Abstract
The choice of using one of many possible neurotransmitter systems is a critical step in defining the identity of an individual neuron type. We show here that the key defining feature of glutamatergic neurons, the vesicular glutamate transporter EAT-4/VGLUT is expressed in 38 of the 118 anatomically defined neuron classes of the C.elegans nervous system. We show that eat-4/VGLUT expression is controlled in a modular manner, with distinct cis-regulatory modules driving expression in distinct glutamatergic neuron classes. We identify 13 different transcription factors, 11 of them homeodomain proteins, that act in specific combinations in 25 different glutamatergic neuron classes to initiate and maintain eat-4/VGLUT expression. We show that the adoption of a glutamatergic phenotype is linked to the adoption of other terminal identity features of a neuron, including cotransmitter phenotypes. Examination of mouse orthologs of these homeodomain proteins resulted in the identification of mouse LHX1 as a regulator of glutamatergic neurons in the brainstem.
INTRODUCTION
A key identity feature of an individual neuron type is its neurotransmitter phenotype. Most classic neurotransmitters are synthesized by specialized enzymes, loaded by specific transporter proteins into synaptic vesicles and taken back into the neuron by specialized plasma membrane transporters. In many cases, the neurotransmitter identity of a specific neuron type is therefore defined by the coordinated expression of genes coding for specific enzymes and transporters. Understanding the regulatory mechanisms that control expression of these enzymes and transporters presents a fruitful “bottom-up” approach that will help explain how a specific neuronal identity is imposed onto a neuron type during development and how this identity is maintained throughout the life of a neuron.
Glutamate is the most broadly employed excitatory neurotransmitter in most vertebrate and invertebrate nervous systems. In contrast to other neurotransmitter systems, the identity of glutamatergic neurons is not defined by the expression of multiple biosynthetic enzymes and transporters. Since glutamate is present in all cells, its utilization as a neurotransmitter critically depends on the ability of a neuron to load glutamate into synaptic vesicles. This is achieved by a vesicular transporter for glutamate of the SLC17 family of solute carriers, called VGLUT (Takamori et al., 2001). Ectopic expression of VGLUT is sufficient to confer the glutamatergic phenotype (i.e. synaptic release of glutamate) onto heterologous neurons (Takamori et al., 2000, 2001). Consistent with the sufficiency of VGLUT to determine the glutamatergic phenotype, there are no pan-glutamatergic markers other than the VGLUT genes (see Supplementary Text).
Given the importance of VGLUT genes in defining the glutamatergic phenotype of a neuron, it is perhaps surprising that very little is known about how VGLUT expression is regulated in the nervous system of any vertebrate or invertebrate species, including mouse, Drosophila and C.elegans. In the mouse, several transcription factors have been described to be involved in the generation of glutamatergic neurons in different areas of the developing central nervous system (Brill et al., 2009; Cheng et al., 2004; Englund et al., 2005; Lou et al., 2013; Ma and Cheng, 2006) but it is not clear whether any of these factors directly initiates and maintains VGLUT expression or whether they act transiently at earlier stages of differentiation and operate through intermediary factors.
The nematode C.elegans contains one well characterized VGLUT-encoding gene, eat-4 (Lee et al., 1999). eat-4/VGLUT enables glutamatergic transmission in various neuronal circuits that control distinct behaviors (e.g. (Chalasani et al., 2007; Lee et al., 1999)) and the eat-4 mutant phenotype can be rescued by human VGLUT (Lee et al., 2008). How C.elegans eat-4/VGLUT expression is regulated in distinct neuronal cell types has not previously been investigated, mirroring the absence of insight into the regulation of Drosophila or vertebrate VGLUT gene expression.
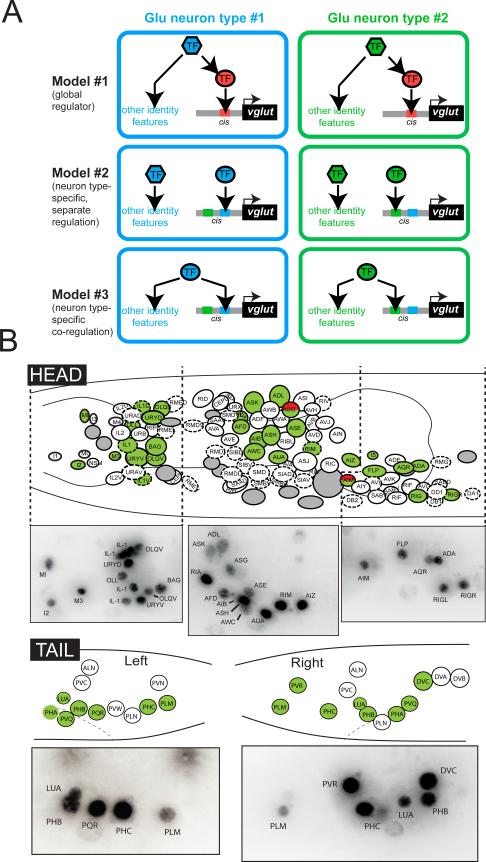
In principle, one could imagine several distinct scenarios by which VGLUT gene expression is controlled in different neuronal cell types. A dedicated regulatory factor (or combination thereof) could exist to control VGLUT expression in all different glutamatergic neuron types (model #1 in Fig.1A). This dedicated factor could be turned on by distinct sets of earlier acting factors in distinct glutamatergic neuron populations. Alternatively, VGLUT gene expression could be regulated in different manners in distinct glutamatergic neuron types (Fig.1A, model #2 and #3). Furthermore, VGLUT expression may be controlled separately from the expression of other identity features of a neuron (Fig.1A, model #2) or its expression could be tightly linked with the expression of other cell-specific identity features of particular glutamatergic neuron types (Fig.1A, model #3). Whether distinct terminal identity features of a neuron are separately regulated or whether they are coregulated via a common (set of) trans-acting factor(s)(model #2 vs. #3) are fundamental but little understood neurodevelopmental problems.
Fig.1. eat-4/VGLUT expression.
(A) Three different model for regulation of VGLUT expression in different glutamatergic neuron types. See text.
(B) Expression of a fosmid-based eat-4 reporter (otIs388) in head and tail ganglia. yfp-based images are lateral projections inverted to sharpen the signal. Green circles indicate eat-4 expressing neurons. Green/red circles indicate co-expression of eat-4 and serotonin. Dashed line indicates motor neuron class. See also Fig.S1.
Here, we approach these questions using two conceptually distinct but converging approaches. First, we elucidate the cis-regulatory logic of eat-4/VGLUT expression by defining cis-regulatory regions in the eat-4 locus that are required for expression of eat-4 in distinct glutamatergic neurons. If one common regulatory mechanism exists (model #1) that controls eat-4/VGLUT expression in all glutamatergic neurons, such cis-regulatory analysis should reveal a specific cis-regulatory element required for expression in all eat-4/VGLUT expressing neurons. Alternatively, if distinct glutamatergic neuron types employ distinct regulatory mechanisms (model #2 and #3), the eat-4/VGLUT regulatory elements should be complex and modular in nature, with different elements driving expression in different neurons types. Second, we analyzed the effect of removal of a number of transcription factors on the expression of not only eat-4/VGLUT but also on the expression of other identity markers of the respective glutamatergic neuron type (hence distinguishing model #2 and #3) and we report evidence of pervasive coregulation. Furthermore, we provide evidence of the conservation of regulatory mechanisms in the mouse. Our analysis reveals a comprehensive picture of the regulation of glutamatergic neuronal identity in the nervous system.
RESULTS
eat-4/VGLUT expression defines 38 glutamatergic neuron classes
We defined the complete glutamatergic nervous system of C.elegans by examining the expression of a fosmid-based eat-4 reporter construct (Fig.1B, 2). This reporter is expressed in 78 of the 302 neurons of the adult hermaphrodite, which fall into 38 neuron classes (out of a total of 118 anatomically defined neuron classes in the hermaphrodite; Table 1; Fig.1B,2). Most of these neurons are either sensory- or interneurons; only two motorneurons utilize glutamate, both located in the pharynx.
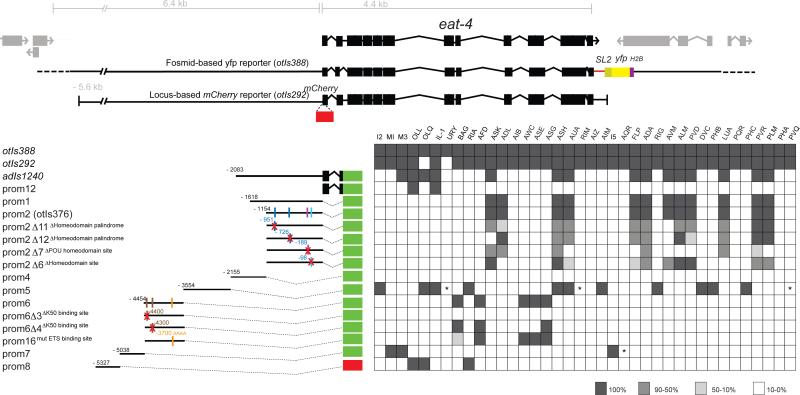
Fig.2. Modular composition of the eat-4 cis-regulatory control regions.
Overview of expression of different eat-4 reporter gene fusions, as indicated. The adIs1240 transgene contains the reporter from the previous Lee et al. (1999) study. For the dissection of 5’ regulatory information between 2 and 3 lines (n≥10) were scored for expression and lines commonly showed very similar expression pattern and penetrances of expression (the penetrance of expression shown in different shades of grey were derived by averaging multiple lines). Cell identifications were done based on either characteristic cell body position and/or through labeling specific, known neuron types with a red fluorescent marker (DiI staining or reporter lines). “*” indicates: DIC identification, not confirmed with reporter and “nd” indicates: not determined. For a list of transgenic strains, see Supplementary Information. See also Fig.S4.
Table 1.
eat-4 expressing neurons, their regulators and postsynaptic targets
| eat-4-expressing neuron class | eat-4 regulator identified in this study | postsynaptic target based on EM analysis1 (in bold: postsynaptic target expresses ionotropic GluR2) | Co-transmitter |
|---|---|---|---|
| ADA interneuron | * | AVB, AVJ, RIM, SMD, RIP | |
| ADL sensory neuron | lin-11 Lim homeobox gene | AIA, AIB, AVD, AVB, AVA | |
| AFD sensory neuron | ttx-1/ceh-14 Otx-type/LIM homeodomain | AIY | |
| AIB interneuron | RIM, RIB, AVB, SAAD | ||
| AIM interneuron | unc-86 POU homeodomain | AIA, ASG, ASK, ASJ, AVF | Serotonin |
| AIZ interneuron | unc-86 POU homeodomain3 | RIA, SMB, AIB, RIM, AIY, AVE | |
| ALM sensory neuron | unc-86/mec-3 POU/LIM homeodomain | BDU, PVC, CEP | |
| AQR sensory neuron | unc-86 POU homeodomain | AVA, AVB, RIA, BAG, PVC, AVD | |
| ASE sensory neuron | che-1 Zn finger & ceh-36 Otx-type homeodomain | AIY, AIA, AIB, RIA | |
| ASG sensory neuron | lin-11 LIM homeodomain + ceh-37 Otx-type homeodomain | AIA, AIB | Serotonin5 |
| ASH sensory neuron | unc-42 Prd-type homeodomain | AIA, AIB, RIA, AVA, AVB, AVD | |
| ASK sensory neuron | ttx-3 LIM homeodomain | AIA, AIB, AIM | |
| AUA interneuron | ceh-6 POU homeodomain | RIA, RIB, AVA, AVE | |
| AVM sensory neuron | unc-86/mec-3 POU/LIM homeodomain | AVB, PVC, BDU, ADE, PVR | |
| AWC sensory neuron | ceh-36 Otx-type homeodomain | AIY, AIA, AIB | |
| BAG sensory neuron | ets-5 Ets + ceh-37 Otx-type homeodomain | RIA, RIB, AVE, RIG | |
| DVC interneuron | ceh-14 LIM homeodomain | RIG, AVA, AIB, RMF | # |
| FLP sensory neuron | mec-3 LIM homeodomain3 | AVA, AVD, AVB, AIB, ADE | |
| IL1 sensory neuron | RMD, RIP | ||
| LUA interneuron | Unknown homeodomain protein4 | AVA, AVD, PVC, AVJ | |
| OLL sensory neuron | vab-3 Pax homeodomain | SMD, AVE, RIB, RMD, CEP | |
| OLQ sensory neuron | RMD, RIC, SIB, RIH | ||
| PHA sensory neuron | ceh-14 LIM homeodomain | PHB, AVG, PVQ, DVA, AVF, AVH | |
| PHB sensory neuron | ceh-14 LIM homeodomain | AVA, PVC | |
| PHC sensory neuron | ceh-14 LIM homeodomain3 | DVA, PVC, DA9 | |
| PLM sensory neuron | unc-86/mec-3 POU/LIM homeodomain | AVA, AVD, DVA, PDE | |
| PQR sensory neuron | unc-86 POU homeodomain | AVA, AVD | |
| PVD sensory neuron | mec-3 LIM homeodomain | AVA, PVC | |
| PVQ interneuron | * | AIA | |
| PVR interneuron | unc-86/ceh-14 POU/LIM homeodomain | AVB, RIP, AVJ | |
| RIA interneuron | SMD, RMD, RIV | ||
| RIG interneuron | AVE, AIZ, AVK, RIB, BAG, RIR, RMH | ||
| RIM motor neuron | head muscle, SMD, RMD, SAA, AVB | Tyramine | |
| URY sensory neuron | vab-3 Pax homeodomain | SMD, RMD, RIB, AVE | |
| Pharyngeal neurons: | |||
| M3 motor neuron | * | pharyngeal muscle, M3 | |
| I2 interneuron | NSM, I4, I6, M1 | ||
| MI motor neuron | pharyngeal muscle, NSM, M1, M2, M3, MC, I4, I5 | ||
| I5 interneuron | M1, M3, M4 | Serotonin |
A recent report using a different, smaller eat-4 reporter also identified AVA, AVE, SIB, RMD and ASJ neurons as eat-4-expressing (Ohnishi et al., 2011). We found that expression of this reporter perfectly overlaps with our fosmid-based reporter, but we could not confirm expression in AVA, SIB, RMD and ASJ.
Based on White et al., 1986.
glc/avr (glutamate-gated ion channel), glr (AMPA/Kainate-type), nmr (NMDA-type glutamate receptor)(Brockie and Maricq, 2006).
Apart from earlier functions of unc-86 in this lineage, unc-86 may also have late roles in terminal differentiation. See Fig.S6 for more information on this subject.
Inferred from requirement of TAAT homeodomain consensus site for eat-4 expression in LUA.
Under hypoxic conditions (Pocock and Hobert, 2011)
Reported to be cholinergic (Duerr et al., 2008) but based on unc-17 and cho-1 reporter expression, we believe that DVA and not DVC is cholinergic.
Candidate regulators expressed in this cell, but found to have no effect on eat-4 expression: unc-86 (ADA), ceh-2 (M3), ceh-14 (PVQ).
If the eat-4/VGLUT expressing neurons that we describe here indeed use glutamate as neurotransmitter, one would expect that synaptically connected neurons should express ionotropic glutamate receptors. Based on the complete synaptic connectivity diagram of the C.elegans hermaphrodite (White et al., 1986) and previously described expression patterns of all known glutamate-gated ion channels (Brockie and Maricq, 2006), we infer that each of the eat-4/VGLUT- expressing cells is presynaptic to at least one neuron (or pharyngeal muscle in the case of the pharyngeal motor neurons) that expresses glutamate-gated ion channels (Table 1). This corroborates the glutamatergic identity of the eat-4/VGLUT expressing neurons. Similar to vertebrates, we found that the expression of C.elegans glutaminases does not track with glutamatergic neuronal identity (Supplemental Text and Fig.S1).
The identity of other neurotransmitter systems (cholinergic, GABAergic, dopaminergic, serotonergic, tyraminergic, octopaminergic) has been described in great detail in C.elegans (www.wormatlas.org). The pattern of eat-4/VGLUT expression that we describe here is complementary and not overlapping with the expression of markers for these other, previously described neurotransmitter identities, with four exceptions (Table 1). The glutamatergic AIM and I5 interneurons are also serotonergic (Jafari et al., 2011; Sawin et al., 2000) and so is the glutamatergic ASG sensory neuron under hypoxic conditions (Pocock and Hobert, 2010) and the eat-4/VGLUT-expressing RIM motorneurons also employ the monoamine tyramine (Alkema et al., 2005).
Dissecting cis-regulatory control regions of eat-4/VGLUT reveals a modular logic of expression
To dissect the cis-regulatory information content of the eat-4 locus, we first compared expression of the eat-4 fosmid reporter and a reporter containing most of the 5’ intergenic region upstream of eat-4, the eat-4 locus and 500 bp of downstream sequences (Fig.2). This reporter is still expressed in all but two neurons classes compared to the fosmid reporter (Fig.2). A previously described, much smaller transcriptional reporter (adIs1240) is expressed in a much more restricted manner (Fig.2)(Lee et al., 1999).
We generated a series of reporter genes that encompasses various overlapping and non-overlapping pieces of the eat-4 locus and examined their expression pattern in transgenic animals. We find that the broad expression generated from the upstream cis-regulatory region can be broken into much smaller elements that direct expression to very small numbers of specific glutamatergic neuron classes (Fig.2). The modularity of the cis-regulatory control logic is further underscored by a more fine-grained mutational dissection in which we mutated conserved small motifs that constitute predicted binding sites for transcription factors whose identity we will describe further below. Mutations of such motifs abrogate expression in even smaller numbers of neuron classes, in some cases single neuron classes (Fig.2). The modular structure of cis-regulatory control regions of the eat-4 locus, with individual cis-regulatory elements driving expression in distinct glutamatergic neuron types, rules out the master regulatory model #1 (Fig.1A) and argues for neuron-type specific control mechanisms (model #2 or model #3 in Fig.1A).
We furthermore note that our mutational analysis did not reveal derepression in other neuronal or non-neuronal cell-types, strongly indicating that eat-4 expression is sculpted by activating rather than repressive regulatory inputs. Our previous analysis of the regulation of other neurotransmitter systems (e.g. genes controlling dopamine or acetylcholine biosynthesis) derived similar conclusions (Flames and Hobert, 2009; Kratsios et al., 2011), thereby corroborating the previously proposed concept that gene activation, rather than gene repression is the predominant mode of controlling terminal identity features of a neuron (Hobert, 2011).
Known terminal selector-type transcription factors control eat-4/VGLUT expression
To identify the trans-acting factors that operate through the modular cis-regulatory elements in the eat-4 locus we first turned to eight distinct, terminal selector-type transcription factors that have previously been shown to define the identity of distinct neuron types that we determine here to be glutamatergic: the che-1 Zn finger transcription factor (controls ASE gustatory neuron differentiation; (Etchberger et al., 2007)), the unc-86 POU homeodomain and mec-3 LIM homeobox genes (light touch receptor differentiation; (Duggan et al., 1998)), the ceh-36 Otx-type homeobox gene (ASE and AWC chemosensory neuron differentiation (Chang et al., 2003; Lanjuin et al., 2003)), the ttx-1 Otx-type and ceh-14 LIM-type homeodomain transcription factors (AFD thermosensory neuron identity; (Cassata et al., 2000; Satterlee et al., 2001)(H. Kagoshima, pers. comm.)), the lin-11 LIM homeodomain transcription factor (ASG chemosensory neuron identity; (Sarafi-Reinach et al., 2001)) and the ets-5 ETS domain transcription factor (BAG CO2/O2 sensory neurons (Brandt et al., 2012; Guillermin et al., 2011)). Each of these transcription factors is continually expressed in mature neuron types to control the expression of various terminal identity features of these individual neurons (e.g. neurotransmitter receptors, neuropeptides, ion channels, sensory receptors etc.), but in none of these cases has it is known whether the glutamatergic phenotype, i.e. expression of eat-4/VGLUT is affected. We crossed eat-4 reporter genes into the respective mutant backgrounds and found that each of the eight terminal selector transcription factors is required for eat-4 expression in the neuron types in which these factors were known to act as terminal selectors (Fig.3; Table 1). Using mec-3 as example, we confirmed through temporally controlled addition and removal of mec-3 that mec-3 is required continuously to maintain eat-4 expression (Fig.S2A). These findings demonstrate that the induction and maintenance of the glutamatergic phenotype is linked to the induction and maintenance of other terminal identity features of specific glutamatergic neuron types.
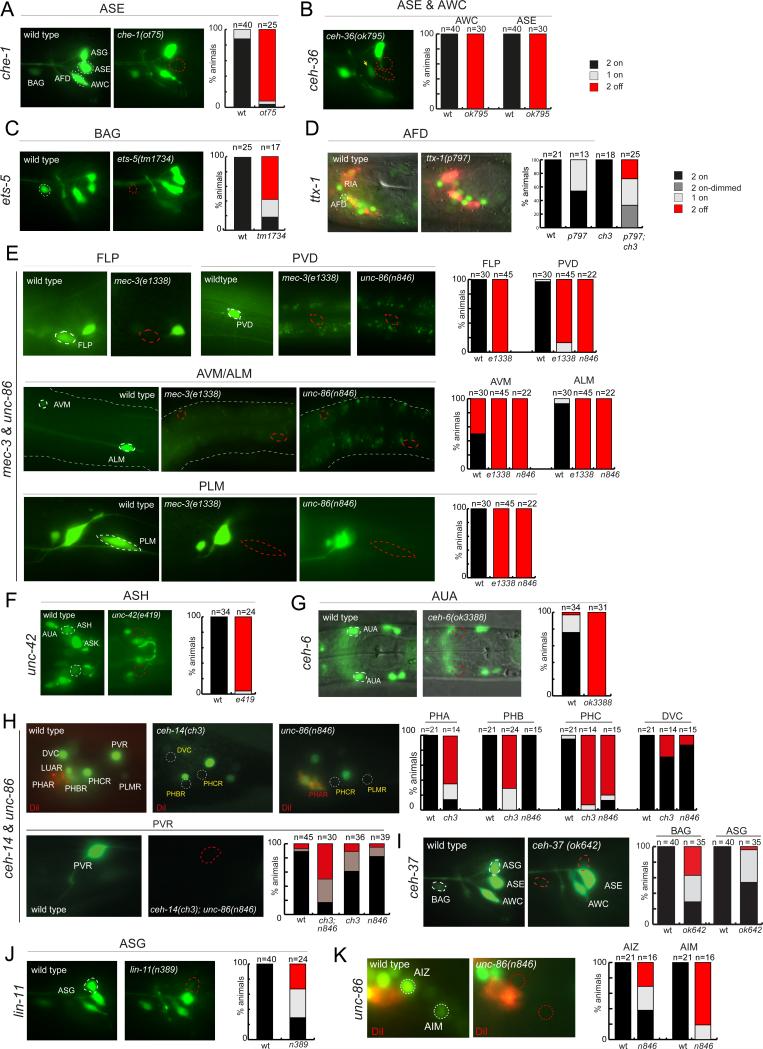
Fig.3. Regulators of eat-4/VGLUT expression.
eat-4 reporter lines where crossed into the null mutant backgrounds indicated. % animals (n=20-40) that express the reporter in both cells of the respective left/right neuron pair (“2 on”) or one of the two neurons of a neuron pair (“1 on”) or in neither (“0 on”) are indicated. (A-E): Known terminal selector of individual neuronal identities control eat-4 expression. See also Fig.S2.
(A) che-1 affects eat-4 expression (assayed with eat-4prom6 reporter transgene otIs392) in the ASE neurons.
(B) ceh-36 affects eat-4 expression (assayed with eat-4prom6 reporter transgene otIs392) in the ASE and AWC neurons. The control for the ceh-36 panel is shown in panel A.
(C) ets-5 affects eat-4 expression (assayed with eat-4prom6 reporter transgene otIs392) in the BAG neurons.
(D) ttx-1 affects eat-4 expression (assayed with eat-4 fosmid reporter transgene otIs388) in the AFD neurons. The animals shown were also stained with DiI, which labels other sensory neurons to facilitate cell identification.
(E) The collaborating mec-3/unc-86 genes affect eat-4 expression (assayed with the reporter transgene adIs1240) in different types of touch receptor neurons. The effect of unc-86 was not examined in FLP neurons do to previously described lineage transformation that eliminate the FLP neuron. See also Fig.S3.
(F-K) Novel regulators of specific neuronal identities.
(F) unc-42 controls eat-4 expression (assayed with eat-4 prom2 reporter transgene otIs376).
(G) ceh-6 controls eat-4 expression (assayed with eat-4 prom2 reporter transgene otIs376).
(H) The LIM homeobox gene ceh-14 and the POU homeobox gene unc-86 regulate glutamatergic identity of several tail sensory neurons. The effect of unc-86 in PHC could be the result of unc-86 function at earlier stages in the PHC-producing lineage (see Fig.S3). The effect of ceh-14 and unc-86 either alone or in combination on eat-4 expression in PVR is assayed using the eat-4prom10 reporter transgene otEx5301.
(I) ceh-37 affects eat-4 expression (assayed with eat-4prom6 reporter transgene otIs392) in the BAG and ASG neurons.
(J) lin-11 affects eat-4 expression (eat-4prom6 transgene otIs392) in the ASG neurons.
(K) unc-86 affects eat-4 expression (eat-4 fosmid reporter transgene otIs388) in the AIZ and AIM neurons. See Fig.S3 for more notes on unc-86 function in AIZ.
Ectopic misexpression of terminal selector-type transcription factors has been shown to impose specific neuronal identities on other cell types (e.g.(Flames and Hobert, 2009; Kratsios et al., 2011)). Using che-1, mec-3 and ceh-36 as examples, we confirmed that misexpression of these terminal selectors is also able to induce ectopic eat-4/VGLUT expression (Fig.S2D-F).
Dual neurotransmitter identity is coregulated via common trans-acting factors
The glutamatergic ASG neuron pair displays the intriguing property of upregulating an additional neurotransmitter system under specific environmental conditions in order to improve chemosensory acuity (Pocock and Hobert, 2010). Specifically, in hypoxic conditions, 5HT antibody staining and expression of tryptophan hydroxylase (tph-1), the rate-limiting enzyme of 5HT biosynthesis, is significantly upregulated in ASG. We find that in addition to regulating eat-4/VGLUT and other terminal features of ASG, lin-11 is also required for the upregulation of tph-1 in ASG under hypoxic conditions (Fig.4A). Therefore, both glutamatergic and serotonergic identity of the ASG neurons are coregulated by a common trans-acting factor.
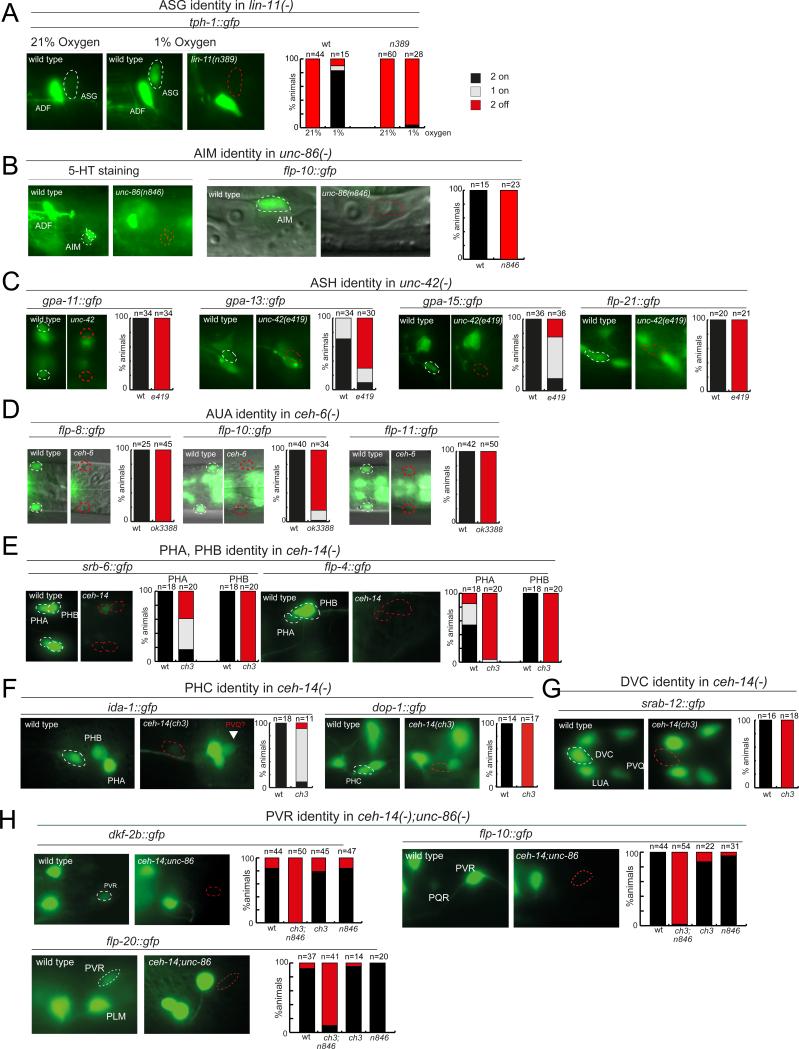
Fig.4. Adoption of glutamatergic identity is linked to the adoption of other neuronal identity features.
(A) In the ASG sensory neurons, lin-11 affects expression of the serotonergic marker tph-1 that is induced under hypoxic conditions in ASG (tph-1::gfp transgene zdIs13).
(B) unc-86 affects serotonin staining and another identity marker of AIM (flp-10::gfp transgene otIs92).
(C) unc-42 controls several features of ASH identity (gpa-11prom2::gfp transgene otEx5336, gpa-13::gfp transgene ofEx213, gpa-15::gfp transgene pkIs591, flp-21::gfp transgene ynIs80). See also Fig.S2.
(D) ceh-6 controls several features of AUA identity (flp-8::gfp transgene ynIs2022, flp-10::gfp transgene otIs92, flp-11::gfp transgene ynIs40).
(E,F,G) ceh-14 controls several features identity features of the PHB, PHC and DVC neurons (ida-1::gfp transgene inIs179, dop-1::gfp transgene vsIs28, srab-12::gfp transgene sEx12012).
(H) ceh-14 and unc-86 redundantly control the identity of the PVR neuron (flp-10::gfp transgene otIs92, flp-20::gfp transgene ynIs54, dkf-2b::gfp transgene otEx5323).
A similar coregulation of dual neurotransmitter identities is observed in the AIM neurons. These neurons were previously reported to be serotonergic and to require the POU homeobox gene unc-86 to acquire their serotonergic identity (Jafari et al., 2011)(Fig.4B). We find that eat-4/VGLUT expression in AIM is also abolished in unc-86 mutant animals (Fig.4B). Additionally, we observe loss of the flp-10 neuropeptide encoding gene in AIM in unc-86 mutants, mirroring the previously reported impact of unc-86 on specific morphological features of AIM (Kage et al., 2005)(Fig.4B). As in the case of lin-11 and ASG, these findings indicate co-regulation of distinct neurotransmitter identities within a single neuron class by a common trans-acting factor.
Identification of new regulators of glutamatergic neuronal identity
We next examined the function of three transcription factors that previous studies had found to be expressed postmitotically and continually in what we define here as eat-4(+), glutamatergic neurons but whose impact on the identity of these neurons has either not been studied in detail or not studied at all.
unc-42 controls the identity of the ASH sensory neurons
unc-42 encodes a paired-type transcription factor related to mammalian Prop1 (Baran et al., 1999). unc-42 is expressed in the ASH nociceptive sensory neurons and its expression persists throughout adulthood due to autoregulation (Baran et al., 1999). unc-42 is required for the expression of two orphan seven transmembrane receptors of the odorant receptor family, sra-6 and srb-6, in ASH (Baran et al., 1999) but since the expression of odorant receptor family members in sensory neurons is strongly activity-dependent (Nolan et al., 2002; Peckol et al., 2001), it was unclear if unc-42 broadly affects neuronal identity or whether it has a narrower role in controlling sensory responsiveness or neuronal activity. We find that unc-42 controls eat-4/VGLUT expression in ASH (Fig.3F). Through postembryonic removal of unc-42, we found that unc-42 is continuously required to maintain eat-4 expression (Fig.S2B). unc-42 also affects the expression of all other terminal identity markers tested, including the three Gα-encoding genes gpa-11, gpa-13, gpa-15 and the neuropeptide flp-21 (Fig.4C). The loss of terminal identity features is not indicative of a loss of these neurons since ASH neurons still take up dye in unc-42 mutants (Baran et al., 1999).
ceh-6 controls the identity of the AUA interneurons
ceh-6 encodes a POU homeobox gene expressed in a small number of head neuron classes (Burglin and Ruvkun, 2001), one of them the AUA interneuron class, which regulates aggregation behavior (Coates and de Bono, 2002). ceh-6 expression in AUA and other head neurons is maintained throughout adulthood (Burglin and Ruvkun, 2001). We find that in ceh-6 null mutant animals eat-4/VGLUT expression in the AUA neurons is abrogated (Fig.3G). We tested a number of additional terminal markers of AUA identity (neuropeptide-encoding genes flp-8, flp-10 andflp-11) and found that the expression of each of them was also strongly affected in the AUA neuron class of ceh-6 mutants (Fig.4D). The AUA neuron is not absent in ceh-6 mutants, since we find that expression of the panneuronal marker rab-3 is unaffected in AUA (Fig.2G). Like other terminal selectors (Hobert, 2011), ceh-6 therefore affects the adoption of a specific identity while panneuronal identity is unaffected.
ceh-14 controls the identity of phasmid sensory neurons
ceh-14 encodes a LIM homeobox gene that is expressed in a small number of head and tail neurons, including the PHA, PHB and PHC tail phasmid sensory neurons (Cassata et al., 2000). Using a fosmid-based reporter, we confirmed that ceh-14 expression is maintained throughout the life of these neurons (data not shown). We find that eat-4 expression is affected in all three phasmid sensory neurons of ceh-14 null mutants (Fig.3H). The expression of additional identity markers for PHA and PHB (GPCR srb-6 and neuropeptide flp-4) and PHC (tyrosine phosphatase ida-1 and the dopamine receptor dop-1) is also affected in ceh-14 mutants (Fig.4E,F). PHA and PHB also display dye filling defects in ceh-14 mutants (Kagoshima et al., 2013). Panneuronal features (rab-3 expression) of the phasmid neurons are unaffected in ceh-14 null mutants (Fig.S2H).
We also observed an effect of loss of ceh-14 on eat-4 expression in the DVC interneurons (Fig.4H). Another DVC cell fate marker, the GPCR srab-12, also displays defective expression in DVC (Fig.5G). We observed no defects in eat-4 expression in the normally ceh-14 expressing PVQ neurons of ceh-14 mutant animals. Joint removal of ceh-14 and ttx-1, a previously described regulatory of AFD neuron identity (Satterlee et al., 2001), results in loss of eat-4 expression in AFD (Fig.3D). ceh-14 and ttx-1 also collaborate in the activation of other AFD-expressed terminal effector genes (H. Kagoshima, pers. comm.).
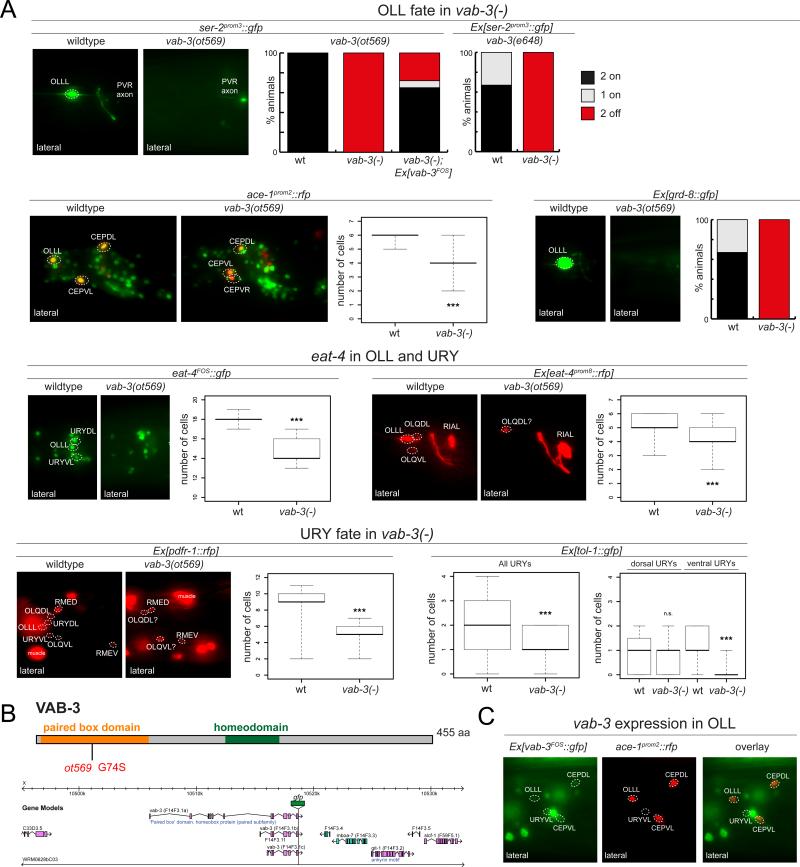
Fig.5. vab-3 is a novel candidate terminal selector for the glutamatergic OLL and URY neurons.
(A) vab-3(ot569) animals affect eat-4 expression in the OLL and URY neurons, as well as other identity markers of OLL and URY. Standard whisker-and-box plots of total cell counts in the anterior ganglia are presented. The vab-3(ot569) allele was used except where indicated otherwise.
(B) Diagram of vab-3 protein structure and the genomic locus encompassed by the rescuing fosmid WRM0628bC03.
(C) Expression of vab-3 in adult OLL and URY neurons. The OLL neurons were identified by overlap with ace-1::tagrfp and the URY neurons were identified by position and cell body morphology.
vab-3 controls the identity of the OLL and URYV neurons
In addition to pursuing candidate genes, we identified an additional regulator of glutamatergic neuron identity through an unbiased screen for EMS-induced mutants in which the glutamatergic OLL neurons, sensors of bacterial pathogens (Chang et al., 2011), do not adopt their identity. This screen identified the vab-3 Paired homeobox gene (Fig.5; see Supplementary Experimental Procedures). In vab-3 mutants, the expression of eat-4, as well as the tyramine receptor ser-2, the acetylcholinesterase ace-1 and the groundhog gene grd-8 fail to be expressed in the OLL neurons (Fig.5A). The OLL neurons are generated in these animals as assessed by unaffected expression of both ift-20 and the panneuronal rab-3 marker in the anterior ganglion (Fig.S2I).
A fosmid-based vab-3 reporter is expressed in the OLL neurons, and this expression is maintained throughout the life of these neurons, consistent with a role of vab-3 in not only initiating but also maintaining the OLL differentiation program (Fig.5B,C). Apart from OLL, we also observe expression of vab-3 in the glutamatergic ventral URY neuron class, which are lineally unrelated sensory neurons of unknown function. Expression of eat-4/VGLUT expression, as well as the expression of two additional markers of ventral URY fate, the pdfr-1 neuropeptide receptor and the Toll receptor tol-1 is lost in vab-3 mutants (Fig.5A).
Taken together, our candidate as well as genetic screening approach has revealed four additional regulators of the identity of multiple distinct glutamatergic neuron types. Each factor is expressed in the respective neuron class during its entire postmitotic life span. Each of these factors not only controls eat-4 expression but also several additional identity features of the respective glutamatergic neuron type. Due to their broad effect on distinct terminal identity features, each of these factors can therefore be considered a terminal selector-type transcription factor.
Redundancy of glutamatergic identity regulators
The expression of a number of the transcriptional regulators of glutamatergic identity that we have identified here is restricted to one class of glutamatergic neurons (che-1 in ASE, ets-5 in BAG, ttx-1 in AFD and mec-3 in touch neurons). In contrast, unc-86, lin-11 and ceh-14 are expressed in multiple very distinct glutamatergic neuron classes and we examined whether these regulators generally affect glutamatergic identity in all neurons in which they are expressed. Genetic elimination of unc-86 results in no effect on eat-4/VGLUT expression in the URY sensory neurons, the I2 pharyngeal interneurons or the tail sensory neuron PVR (Fig.S3). Similarly, the LIM homeobox gene ceh-14 affects glutamatergic identity in PHA, PHB and PHC, but not in PVQ or PVR neurons (Fig.3H and data not shown). Likewise, the LIM homeobox gene lin-11 affects ASG glutamatergic identity, but not AIZ glutamatergic identity (data not shown).
We considered the possibility that potential functions of these regulators may be masked by redundancies with other glutamatergic regulators. To address this possibility, we specifically focused on the PVR tail sensory neuron because it co-expresses unc-86 and ceh-14, each of which acts as a glutamatergic identity regulator in other neuron types, as described above. PVR co-expresses unc-86 and ceh-14 throughout the life of the neuron, consistent with a role for these factors in controlling the terminal differentiation state of PVR (data not shown). While neither single unc-86 or ceh-14 null mutant affects eat-4 expression, unc-86;ceh-14 double mutants show a dramatic loss of eat-4 expression in PVR (Fig.3H). Three additional terminal markers of PVR identity (flp-10, flp-20, dkf-2) are also redundantly controlled by ceh-14 and unc-86 (Fig.3H). The loss of marker gene expression in unc-86; ceh-14 double mutants is not due to loss of the neurons, since we were able to detect generation of the cell by lineage analysis using Nomarski optics. Taken together, the redundancy of unc-86 and ceh-14 function in PVR suggests that glutamatergic neurons not affected by a transcription factor that has a glutamatergic control function in other neuron types may be masked by redundantly acting factors.
Requirement for specific cis-regulatory motifs in the eat-4/VGLUT locus argues for direct regulation by specific transcription factors
We next asked whether any of the glutamatergic identity regulators may control eat-4/VGLUT expression directly. To this end, we made use of a knowledge of the binding sites for six of the above mentioned transcription factors, ETS-5, UNC-86, CEH-6, CEH-36, TTX-1 and CEH-14 (Baird-Titus et al., 2006; Brandt et al., 2012; Duggan et al., 1998; Etchberger et al., 2007; Kim et al., 2010; Wingender et al., 1996). We indeed found binding sites for each of these factors in the eat-4 modular elements that drive expression in the respective neurons types and we found that deletion of these binding sites resulted in a loss of expression in the respective neuron types (Fig.2), thereby providing a strong indication that these transcription factors directly control eat-4 expression. For example, deletion of an Otx/K50-type homeodomain site (TAATCC) affects expression exclusively in the ASE and AWC neuron classes (Fig.2), which both require ceh-36/Otx for correct eat-4 expression (Fig.3A, B). Mutation of another K50-type homeodomain binding also affected eat-4 expression in the ASE and AWC neurons, but additionally in the AFD neurons, which require the K50-type ttx-1/Otx homeobox gene for correct eat-4 expression (Fig.2,3). Mutation of an ETS domain binding site affected expression exclusively in the BAG neurons which require the ETS-5 transcription factor for eat-4 expression (Fig.2). Mutation of a predicted POU homeodomain sites affected unc-86- and ceh-6-dependent eat-4 expression in touch neurons, PVR and AUA (Fig.2). Lastly, ceh-14-dependent eat-4 expression in PHA, PHB, PHC and DVC expression is controlled by a cis-regulatory module, eat-4prom5, and ModEncode data (Niu et al., 2011) reveals several CEH-14 binding peaks that map onto this module (Fig.S4), providing additional support for ceh-14 directly regulating eat-4/VGLUT.
Redeployment of the same cis-regulatory motif in distinct neuron types
Our cis-regulatory analysis also revealed that distinct transcription factor family members in distinct neuron types apparently use the same cis-regulatory sites to control eat-4 expression. For example, deletion of the putative POU homeobox site in the eat-4prom2module affects expression of eat-4/VGLUT in the light and harsh touch receptor neurons that require the POU homeobox gene unc-86 for correct eat-4 expression. This deletion also affects eat-4/VGLUT expression in the AUA neurons that require the POU homeobox gene ceh-6 for correct eat-4 expression (Fig.2,3). The same scenario applies for TAATCC K50-homeodomain sites in the eat-4prom6 module: as mentioned above, deletion of one of the two sites abrogates ceh-36K50-homeodomain-dependent eat-4 expression in the ASE and AWC neurons, while a mutation in the other site also abrogates ttx-1K50-homeodomain-dependent expression in AFD (Fig.2). Intriguingly, mutation of this second TAATCC site also abrogates eat-4 expression in the BAG and ASG neurons, suggesting that these neurons utilize another K50 homeodomain protein.
We tested whether ceh-37, the third Otx/K50 ortholog in C.elegans besides ceh-36 and ttx-1, could be involved in controlling eat-4 expression in ASG and BAG. ceh-37 is expressed in the non-glutamatergic AWB neurons as well as ASG and BAG (Lanjuin et al 2003; T. Burglin, pers comm.). We find that ceh-37 is indeed required for eat-4 expression in both the ASG and BAG neuron classes (Fig.3I), likely through its cognate binding site TAATCC. Hence, the glutamatergic identity of five different neuron classes is controlled by three distinct Otx-type homeodomain transcription factors (ceh-36: AWC and ASE, ttx-1: AFD, ceh-37: ASG and BAG), operating through at least partially shared TAATCC sites. Each of these Otx factors may collaborate with distinct cofactors in these distinct cell types (e.g. ceh-37 with ets-5 in BAG and with lin-11 in ASG; Fig.S5).
Additional homeodomain regulators of eat-4 expression
The striking preponderance of homeodomain transcription factors in the collection of transcription factors that we found to control eat-4 expression prompted us to search for additional predicted homeodomain binding sites that may be required for eat-4 expression in neurons for which we had not yet identified regulatory factors. We focused on the eat-4prom2module, which drives expression in a number of distinct neuron classes (Fig.2). This module contains two sets of palindromic, predicted homeodomain binding sequences (ATTAN2-3TAAT). Mutation of one of these homeodomain binding palindromes resulted in complete elimination of eat-4 reporter expression in the ASK neurons, while mutation of the other homeodomain binding palindrome (located ~200 bp away) resulted in very strong reduction of eat-4 expression in the ADL sensory neurons (Fig.2). ASK and ADL express distinct LIM homeobox genes that may operate through these sites. Based on a fosmid reporter gene provided by the ModEncode consortium, the ASK neurons express and maintain the LIM homeobox gene ttx-3, the worm ortholog of the vertebrate Lhx2/9 gene (Zhang et al., submitted). eat-4/VGLUT expression is abolished in the ASK neurons of ttx-3 mutants (Fig.S6). The ADL neurons express the Lhx1/5 ortholog lin-11 (Hobert et al., 1998) and we find eat-4/VGLUT expression in this neuron class to be reduced in lin-11 mutants (Fig.S6). In the case of ASK and ttx-3, we examined four additional terminal identity features of ASK in ttx-3 mutants (neuropeptides flp-13 and nlp-14, rGC gcy-27 and dye filling of the neuron) and found expression of each identity feature to be also defective (Fig.S6). Hence, neurotransmitter identity is also linked in this case to the adoption of other identity features through coregulation of these distinct features by ttx-3.
Mutation of yet another predicted homeodomain binding site in the eat-4prom2 cis-regulatory module eliminated expression of eat-4 exclusively in the LUA tail interneurons (Fig.2). It is not yet known which homeobox genes are expressed in LUA.
Potential conservation of homeodomain regulation of glutamatergic identity in mouse
The preponderance of homeodomain transcription factors among the glutamatergic identity regulators in C.elegans is remarkable. Every single glutamatergic neuron differentiation program that we have described here depends on at least one homeodomain protein. 11 out of the 13 factors (85%) described here to control terminal neuron identity are homeodomain transcription factors. This proportion far exceeds the proportions (~11%) of homeodomain transcription factors (~100) relative to other types of transcription factors (~900) in the C.elegans genome. This preponderance of homeodomain regulators of glutamatergic identity in C.elegans prompted us to ask whether homeodomain proteins may also control glutamatergic identity in vertebrates. Indeed, the Otx-type protein CRX is known to control the identity of glutamatergic photoreceptors in the mouse retina (Furukawa et al., 1997). To show that homeodomain proteins may be more commonly employed as regulatory of glutamatergic neurons, we examined the expression of mouse homologs of the two most prominent homeodomain subtypes that we identified as glutamatergic regulators in C.elegans, Brn-type POU homeodomain proteins (unc-86 and ceh-6 orthologs) and LIM homeodomain proteins (ceh-14, mec-3, lin-11 and ttx-3 orthologs). We focused on examining expression in adult neurons because we aimed to avoid identifying factors with only transient roles in glutamatergic neurons and because our C.elegans studies suggested that glutamatergic identity regulators are continuously expressed throughout the life of the neuron to maintain their identity.
We stained adult mice with antibodies directed against three Brn-type POU (BRN3a,b,c) and four LIM-type homeodomain proteins (LHX1, LHX2, LHX3 and LHX5). We found expression of five of these seven proteins in adult CNS neurons (data not shown). We then made use of the observation made in C.elegans that in several cases LIM and POU homeodomain work together to determine glutamatergic identity (unc-86/mec-3 in touch neurons; unc-86/ceh-14 in PVR neurons) and sought to identify regions of overlap of POU and LIM homeodomain expression in adult glutamatergic CNS neurons. Antibody costaining revealed that the BRN3a and LHX1 are coexpressed in two sets of adult glutamatergic neuron types, one set in the supramammillary nucleus of the hippocampus (data not shown) and one set in the largest nucleus of the olivary body, the inferior olive (Fig.6A). To examine whether one of these genes may indeed have a role in these glutamatergic neurons, we used a floxed, tamoxifen-inducible LHX1 knockout allele (Kwan and Behringer, 2002) to conditionally remove LHX1 in 8 week old mice. We find that 10 days after tamoxifen treatment, BRNA3a and VGLUT2-expressing glutamatergic neurons have disappeared from the inferior olive (Fig.6B). Whether these cells have died because LHX1 is directly involved in controlling survival or whether they have died due to a neuronal identity loss is not clear at present. In either case, this data strongly implies an important function of LHX1 in defining the existence of this glutamatergic neuron type.
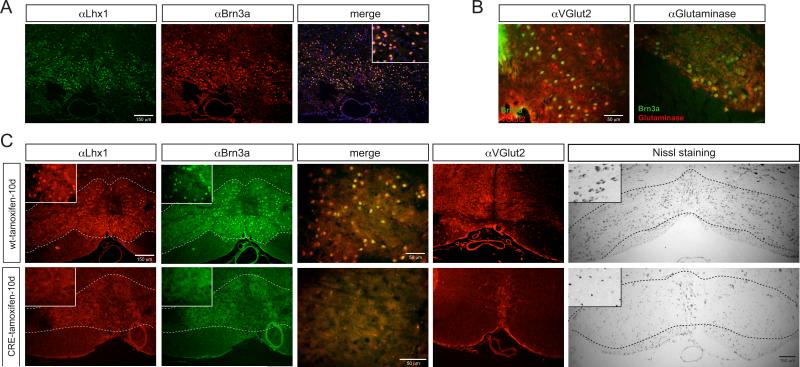
Fig.6. The loss of Lhx1 gene in the adult mouse brain affects the survival of glutamatergic neurons in the Inferior olive.
(A) LHX1 and BRN3A colocalize in neurons of the inferior olive in the brain stem. See also Fig.S7.
(B) Double immunostaining of VGLUT2 and glutaminase with BRN3A demonstrates the glutamatergic identity of the BRN3A/LHX1-positive neurons in the inferior olive (yellow overlap).
(C) Loss of glutamatergic inferior olive neurons upon postdevelopmental removal of Lhx1. Immunostaining analysis and Nissl staining of the inferior olive nucleus in Lhx1flox;R26CreER animals compared to Lhx1flox siblings. 8-10 week old adult animals were administered tamoxifen for 10 days and analyzed shortly thereafter. In the wild-type, tamoxifen-treated animals the neurons show LHX1, BRN3a and VGLUT2 staining (upper panels), but Lhx1flox;R26CreER animals do not. Nissl staining reveals that in these animals there is a massive loss of cells in the inferior olive nucleus compared with the wild-type brains. 5 animals were analyzed for each group.
As it is the case for C.elegans LIM homeobox genes, there are glutamatergic CNS neurons that do not express LHX1 and, vice versa, LHX1 is also expressed in non-glutamatergic neurons (Fig.S7)(Lein et al., 2007; Zhao et al., 2007). This supports the existence of neuron-type specific mechanisms for the regulation of glutamatergic identity in the vertebrate nervous system akin to what we observe in the C.elegans system.
DISCUSSION
We have revealed novel insights into how glutamatergic neuron identity is controlled. Through mutational analysis of the cis-regulatory control regions of eat-4/VGLUT as well as through genetic loss of function analysis of trans-acting factors, we have ruled out the possibility that eat-4/VGLUT expression is controlled by one global, glutamatergic regulator in C.elegans (see model #1 in Fig.1A). Instead, the eat-4/VGLUT locus operates as an integrator device that samples distinct regulatory inputs in distinct cellular contexts through a modular arrangement of cis-regulatory elements.
We have described here more than a dozen terminal selector-type transcription factors that act through these modular cis-regulatory elements to control VGLUT expression in two thirds of all glutamatergic neurons (Table 1). All of these transcription factors are continuously expressed throughout the life of the respective neuron type suggesting they do not only initiate but also maintain VGLUT expression. Moreover, all the transcription factors described here affect not only VGLUT expression, and hence the glutamatergic phenotype, but also a number of additional terminal differentiation genes that define the specific identify features of distinct glutamatergic neurons. This is in line with model #3 described in the introductory Fig.1A and further supports the terminal selector concept which posits the existence of master regulatory-type transcription factors that coregulate a multitude of terminal identity feature of a mature neuron (Hobert, 2011). In the absence of these terminal selectors, neurons appear to remain in an undifferentiated neuronal ground state and do not display obvious switches in identity (see Supplementary Text).
Coregulation of neuronal identity features also extends to multiple transmitter identities of an individual neuron type, as exemplified by lin-11 and unc-86, which control the glutamatergic/serotonergic identity of the ASG sensory and AIM interneurons, respectively. Neurons have long thought to be only employing one neurotransmitter system (“Dale's principle”), but over the last several years more than one transmitter system has been found to be employed simultaneously in a number of distinct neuron types in vertebrates and invertebrates, including the co-usage of glutamate and serotonin in some regions of the vertebrate CNS (Seal and Edwards, 2006). However, in none of these co-transmitter cases has it been reported whether the two neurotransmitter phenotypes of a given neuron type are independently regulated or controlled by the same trans-acting factor. We have provided here two examples of co-regulation of two distinct neurotransmitter identities by a common trans-acting factor.
Our analysis of trans-acting factors also illustrates the combinatorial coding nature of neuronal identity control. This is illustrated with the POU homeobox gene unc-86 and the LIM homeobox genes mec-3 and ceh-14. unc-86 and mec-3 cooperate to control glutamatergic touch neurons, such as the ALM neurons. In the PVR neuron and possibly also in the PHC neurons, unc-86 cooperates with another LIM homeobox gene, ceh-14. In yet other neurons (phasmid neurons), ceh-14 controls glutamatergic identity independently of unc-86 (which is not expressed in phasmid neurons).
The observation that mutation of the same cis-regulatory motif can affect eat-4 expression in multiple distinct neuron types indicates that there are limits to modularity. That is, cis-regulatory information is not always encoded by distinct cis-regulatory modules but it may be encoded by a similar grammar to be read out by different trans-acting factors in different neuron types. For example, the same POU homeobox site is apparently recognized by unc-86 in light touch receptor neurons and by ceh-6 in the AUA neurons. Three distinct Otx-type transcription factors read out the same cis-regulatory motif to control eat-4 expression in distinct sensory neuron classes. Each of these Otx genes appear to cooperate with distinct cofactors in different neuron types (Fig.S5). Otx genes are expressed in several distinct sensory neuron structures in the mouse (Acampora et al., 2001), most of which likely use glutamate as neurotransmitter.
Every single C.elegans glutamatergic neuron differentiation program that we have described here depends on at least one homeodomain protein. This remarkable preponderance of homeodomain transcription factors led us to explore the expression and function of homeodomain transcription factors in terminal differentiation of mouse glutamatergic neurons. Following the C.elegans lead of POU and LIM homeodomain proteins working together in distinct glutamatergic neuron types, we identified a glutamatergic neuron type in the inferior olive of the brainstem that coexpresses POU and LIM homeobox genes and requires the LIM homeobox genes LHX1 for its continuous presence. Even though our mouse findings do not yet prove that vertebrate POU/LIM homeodomain proteins directly activate VGLUT expression, they are consistent with such a notion. The selective loss of inferior olive glutamatergic neurons and the restricted expression of LHX1 in only some glutamatergic neuron types in the CNS is also consistent with a conservation of the modular, piece-meal regulatory logic of VGLUT regulation.
Previous work on terminal selector-type transcription factors, further extended here, has shown that they operate through simple cis-regulatory motifs (Hobert, 2011). Terminal differentiation genes that are expressed in multiple distinct neuron types, such as the eat-4/VGLUT gene described here, contain a modular assembly of simple terminal selector motifs that are read out in individual neuron types by specific terminal selectors. Gene expression profiles may be able to rapidly evolve through the gain and loss of terminal selector motifs. In the context of eat-4/VGLUT this means that on an evolutionary time scale the glutamatergic phenotype of a neuron can be rapidly gained (or lost) through the acquisition (or loss) of terminal selector motifs in eat-4/VGLUT. Compared to other neurotransmitters, glutamate is different because its employment as a neurotransmitter does not require the presence of a specialized synthesis and recycling machinery; rather the only determinant of the glutamatergic phenotype is the expression of VGLUT (Takamori et al., 2000, 2001). Hence, to gain a glutamatergic phenotype, only the VGLUT locus rather than an entire pathway of neurotransmitter synthesizing enzymes and transporters needs to acquire responsiveness to a neuron-type specific terminal selector. Since glutamate is a very widely employed neurotransmitter in many different nervous systems, our findings - and the terminal selector gene concept in general - provide a straight-forward conceptual framework how neurotransmitter phenotypes and neuronal gene expression patterns in general can rapidly evolve to generate the enormous diversity of neuronal cell types.
EXPERIMENTAL PROCEDURES
C.elegans strains and transgenes
A list of strains and transgenes can be found in the Supplementary Information.
eat-4/VGLUT reporter transgenes
The eat-4 fosmid reporter was generated by fosmid recombineering using fosmid WRM0623aF12 and an SL2-based, nuclear localized yfp reporter (Tursun et al., 2009). The eat-4 locus reporter was generated by in vivo recombination (Boulin et al., 2006), using two overlapping fragments of the eat-4 locus (see Supplementary Experimental Procedures).
The 5’ eat-4 reporter constructs were generated by PCR and subcloning into the standard pPD95.75-based expression vectors and mutagenized with the QuickChangeII XL Site-Directed Mutagenesis Kit (Stratagene). Constructs were injected at 50ng/ul with rol-6(su1006) or ttx-3::dsRed as co-injection marker. The resulting transgenic arrays are listed in the Supplementary information. All strains were scored as young adults.
Genetic screen
otIs138 transgenic animals (ser-2prom::gfp) were EMS-mutagenized and animals with loss of expression in OLL were isolated a Copas Biosort machine (Doitsidou et al. 2008). Whole genome sequencing followed by data analysis with MAQGene (Bigelow et al. 2009) was used to determine the molecular identity of ot569, a new vab-3 allele. See Supplementary Information.
Analysis of serotonergic fate of ASG neurons
Young adult worms were incubated at 1% oxygen for 24 hours at 25°C in a hypoxic semi-sealed chamber (oxygen levels were controlled by a ProOx P110 compact oxygen controller (BioSpherix)) and compared to 21% oxygen incubated worms at 25°C. Antibody staining was performed using a tube fixation protocol described in more detail in the Supplementary Information.
Mouse genetics
We used two previously generated mouse lines, Lhx1flox (Kwan and Behringer, 2002) and ROSA26CreER (Badea et al., 2003). Tamoxifen was administrated orally in the diet at a dose of 80 mg/kg/day (Harlan tamoxifen diet). Animals were treated when they reached 8-10 weeks of age. After 10 days they were perfused and analyzed. For each condition, 5 animals were used.
Mouse brain sections were stained with antibodies, in situ hybrization probes or other strains using standard procedure, described in the Supplementary Information which also contain details on antibodies and probes.
Supplementary Material
Article Highlights.
Regulation of expression of the vesicular glutamate transporter is highly modular
Homeodomain proteins initiate and maintain VGLUT expression in distinct neuron types
Distinct terminal neuronal features including cotransmitter identity are coregulated
Homeodomain regulation of glutamatergic identity is phylogenetically conserved
ACKNOWLEDGEMENTS
We thank Q. Chen for expert assistance in generating transgenic strains, A. Goldsmith for generating the eat-4 locus reporter, members of the C.elegans community for providing strains, A. Boyanov for expert assistance in whole genome sequencing, G. Minevich for initial identification of three mutants that affect ser-2 expression in OLL, J. Crissman, L. Khrimian, M. Peterson, J. Hou, B. Wu and U. Aghayeva for help with generating strains, M. Oren for examining eat-4 expression in the male tail, J. Dodd for providing Lhx1 antibody, T. Buerglin and H. Kagoshima for communicating unpublished results and members of the Hobert lab, Iva Greenwald and Piali Sengupta for comments on the manuscript. This work was funded by the NIH (R01NS039996-05; R01NS050266-03). O.H. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acampora D, Gulisano M, Broccoli V, Simeone A. Otx genes in brain morphogenesis. Prog Neurobiol. 2001;64:69–95. doi: 10.1016/s0301-0082(00)00042-3. [DOI] [PubMed] [Google Scholar]
- Alkema MJ, Hunter-Ensor M, Ringstad N, Horvitz HR. Tyramine Functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron. 2005;46:247–260. doi: 10.1016/j.neuron.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird-Titus JM, Clark-Baldwin K, Dave V, Caperelli CA, Ma J, Rance M. The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site. J Mol Biol. 2006;356:1137–1151. doi: 10.1016/j.jmb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Baran R, Aronoff R, Garriga G. The C.elegans homeodomain gene unc-42 regulates chemosensory and glutamate receptor expression. Development. 1999;126:2241–2251. doi: 10.1242/dev.126.10.2241. [DOI] [PubMed] [Google Scholar]
- Boulin T, Etchberger JF, Hobert O. Reporter gene fusions. WormBook. 2006:1–23. doi: 10.1895/wormbook.1.106.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt JP, Aziz-Zaman S, Juozaityte V, Martinez-Velazquez LA, Petersen JG, Pocock R, Ringstad N. A single gene target of an ETS-family transcription factor determines neuronal CO2-chemosensitivity. PLoS ONE. 2012;7:e34014. doi: 10.1371/journal.pone.0034014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill MS, Ninkovic J, Winpenny E, Hodge RD, Ozen I, Yang R, Lepier A, Gascon S, Erdelyi F, Szabo G, et al. Adult generation of glutamatergic olfactory bulb interneurons. Nat Neurosci. 2009;12:1524–1533. doi: 10.1038/nn.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Maricq AV. Ionotropic glutamate receptors: genetics, behavior and electrophysiology. WormBook. 2006:1–16. doi: 10.1895/wormbook.1.61.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR, Ruvkun G. Regulation of ectodermal and excretory function by the C.elegans POU homeobox gene ceh-6. Development. 2001;128:779–790. doi: 10.1242/dev.128.5.779. [DOI] [PubMed] [Google Scholar]
- Cassata G, Kagoshima H, Andachi Y, Kohara Y, Durrenberger MB, Hall DH, Burglin TR. The LIM homeobox gene ceh-14 confers thermosensory function to the AFD neurons in Caenorhabditis elegans. Neuron. 2000;25:587–597. doi: 10.1016/s0896-6273(00)81062-4. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- Chang HC, Paek J, Kim DH. Natural polymorphisms in C.elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature. 2011;480:525–529. doi: 10.1038/nature10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr., Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C.elegans. Genes Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Coates JC, de Bono M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature. 2002;419:925–929. doi: 10.1038/nature01170. [DOI] [PubMed] [Google Scholar]
- Duerr JS, Han HP, Fields SD, Rand JB. Identification of major classes of cholinergic neurons in the nematode Caenorhabditis elegans. The Journal of comparative neurology. 2008;506:398–408. doi: 10.1002/cne.21551. [DOI] [PubMed] [Google Scholar]
- Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C.elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Guillermin ML, Castelletto ML, Hallem EA. Differentiation of carbon dioxide-sensing neurons in Caenorhabditis elegans requires the ETS-5 transcription factor. Genetics. 2011;189:1327–1339. doi: 10.1534/genetics.111.133835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- Hobert O, D'Alberti T, Liu Y, Ruvkun G. Control of neural development and function in a thermoregulatory network by the LIM homeobox gene lin-11. J Neurosci. 1998;18:2084–2096. doi: 10.1523/JNEUROSCI.18-06-02084.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari G, Xie Y, Kullyev A, Liang B, Sze JY. Regulation of extrasynaptic 5-HT by serotonin reuptake transporter function in 5-HT-absorbing neurons underscores adaptation behavior in Caenorhabditis elegans. J Neurosci. 2011;31:8948–8957. doi: 10.1523/JNEUROSCI.1692-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kage E, Hayashi Y, Takeuchi H, Hirotsu T, Kunitomo H, Inoue T, Arai H, Iino Y, Kubo T. MBR-1, a novel helix-turn-helix transcription factor, is required for pruning excessive neurites in Caenorhabditis elegans. Curr Biol. 2005;15:1554–1559. doi: 10.1016/j.cub.2005.07.057. [DOI] [PubMed] [Google Scholar]
- Kagoshima H, Cassata G, Tong YG, Pujol N, Niklaus G, Burglin TR. The LIM homeobox gene ceh-14 is required for phasmid function and neurite outgrowth. Dev Biol. 2013;380:314–323. doi: 10.1016/j.ydbio.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim R, Sengupta P. The HMX/NKX homeodomain protein MLS-2 specifies the identity of the AWC sensory neuron type via regulation of the ceh-36 Otx gene in C.elegans. Development. 2010;137:963–974. doi: 10.1242/dev.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P, Stolfi A, Levine M, Hobert O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci. 2011;15:205–214. doi: 10.1038/nn.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Behringer RR. Conditional inactivation of Lim1 function. Genesis. 2002;32:118–120. doi: 10.1002/gene.10074. [DOI] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd Homeobox Genes Specify Distinct Sensory Neuron Identities in C.elegans. Dev Cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lee D, Jung S, Ryu J, Ahnn J, Ha I. Human vesicular glutamate transporters functionally complement EAT-4 in C.elegans. Mol Cells. 2008;25:50–54. [PubMed] [Google Scholar]
- Lee RY, Sawin ER, Chalfie M, Horvitz HR, Avery L. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in caenorhabditis elegans. J Neurosci. 1999;19:159–167. doi: 10.1523/JNEUROSCI.19-01-00159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lou S, Duan B, Vong L, Lowell BB, Ma Q. Runx1 controls terminal morphology and mechanosensitivity of VGLUT3-expressing C-mechanoreceptors. J Neurosci. 2013;33:870–882. doi: 10.1523/JNEUROSCI.3942-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Cheng L. Roles of Tlx1 and Tlx3 and Neuronal Activity in Controlling Glutamatergic over GABAergic Cell Fates. In: Thiel G, editor. Transcription Factors in the Nervous System. Wiley-VCH; Weinheim: 2006. [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C.elegans. Genome Res. 2011;21:245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KM, Sarafi-Reinach TR, Horne JG, Saffer AM, Sengupta P. The DAF-7 TGF-beta signaling pathway regulates chemosensory receptor gene expression in C.elegans. Genes Dev. 2002;16:3061–3073. doi: 10.1101/gad.1027702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi N, Kuhara A, Nakamura F, Okochi Y, Mori I. Bidirectional regulation of thermotaxis by glutamate transmissions in Caenorhabditis elegans. Embo J. 2011;30:1376–1388. doi: 10.1038/emboj.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckol EL, Troemel ER, Bargmann CI. Sensory experience and sensory activity regulate chemosensory receptor gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:11032–11038. doi: 10.1073/pnas.191352498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock R, Hobert O. Hypoxia activates a latent circuit for processing gustatory information in C.elegans. Nat Neurosci. 2010;13:610–614. doi: 10.1038/nn.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafi-Reinach TR, Melkman T, Hobert O, Sengupta P. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C.elegans. Development. 2001;128:3269–3281. doi: 10.1242/dev.128.17.3269. [DOI] [PubMed] [Google Scholar]
- Satterlee JS, Sasakura H, Kuhara A, Berkeley M, Mori I, Sengupta P. Specification of Thermosensory Neuron Fate in C.elegans Requires ttx-1, a Homolog of otd/Otx. Neuron. 2001;31:943–956. doi: 10.1016/s0896-6273(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR. C.elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Seal RP, Edwards RH. Functional implications of neurotransmitter co-release: glutamate and GABA share the load. Curr Opin Pharmacol. 2006;6:114–119. doi: 10.1016/j.coph.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Takamori S, Rhee JS, Rosenmund C, Jahn R. Identification of a vesicular glutamate transporter that defines a glutamatergic phenotype in neurons. Nature. 2000;407:189–194. doi: 10.1038/35025070. [DOI] [PubMed] [Google Scholar]
- Takamori S, Rhee JS, Rosenmund C, Jahn R. Identification of differentiation-associated brain-specific phosphate transporter as a second vesicular glutamate transporter (VGLUT2). J Neurosci. 2001;21:RC182. doi: 10.1523/JNEUROSCI.21-22-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C.elegans. PLoS ONE. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London B Biological Sciences. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR, Westphal H. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci U S A. 2007;104:13182–13186. doi: 10.1073/pnas.0705464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.