been shown for the mRNA-binding protein hematopoietic zinc finger (Hzf). In a study aimed mainly at dissecting its role in the translational control of adipogenesis, it was observed that Hzf-null mice show impaired glucose tolerance and increased insulin resistance [72]. Interestingly, the authors note that the human HZF gene resides in a genomic locus involved in susceptibility to type 2 diabetes. Finally, insulin-like growth factor 2 (IGF2) mRNA-binding protein 2 (IGF2BP2, also known as IMP2) has also been implicated in type 2 diabetes. This mRNA-binding protein is known to regulate the IRES-mediated translation of IGF2 by binding to its mRNA 5′-UTR [73]. A link between IGF2BP2 genetic variations and type 2 diabetes was found in a series of genome-wide association studies and verified in subsequent replication studies in different populations and meta-analyses [74]. Although these studies have indicated that IGF2BP2 variants are more likely associated with reduced β-cell function (i.e. insulin secretion) than with reduced insulin sensitivity or fasting glucose, the specific role of IGF2BP2 in islet function and type 2 diabetes has not yet been defined.
7. Outlook on RBPs and diabetes
The importance of RBPs in controlling β-cell function and glycemia is now broadly appreciated, and these proteins and their regulatory mechanisms are under extensive investigation. Indeed, a better understanding of the molecular mechanisms of RBPs and the upstream factors regulating their expression could provide opportunities for the design of additional anti-diabetic strategies.
A growing body of evidence supports a key role of RBPs particularly in the post-transcriptional control of glucose homeostasis. To further understand the role of RBPs in post-transcriptional and translational control and RNA metabolism in general, it is crucial to identify their target transcripts. Since RBPs typically regulate a large number of mRNA targets in a functionally coordinated fashion, genome-wide methods are emerging as powerful tools for these studies. In combination with microarrays and RNA sequencing, immunoprecipitation-based methods such as CLIP (UV crosslinking and immunoprecipitation) and its variants (reviewed in [75]) allow for the isolation of RBP–mRNA complexes and identification of the specific sequences to which RBPs bind. Techniques that provide unbiased insights into the effect of RBPs on translational control are also available. For instance, polysome profiling and other refined methods to assess the association of RBPs with the ribosomal machinery, such as ribosome profiling, have the potential to map the precise position of ribosomes within the transcriptome [76–78]. Moreover, polysome profiling of specific cell types in vivo is now facilitated by two recently developed methods which use genetically modified mice expressing epitope-tagged versions of ribosome protein subunits [79,80].
Despite the high number of RBP-mediated regulatory mechanisms unveiled in the recent years, a large proportion of potentially relevant RBPs remain to be characterized regarding their impact on β-cell function and glucose homeostasis. This observation, together with the many tools available to investigate RBPs and their function both in vitro and in genetically modified organisms, identifies this as a rapidly expanding and exciting research topic in the field of diabetes and metabolism.
Conflict of interest
None declared.
Acknowledgements
We thank Christin Süss and Klaus-Peter Knoch for sharing unpublished results, Klaus-Peter Knoch and Ronald Dirkx Jr. for critical reading of the manuscript, and Katja Pfriem for administrative help. MS is supported by the German Center for Diabetes Research (DZD e.V.), which is funded by the German Ministry for Education and Research (BMBF). MGM has been awarded a fellowship from the Dresden International Graduate School for Biomedicine and Bioengineering (DIGS-BB) and is the recipient of a MeDDrive Grant from the Medical Faculty at TU Dresden.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Maria Grazia Magro, Email: maria-grazia.magro@mailbox.tu-dresden.de.
Michele Solimena, Email: michele.solimena@tu-dresden.de.
References
- 1.Dodson G., Steiner D. The role of assembly in insulin's biosynthesis. Current Opinion in Structural Biology. 1998;8:189–194. doi: 10.1016/s0959-440x(98)80037-7. [DOI] [PubMed] [Google Scholar]
- 2.Fava E., Dehghany J., Ouwendijk J., Müller A., Niederlein A., Verkade P. Novel standards in the measurement of rat insulin granules combining electron microscopy, high-content image analysis and in silico modelling. Diabetologia. 2012;55:1013–1023. doi: 10.1007/s00125-011-2438-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rorsman P., Renström E. Insulin granule dynamics in pancreatic beta cells. Diabetologia. 2003;46:1029–1045. doi: 10.1007/s00125-003-1153-1. [DOI] [PubMed] [Google Scholar]
- 4.Suckale J., Solimena M. The insulin secretory granule as a signaling hub. Trends in Endocrinology and Metabolism. 2010;21:599–609. doi: 10.1016/j.tem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Goodge K.A., Hutton J.C. Translational regulation of proinsulin biosynthesis and proinsulin conversion in the pancreatic beta-cell. Seminars in Cell & Developmental Biology. 2000;11:235–242. doi: 10.1006/scdb.2000.0172. [DOI] [PubMed] [Google Scholar]
- 6.Gold G., Ghishizky M.L., Grodsky G.M. Evidence that glucose marks beta cells resulting in preferential release of newly synthesized insulin. Science. 1982;218:56–58. doi: 10.1126/science.6181562. [DOI] [PubMed] [Google Scholar]
- 7.Halban P.A. Differential rates of release of newly synthesized and of stored insulin from pancreatic islets. Endocrinology. 1982;110:1183–1188. doi: 10.1210/endo-110-4-1183. [DOI] [PubMed] [Google Scholar]
- 8.Ivanova, A., Kalaidzidis, J., Dirkx, R., Sarov, M., Gerlach, M., Schroth-Diez B., et al., Age-dependent labeling and imaging of insulin secretory granules, Diabetes. 10.2337/db12-1819, in press [DOI] [PMC free article] [PubMed]
- 9.Jahr H., Schroeder D., Ziegler B., Ziegler M., Zuehlke H. Transcriptional and translational control of glucose-stimulated (pro)insulin biosynthesis. European Journal of Biochemistry. 1980;110:499–505. doi: 10.1111/j.1432-1033.1980.tb04892.x. [DOI] [PubMed] [Google Scholar]
- 10.Permutt M.A. Effect of glucose on initiation and elongation rates in isolated rat pancreatic islets. Journal of Biological Chemistry. 1974;249:2738–2742. [PubMed] [Google Scholar]
- 11.Leibiger B., Wahlander K., Berggren P.O., Leibiger I.B. Glucose-stimulated insulin biosynthesis depends on insulin-stimulated insulin gene transcription. Journal of Biological Chemistry. 2000;275:30153–30156. doi: 10.1074/jbc.M005216200. [DOI] [PubMed] [Google Scholar]
- 12.Perfetti R., Merkel P. Glucagon-like peptide-1: a major regulator of pancreatic beta-cell function. European Journal of Endocrinology. 2000;143:717–725. doi: 10.1530/eje.0.1430717. [DOI] [PubMed] [Google Scholar]
- 13.Permutt M.A., Kipnis D.M. Insulin biosynthesis. II. Effect of glucose on ribonucleic acid synthesis in isolated rat islets. Journal of Biological Chemistry. 1972;247:1200–1207. [PubMed] [Google Scholar]
- 14.Itoh N., Okamoto H. Translational control of proinsulin synthesis by glucose. Nature. 1980;283:100–102. doi: 10.1038/283100a0. [DOI] [PubMed] [Google Scholar]
- 15.Brunstedt J., Chan S.J. Direct effect of glucose on the preproinsulin mRNA level in isolated pancreatic islets. Biochemical and Biophysical Research Communications. 1982;106:1383–1389. doi: 10.1016/0006-291x(82)91267-0. [DOI] [PubMed] [Google Scholar]
- 16.Welsh M., Nielsen D.A., MacKrell A.J., Steiner D.F. Control of insulin gene expression in pancreatic beta-cells and in an insulin-producing cell line, RIN-5F cells. II. Regulation of insulin mRNA stability. Journal of Biological Chemistry. 1985;260:13590–13594. [PubMed] [Google Scholar]
- 17.Wicksteed B., Herbert T.P., Alarcon C., Lingohr M.K., Moss L.G., Rhodes C.J. Cooperativity between the preproinsulin mRNA untranslated regions is necessary for glucose-stimulated translation. Journal of Biological Chemistry. 2001;276:22553–22558. doi: 10.1074/jbc.M011214200. [DOI] [PubMed] [Google Scholar]
- 18.Tillmar L., Carlsson C., Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3′-untranslated region pyrimidine-rich sequence. Journal of Biological Chemistry. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- 19.Tillmar L., Welsh N. Hypoxia may increase rat insulin mRNA levels by promoting binding of the polypyrimidine tract-binding protein (PTB) to the pyrimidine-rich insulin mRNA 3′-untranslated region. Molecular Medicine. 2002;8:263–272. [PMC free article] [PubMed] [Google Scholar]
- 20.Wicksteed B., Uchizono Y., Alarcon C., McCuaig J.F., Shalev A., Rhodes C.J. A cis-element in the 5′ untranslated region of the preproinsulin mRNA (ppIGE) is required for glucose regulation of proinsulin translation. Cell Metabolism. 2007;5:221–227. doi: 10.1016/j.cmet.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Muralidharan B., Bakthavachalu B., Pathak A., Seshadri V. A minimal element in 5′UTR of insulin mRNA mediates its translational regulation by glucose. FEBS Letters. 2007;581:4103–4108. doi: 10.1016/j.febslet.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 22.Uchizono Y., Alarcón C., Wicksteed B.L., Marsh B.J., Rhodes C.J. The balance between proinsulin biosynthesis and insulin secretion: where can imbalance lead? Diabetes, Obesity & Metabolism. 2007;9:56–66. doi: 10.1111/j.1463-1326.2007.00774.x. [DOI] [PubMed] [Google Scholar]
- 23.Kim W., Kyung Lee E. Post-transcriptional regulation in metabolic diseases. RNA Biology. 2012;9:772–780. doi: 10.4161/rna.20091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lunde B.M., Moore C., Varani G. RNA-binding proteins: modular design for efficient function. Nature Reviews. Molecular Cell Biology. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glisovic T., Bachorik J.L., Yong J., Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Letters. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Süss C., Czupalla C., Winter C., Pursche T., Knoch K.-P., Schroeder M. Rapid changes of mRNA-binding protein levels following glucose and 3-isobutyl-1-methylxanthine stimulation of insulinoma INS-1 cells. Molecular & Cellular Proteomics. 2009;8:393–408. doi: 10.1074/mcp.M800157-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giddings S.J., Chirgwin J., Permutt M.A. Effects of glucose on proinsulin messenger RNA in rats in vivo. Diabetes. 1982;31:624–629. doi: 10.2337/diab.31.7.624. [DOI] [PubMed] [Google Scholar]
- 28.Knoch K.-P., Meisterfeld R., Kersting S., Bergert H., Altkrueger A., Wegbrod C. cAMP-dependent phosphorylation of PTB1 promotes the expression of insulin secretory granule proteins in beta cells. Cell Metabolism. 2006;3:123–134. doi: 10.1016/j.cmet.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Shibayama M., Ohno S., Osaka T., Sakamoto R., Tokunaga A., Nakatake Y. Polypyrimidine tract-binding protein is essential for early mouse development and embryonic stem cell proliferation. FEBS Journal. 2009;276:6658–6668. doi: 10.1111/j.1742-4658.2009.07380.x. [DOI] [PubMed] [Google Scholar]
- 30.Suckale J., Wendling O., Masjkur J., Jaeger M., Muenster C., Anastassiadis K. PTBP1 is required for embryonic development before gastrulation. PLoS One. 2011;6:e16992. doi: 10.1371/journal.pone.0016992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dreyfuss G., Kin V.N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nature Reviews Molecular Cell Biology. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 32.Sawicka K., Bushell M., Spriggs K.A., Willis A.E. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochemical Society Transactions. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 33.Knoch K.-P., Bergert H., Borgonovo B., Saeger H.-D., Altkrueger A., Verkade P. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nature Cell Biology. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- 34.Kuwahata M., Tomoe Y., Harada N., Amano S., Segawa H., Tatsumi S. Characterization of the molecular mechanisms involved in the increased insulin secretion in rats with acute liver failure. Biochimica et Biophysica Acta. 2007;1772:60–65. doi: 10.1016/j.bbadis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Fred R.G., Welsh N. The importance of RNA binding proteins in preproinsulin mRNA stability. Molecular and Cellular Endocrinology. 2009;297:28–33. doi: 10.1016/j.mce.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Welsh M., Scherberg N., Gilmore R., Steiner D.F. Translational control of insulin biosynthesis. Evidence for regulation of elongation, initiation and signal-recognition-particle-mediated translational arrest by glucose. Biochemical Journal. 1986;235:459–467. doi: 10.1042/bj2350459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guest P.C., Rhodes C.J., Hutton J.C. Regulation of the biosynthesis of insulin-secretory-granule proteins. Co-ordinate translational control is exerted on some, but not all, granule matrix constituents. Biochemical Journal. 1989;257:431–437. doi: 10.1042/bj2570431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egea P.F., Stroud R.M., Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Current Opinion in Structural Biology. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Wolin S.L., Walter P. Discrete nascent chain lengths are required for the insertion of presecretory proteins into microsomal membranes. Journal of Cell Biology. 1993;121:1211–1219. doi: 10.1083/jcb.121.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenman I.C., Gomez E., Moore C.E.J., Herbert T.P. The selective recruitment of mRNA to the ER and an increase in initiation are important for glucose-stimulated proinsulin synthesis in pancreatic beta-cells. Biochemical Journal. 2005;391:291–300. doi: 10.1042/BJ20050468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guest P.C., Bailyes E.M., Rutherford N.G., Hutton J.C. Insulin secretory granule biogenesis. Co-ordinate regulation of the biosynthesis of the majority of constituent proteins. Biochemical Journal. 1991;274:73–78. doi: 10.1042/bj2740073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alarcon C., Lincoln B., Rhodes C.J. The biosynthesis of the subtilisin-related proprotein convertase PC3, but no that of the PC2 convertase, is regulated by glucose in parallel to proinsulin biosynthesis in rat pancreatic islets. Journal of Biological Chemistry. 1993;268:4276–4280. [PubMed] [Google Scholar]
- 43.Martin S.K., Carroll R., Benig M., Steiner D.F. Regulation by glucose of the biosynthesis of PC2, PC3 and proinsulin in (ob/ob) mouse islets of Langerhans. FEBS Letters. 1994;356:279–282. doi: 10.1016/0014-5793(94)01284-9. [DOI] [PubMed] [Google Scholar]
- 44.Skelly R.H., Schuppin G.T., Ishihara H., Oka Y., Rhodes C.J. Glucose-regulated translational control of proinsulin biosynthesis with that of the proinsulin endopeptidases PC2 and PC3 in the insulin-producing MIN6 cell line. Diabetes. 1996;45:37–43. doi: 10.2337/diab.45.1.37. [DOI] [PubMed] [Google Scholar]
- 45.Ort T., Voronov S., Guo J., Zawalich K., Froehner S.C., Zawalich W. Dephosphorylation of beta2-syntrophin and Ca2+/mu-calpain-mediated cleavage of ICA512 upon stimulation of insulin secretion. EMBO Journal. 2001;20:4013–4023. doi: 10.1093/emboj/20.15.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trajkovski M., Mziaut H., Altkrüger A., Ouwendijk J., Knoch K.-P., Müller S. Nuclear translocation of an ICA512 cytosolic fragment couples granule exocytosis and insulin expression in {beta}-cells. Journal of Cell Biology. 2004;167:1063–1074. doi: 10.1083/jcb.200408172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belsham G.J., Sonenberg N. Picornavirus RNA translation: roles for cellular proteins. Trends in Microbiology. 2000;8:330–335. doi: 10.1016/s0966-842x(00)01788-1. [DOI] [PubMed] [Google Scholar]
- 48.Fraser C.S., Doudna J.A. Structural and mechanistic insights into hepatitis C viral translation initiation. Nature Reviews Microbiology. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 49.Balvay L., Lopez Lastra M., Sargueil B., Darlix J.-L., Ohlmann T. Translational control of retroviruses. Nature Reviews Microbiology. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell S.A., Spriggs K.A., Bushell M., Evans J.R., Stoneley M., Le Quesne J.P. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes & Development. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoneley M., Willis A.E. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 52.Fred R.G., Sandberg M., Pelletier J., Welsh N. The human insulin mRNA is partly translated via a cap- and eIF4A-independent mechanism. Biochemical and Biophysical Research Communications. 2011;412:693–698. doi: 10.1016/j.bbrc.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 53.Bordeleau M.-E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nature Chemical Biology. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 54.Webb G.C., Akbar M.S., Zhao C., Steiner D.F. Expression profiling of pancreatic beta cells: glucose regulation of secretory and metabolic pathway genes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5773–5778. doi: 10.1073/pnas.100126597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fred R.G., Welsh N. Increased expression of polypyrimidine tract binding protein results in higher insulin mRNA levels. Biochemical and Biophysical Research Communications. 2005;328:38–42. doi: 10.1016/j.bbrc.2004.12.147. [DOI] [PubMed] [Google Scholar]
- 56.Xie J., Lee J.-A., Kress T.L., Mowry K.L., Black D.L. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma S., Liu G., Sun Y., Xie J. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochimica and Biophysica Acta. 2007;1773:912–923. doi: 10.1016/j.bbamcr.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Ehehalt F., Knoch K., Erdmann K., Krautz C., Jaeger M., Steffen A. Impaired insulin turnover in islets from type 2 diabetic patients. Islets. 2010;2:30–36. doi: 10.4161/isl.2.1.10098. [DOI] [PubMed] [Google Scholar]
- 59.Heni M., Ketterer C., Wagner R., Linder K., Böhm A., Herzberg-Schäfer S.A. Polymorphism rs11085226 in the gene encoding polypyrimidine tract-binding protein 1 negatively affects glucose-stimulated insulin secretion. PLoS One. 2012;7:e46154. doi: 10.1371/journal.pone.0046154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kulkarni S.D., Muralidharan B., Panda A.C., Bakthavachalu B., Vindu A., Seshadri V. Glucose-stimulated translation regulation of insulin by the 5′ UTR-binding proteins. Journal of Biological Chemistry. 2011;286:14146–14156. doi: 10.1074/jbc.M110.190553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee E.K., Kim W., Tominaga K., Martindale J.L., Yang X., Subaran S.S. RNA-binding protein HuD controls insulin translation. Molecular Cell. 2012;45:826–835. doi: 10.1016/j.molcel.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoo Y.-M. Melatonin-mediated insulin synthesis during endoplasmic reticulum stress involves HuD expression in rat insulinoma INS-1E cells. Journal of Pineal Research. 2013;55:207–220. doi: 10.1111/jpi.12064. [DOI] [PubMed] [Google Scholar]
- 63.Lin J.-C., Yan Y.-T., Hsieh W.-K., Peng P.-J., Su C.-H., Tarn W.-Y. RBM4 promotes pancreas cell differentiation and insulin expression. Mocular and Cellular Biology. 2013;33:319–327. doi: 10.1128/MCB.01266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin J.-C., Tarn W.-Y. Multiple roles of RBM4 in muscle cell differentiation. Frontiers in Bioscience. 2012;4:181–189. doi: 10.2741/260. [DOI] [PubMed] [Google Scholar]
- 65.Szabat M., Kalynyak T.B., Lim G.E., Chu K.Y., Yang Y.H., Asadi A. Musashi expression in β-cells coordinates insulin expression, apoptosis and proliferation in response to endoplasmic reticulum stress in diabetes. Cell Death & Disease. 2011;24:e232. doi: 10.1038/cddis.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malter J.S. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989;246:664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- 67.Roggli E., Gattesco S., Pautz A., Regazzi R. Involvement of the RNA-binding protein ARE/poly(U)-binding factor 1 (AUF1) in the cytotoxic effects of proinflammatory cytokines on pancreatic beta cells. Diabetologia. 2012;55:1699–1708. doi: 10.1007/s00125-011-2399-7. [DOI] [PubMed] [Google Scholar]
- 68.Spriggs K.A., Cobbold L.C., Ridley S.H., Coldwell M., Bottley A., Bushell M. The human insulin receptor mRNA contains a functional internal ribosome entry segment. Nucleic Acids Research. 2009;37:5881–5893. doi: 10.1093/nar/gkp623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sen S., Jumaa H., Webster N.J.G. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nature Communications. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamagata K., Oda N., Kaisaki P.J., Menzel S., Furuta H., Vaxillaire M. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 71.Triggs-Raine B.L., Kirkpatrick R.D., Kelly S.L., Norquay L.D., Cattini P.A., Yamagata K. HNF-1alpha G319S, a transactivation-deficient mutant, is associated with altered dynamics of diabetes onset in an Oji-Cree community. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4614–4619. doi: 10.1073/pnas.062059799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawagishi H., Wakoh T., Uno H., Maruyama M., Moriya A., Morikawa S. Hzf regulates adipogenesis through translational control of C/EBPalpha. EMBO Journal. 2008;27:1481–1490. doi: 10.1038/emboj.2008.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai N., Rapley J., Angel M., Yanik M.F., Blower M.D., Avruch J. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes & Development. 2011;25:1159–1172. doi: 10.1101/gad.2042311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Christiansen J., Kolte A.M., Hansen T.vO., Nielsen F.C. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. Journal of Molecular Endocrinology. 2009;43:187–195. doi: 10.1677/JME-09-0016. [DOI] [PubMed] [Google Scholar]
- 75.König J., Zarnack K., Luscombe N.M., Ule J. Protein–RNA interactions: new genomic technologies and perspectives. Nature Reviews Genetics. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 76.Zong Q., Schummer M., Hood L., Morris D.R. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature Protocols. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heiman M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanz E., Yang L., Su T., Morris D.R., McKnight G.S., Amieux P.S. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]


