Abstract
Candida albicans is a fungal commensal-pathogen that persistently associates with its mammalian hosts. Between the commensal and pathogenic lifestyles, this microorganism inhabits host niches that differ markedly in the levels of bioavailable iron. A number of recent studies have exposed C. albicans specializations for acquiring iron from specific host molecules in regions where iron is scarce, while also defending against iron-related toxicity in regions where iron occurs in surfeit. Together, these results point to a central role for iron homeostasis in the evolution of this important human pathogen.
Introduction
Unlike the majority of pathogenic fungi, C. albicans remains perpetually associated with its mammalian hosts [1]. It typically exists as a commensal of the mammalian microbiome, occupying mucocutaneous surfaces such as skin, the genitourinary tract, and particularly the gastrointestinal tract. C. albicans also functions as an opportunistic pathogen and can disseminate to virtually any internal organ. This ability to adapt to host microenvironments differing markedly in the levels of key micronutrients is a hallmark of C. albicans biology. For example, C. albicans is among the most common pathogens recovered from the human bloodstream [2], a region characterized by extremely low levels of iron (~10−24 M Fe3+) because of low aqueous solubility at neutral pH, combined with active sequestration by the host [3]. In contrast, dietary iron remains abundant throughout its commensal niche in the gastrointestinal tract, and iron may even approach toxic levels in regions where local acidity or hypoxia increases its bioavailability [4]. Recent investigations of C. albicans mechanisms for acquiring necessary iron in the bloodstream and tissues while also defending against iron toxicity in the gut provide insights into the importance of iron as an evolutionary force in the most common human fungal pathogen.
Mechanisms of iron acquisition in the bloodstream
Roughly two-thirds of mammalian total body iron occurs in the bloodstream, mostly in the form of hemoglobin in red blood cells [5]. In addition, the minute amount of extracellular transferrin-bound iron serves as the major mechanism for iron distribution [5]. Such fastidious sequestration of iron protects healthy hosts against iron-catalyzed toxic free radical generation (via Fenton chemistry [6]) and provides “nutritional immunity” against infection with iron-dependent pathogens [7]. Moreover, in the presence of pathogens such as C. albicans, host inflammatory responses further reduce serum iron levels through mechanisms such as decreased intestinal absorption and retention within reticuloendothelial cells [8,9]. People lacking these defenses, such patients with hemochromatosis and other iron overload syndromes, have an excess risk for bloodstream infections with C. albicans and other pathogens [10].
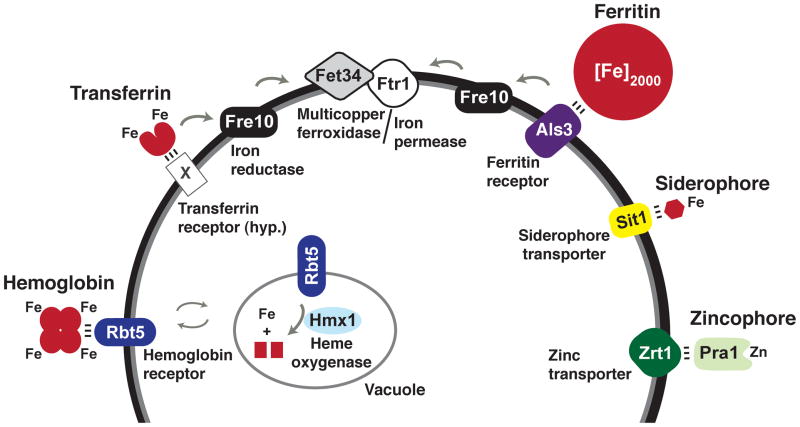
Modern medical practices have nevertheless contributed to an increased incidence of candidemia, worldwide [11–13]. Medical therapies such as intravenous catheters [12,14,15], surgery [12,14,15], antibiotics [15,16], and immunosuppressants [15] promote C. albicans bloodstream infections through disruption of host immune and epithelial barriers and by altering the microbiome. Once introduced into the bloodstream, C. albicans can acquire iron from the very molecules that are used by the host to sequester it [17–20] (Figure 1). For example, several groups have identified C. albicans hemolytic activity capable of releasing hemoglobin from host erythrocytes [21–23]. Free hemoglobin or its heme/hemin metal-porphyrin ring are bound by a hemoglobin receptor, Rbt5, on the fungal cell surface [24], followed by endocytosis of Rbt5-hemoglobin complexes [25] and release of ferrous iron by the heme oxidase, Hmx1 [19,26]. Notably, C. albicans encodes four additional homologs of Rbt5, of which one (Rbt51) has also been demonstrated to bind to hemin and to confer hemin utilization capability when expressed in the nonpathogenic model yeast, Saccharomyces cerevisiae [24].
Figure 1. C. albicans mechanisms for acquiring iron and zinc from the host.
C. albicans has evolved systems targeting host hemoglobin, host transferrin, host ferritin, and siderophores as sources of iron, and the Pra1/Zrt1 system for scavenging zinc. The depicted transferrin receptor (“X”) is hypothetical (hyp.) The depicted variants of the hemoglobin receptor, iron reductase, multicopper ferroxidase, and iron permease are members of larger gene families with demonstrated in vitro activity.
C. albicans can also utilize host transferrin in vitro as a sole source of iron [20]. It is uncertain whether C. albicans expresses a transferrin receptor, similar to certain bacterial pathogens [27], but the observation that it requires direct contact with transferrin in order to utilize it [20] suggests that this may be the case. Ferric iron derived from transferrin is taken up by a reductive iron uptake system that is conserved with the well-described high affinity iron uptake system of S. cerevisiae (reviewed in [27] and [28]). Fe3+ is first reduced to soluble Fe2+ by a cell surface-associated ferric reductase [29,30]. In coupled reactions, Fe2+ is then oxidized and imported into the fungal cytoplasm by a multicopper ferroxidase/iron permease complex [30,31]. C. albicans encodes 17 putative ferric reductases, five putative multicopper ferroxidases, and four putative ferric permeases [32] with potential functions in reductive iron uptake, and different subsets of these enzymes are expressed under different in vitro conditions (e.g.[20,33,34]). Of the two ferric permeases, only Ftr1 is expressed when iron is scarce, and FTR1 is essential in a murine bloodstream infection model of virulence [34].
Strategies for iron acquisition in tissues
Approximately one-third of mammalian total body iron occurs bound to ferritin in tissues and macrophages [5]. A single ferritin heteropolymer binds as many as 4500 iron atoms, and cytoplasmic iron-ferritin complexes are generally extremely stable [35]. Only a few bacterial pathogens such as Neisseria meningitidis have been shown to use ferritin as an iron source [36]. Among fungi, C. albicans can also utilize ferritin when provided under standard in vitro conditions or directly from host epithelial cells in culture [37]. When co-cultured with a human oral epithelial cell line, invading C. albicans hyphae aggregate host ferritin onto their surfaces using a hypha-specific cell surface protein, Als3. In vitro, fungal-mediated acidification of the laboratory culture media is required to dissociate Fe3+ from ferritin [37]. Fe3+ is transported into the fungal cytoplasm via the same reductive iron uptake system [37] described above for transferrin. Intriguingly, Als3 also plays important roles in C. albicans biofilm formation [38,39], adhesion to host epithelial and endothelial cells [40], and induced endocytosis of hyphae [41]. Further, deletion of ALS3 abrogates C. albicans virulence in oral epithelial infection models [37,40] but not in a bloodstream infection model [42]. Thus, Als3 integrates iron uptake and virulence functions, a characteristic displayed by several other key players in C. albicans iron homeostasis, as discussed below.
In common with numerous other fungal and bacterial species, C. albicans possesses a third system of iron uptake that targets siderophores rather than host molecules [43]. Siderophores are small ferric iron chelators that bind with extremely high affinity (iron formation constants Kd range from 10−20–10−50 M), some of which can extract iron from transferrin and lactoferrin [27,44–46]. It is unclear whether C. albicans synthesizes its own siderophores: siderophore activity has been reported for this species [47,48] but its genome does not encode the known fungal biosynthetic enzymes [44]. Regardless, C. albicans has been demonstrated to utilize exogenous ferrichrome-type siderophores via the Sit1 siderophore importer [43], an ortholog of S. cerevisiae Arn1 [49]. Similar to ALS3, deletion of SIT1 abrogates C. albicans virulence in a reconstituted human epithelial infection model [43] but not in a bloodstream infection model [43,50]. Future studies will be required to determine whether Sit1 plays a role in mixed infection models that include additional siderophore-producing microorganisms.
Zinc acquisition via a “zincophore”
Iron is not the only essential micronutrient, and host sequestration of manganese and zinc also contributes to nutritional immunity [51]. C. albicans was recently reported to scavenge zinc by means of Pra1, a small, secreted protein dubbed a “zincophore,” by analogy with iron-scavenging siderophores [52]. Similar to the story with ferritin utilization, during infection of human endothelial cell monolayers, invading C. albicans hyphae are able to aggregate host zinc onto their cell surfaces. The aggregation activity requires both Pra1, which exhibits zinc-binding activity in vitro, and a predicted cell surface zinc transporter, Zrt1, which binds to Pra1 and is thought to recruit soluble Pra1-zinc complexes to the fungal cell surface [52]. Pra1 was previously shown to have multiple interactions with components of the host innate immune system, including engagement of a leukocyte receptor that promotes neutrophil migration and fungal killing [53] and interactions with host complement regulators that favor fungal escape [54]. Notably, PRA1 and ZRT1 are adjacent on C. albicans chromosome 4, and the conservation of synteny among evolutionarily distant fungi (including nonpathogens) suggests that zinc acquisition is the original function of this system [52].
Defense against iron excess in the gut, a major site of commensalism
Topologically, the mammalian gut exists outside of the body, and dietary iron is not included in estimates of total body iron. A typical Western diet contains approximately 15 mg of daily iron, of which less than 10% is absorbed [4]. Net iron remains relatively high throughout the GI tract, with large boosts in iron bioavailability predicted in regions of acidity, such as the stomach [4], and oxygen-depletion, such as the large intestine [55]. Gastrointestinal commensals thus face the risk of potential iron-associated toxicity (toxic free radicals generated in the Fenton reaction [56]), at least in some regions of the gut.
How does C. albicans defend against iron-related toxicity in areas of iron excess, while retaining the capacity for aggressive iron uptake in host niches of iron depletion? One answer lies in its evolution of unique transcriptional regulatory circuit for maintaining iron homeostasis (Figure 2). In most characterized fungi (apart from S. cerevisiae), a highly conserved GATA family transcription factor represses genes for iron uptake factors when environmental iron is replete (see Box 1) [57–64]. C. albicans maintains an ortholog of this factor, Sfu1 [65,66], which directly represses genes for iron uptake factors, including components of the hemoglobin uptake system, the reductive iron uptake system, and the Sit1 siderophore transporter [67]. Indeed, SFU1 is essential for defense against high iron in vitro and for normal commensal fitness in the mammalian gut [67].
Figure 2. C. albicans regulation of iron homeostasis, virulence, and commensalism.
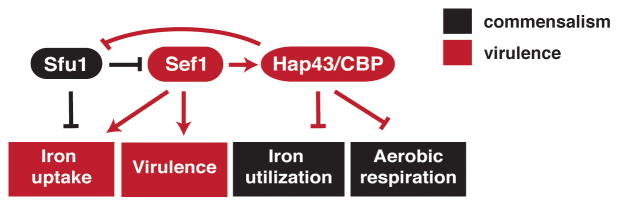
In C. albicans, the transcription factors controlling iron homeostasis are also required for virulence or commensalism. SEF1 is induced in the bloodstream, where iron is scarce, and is required for virulence in a murine bloodstream infection model. Its direct binding targets include genes for all three of its iron uptake systems, as well as known virulence factors, similar to the responses of certain bacterial pathogens that use iron depletion as a cue for virulence gene expression. By contrast, C. albicans SFU1 is induced in iron-replete environments such as the GI tract and represses genes for iron uptake factors. It remains to be determined whether the iron surfeit serves as a cue for the expression of commensalism factors.
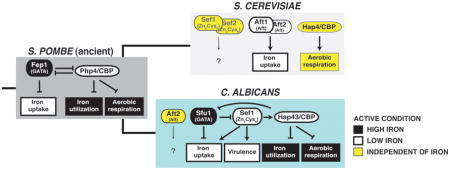
Box 1 (with Figure). Evolution of iron homeostasis in fungi.
Most characterized fungal species maintain cellular iron homeostasis by regulating the expression of iron uptake and iron utilization via a simple transcriptional switch. As depicted for the prototypical model yeast, S. pombe, a GATA family transcription factor with iron-sensing activity (Fep1) is expressed when environmental iron is replete. Fep1 directly represses iron uptake genes and PHP4, the gene for the regulatory subunit of the CCAAT binding complex. When environmental iron is low, Php4 directly represses genes for nonessential iron-utilizing processes as well as FEP1. A common ancestor to the Candida and Saccharomyces lineages gained two transcription factors (Sef1 and Aft1 precursors), leading to rewiring of the circuit. In C. albicans, Sef1 was intercalated into the existing circuit and associated with virulence genes in addition to iron uptake genes. The coregulation of iron uptake genes by both Sfu1 and Sef1 creates a feed forward loop predicted to buffer the expression of coregulated genes against transient fluctuations in environmental iron. In S. cerevisiae, the GATA factor was lost, and regulation of iron uptake genes was transferred to Aft1 and Aft2 (produced by a whole genome duplication event in the Saccharomyces lineage).
SFU1 is not required for virulence in the bloodstream, however; rather, an sfu1 knockout mutant exhibits enhanced fitness in competitive infections in this environment, presumably because of enhanced expression of iron uptake factors [67]. At least three different transcription factors mediate the fungal response to iron sequestration in the bloodstream. Rim101 is a Cys2His2 zinc finger transcription factor required for expression of iron uptake genes under neutral or alkaline conditions [68]. Rim101 also activates known virulence genes [69,70] and is required for virulence in bloodstream [71] and oropharyngeal [72] infection models. Similarly, two transcription factors that form a circuit with Sfu1 are also required for virulence in the bloodstream (Figure 2): Hap43, a highly conserved regulatory component of the CCAAT binding protein (CBP) complex [73,74] (whose Aspergillus fumigatus and Cryptococcus neoformans orthologs likewise promote virulence in these species [75,76]) and Sef1, a Zn2Cys6 zinc knuckle transcriptional activator that was introduced relatively recently into the C. albicans lineage [67,77]. Under iron-depleted conditions, the Hap43/CBP complex directly represses SFU1 and genes for nonessential iron-utilizing processes, such as aerobic respiration and iron-sulfur cluster assembly [67,73,74,78]. Under these same conditions, Sef1 directly activates HAP43, genes for all three modes of iron uptake, as well as virulence genes thought to act independently of iron [67]. Sef1 also affects fitness in a gastrointestinal infection model [67], suggesting that some at least some regions of the gut are effectively depleted for iron.
Reminiscent of Rim101, Sef1 integrates the expression of iron uptake and virulence genes, and this transcriptional regulator is itself regulated at multiple levels. When environmental iron is replete, SEF1 transcription is directly inhibited by Sfu1, as described above. Remarkably, Sfu1 also directly inhibits Sef1 protein activity by sequestering it in the cytoplasm, where it is rapidly degraded [79]. Under iron-depleted conditions, however, Sef1 is phosphorylated by the protein kinase, Ssn3, and transported into the nucleus, where it induces the transcription of iron uptake genes [79]. We hypothesize that the introduction of Sef1 into C. albicans iron homeostasis, along with mechanisms controlling its expression and protein activity, allowed for finer control of a critical gene regulon in the context of fluctuating host iron levels.
From iron homeostasis to developmental switches
Similar to free-living fungi, C. albicans must adapt to stark differences in the abundance of iron and other essential micronutrients within different niches of the mammalian host. Where this metal is scarce, C. albicans succeeds by extracting iron from host iron-sequestering molecules. Ferritin and transferrin are utilized by means of an ancient reductive iron uptake system, which C. albicans has customized with the introduction of additional homologs of each component and a novel ferritin receptor, Als3. Specific roles for the ferric reductase, multicopper ferroxidase, and iron permease homologs remain to be defined, but a plausible hypothesis is that different alleles are optimized for uptake of iron from different host compartments. Homologs of Rbt5 may serve analogous roles in hemoglobin utilization. Also, the recent discovery of a role for Pra1 as a “zincophore” speaks to the importance of micronutrients other than iron and implies the existence of as yet undiscovered mechanisms targeting these alternative nutrients..
Some bacterial pathogens use iron depletion as a kind of location marker, signifying entry into the mammalian host and triggering the expression of virulence factors [46]. C. albicans appears to use a similar logic, such that key transcriptional regulators of iron uptake genes such as Sef1 and Rim101 also activate the expression of virulence factors. Moreover, the coupling of virulence with nutrient uptake functions extends to the iron and zinc uptake effectors, Als3 and Pra1, which play additional, direct roles in virulence.
Finally, in its commensal role within the mammalian gastrointestinal tract, C. albicans encounters much higher levels of iron. It is possible that, by the converse of the logic described above, high iron levels may signify the gastrointestinal milieu and the need to express commensalism factors. My research group recently discovered that passage of C. albicans through the host digestive tract induces a developmental switch to a novel commensal cell type (Box 2) [80]. “GUT” (gastrointestinally induced transition) cells strongly outcompete previously defined virulent (“white”) and sexually-competent (“opaque”) cell types within a mouse gastrointestinal infection model and express a distinct transcriptome [80]. Compared to white and opaque cells, GUT cells upregulate SFU1 and downregulate SEF1 and iron uptake genes, as well as altering the expression of metabolic genes to match the nutrient composition of the distal mammalian GI tract [80]. Future experiments will be required to determine what additional factors promote the commensal lifestyle and whether they are triggered by levels of iron, zinc, and other critical nutrients.
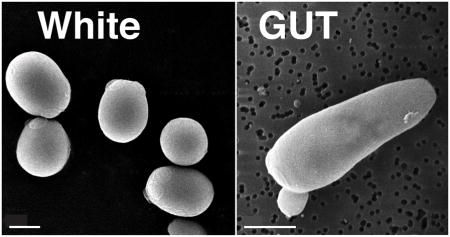
Box 2 (with Figure). C. albicans white and GUT cell types differ in the expression of iron uptake genes.
White cells (left) exhibit a round-to-oval cell morphology and are the default C. albicans cell type. White cells are virulent in murine bloodstream models of virulence. GUT (Gastrointestinally IndUced Transition) cells (right) correspond to a recently described developmental state that is triggered by passage of C. albicans yeasts through the mammalian gastrointestinal tract. GUT cells exhibit elongated cell morphology and enhanced commensal fitness but are attenuated for virulence in the bloodstream. Consistent with these functional specializations, white and GUT cells express inverse sets of iron-related genes, with SEF1 and iron uptake genes induced in white cells and SFU1 induced in GUT cells. Shown are scanning electron micrographs of white and GUT cells, with scale bars corresponding to 2μ. (Images from Chen and Noble, unpublished).
Highlights.
The yeast C. albicans is a commensal and pathogen of humans and other mammals.
Iron and zinc are sequestered in mammalian blood and tissues but not in the gut.
C. albicans virulence requires specialized mechanisms for acquiring host iron and zinc.
Limitation of iron uptake promotes C. albicans commensalism in the gut.
Acknowledgments
The author apologizes to colleagues whose work could not be described in this brief review. I am grateful to HD Madhani for helpful comments. Work in the author’s laboratory is supported by the Burroughs Wellcome Fund, the Pew Charitable Trust, and NIH R21AI099659-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Odds FC. Candida and Candidosis, a Review and Bibliography. 2. London: W.B. Saunders; 1988. [Google Scholar]
- 2.Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis. 2005;41:1232–1239. doi: 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 3.Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miret S, Simpson RJ, McKie AT. Physiology and molecular biology of dietary iron absorption. Annu Rev Nutr. 2003;23:283–301. doi: 10.1146/annurev.nutr.23.011702.073139. [DOI] [PubMed] [Google Scholar]
- 5.Andrews NC. Disorders of iron metabolism. N Engl J Med. 1999;341:1986–1995. doi: 10.1056/NEJM199912233412607. [DOI] [PubMed] [Google Scholar]
- 6.Pierre JL, Fontecave M. Iron and activated oxygen species in biology: the basic chemistry. Biometals. 1999;12:195–199. doi: 10.1023/a:1009252919854. [DOI] [PubMed] [Google Scholar]
- 7.Weinberg ED. Nutritional immunity. Host’s attempt to withold iron from microbial invaders. JAMA. 1975;231:39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- 8.Yang F, Liu XB, Quinones M, Melby PC, Ghio A, Haile DJ. Regulation of reticuloendothelial iron transporter MTP1 (Slc11a3) by inflammation. J Biol Chem. 2002;277:39786–39791. doi: 10.1074/jbc.M201485200. [DOI] [PubMed] [Google Scholar]
- 9.Prentice AM, Doherty CP, Abrams SA, Cox SE, Atkinson SH, Verhoef H, Armitage AE, Drakesmith H. Hepcidin is the major predictor of erythrocyte iron incorporation in anemic African children. Blood. 2012;119:1922–1928. doi: 10.1182/blood-2011-11-391219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullen JJ, Rogers HJ, Spalding PB, Ward CG. Natural resistance, iron and infection: a challenge for clinical medicine. J Med Microbiol. 2006;55:251–258. doi: 10.1099/jmm.0.46386-0. [DOI] [PubMed] [Google Scholar]
- 11.Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 12.Hajjeh RA, Sofair AN, Harrison LH, Lyon GM, Arthington-Skaggs BA, Mirza SA, Phelan M, Morgan J, Lee-Yang W, Ciblak MA, et al. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J Clin Microbiol. 2004;42:1519–1527. doi: 10.1128/JCM.42.4.1519-1527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 14.Blumberg HM, Jarvis WR, Soucie JM, Edwards JE, Patterson JE, Pfaller MA, Rangel-Frausto MS, Rinaldi MG, Saiman L, Wiblin RT, et al. Risk factors for candidal bloodstream infections in surgical intensive care unit patients: the NEMIS prospective multicenter study. The National Epidemiology of Mycosis Survey. Clin Infect Dis. 2001;33:177–186. doi: 10.1086/321811. [DOI] [PubMed] [Google Scholar]
- 15.Komshian SV, Uwaydah AK, Sobel JD, Crane LR. Fungemia caused by Candida species and Torulopsis glabrata in the hospitalized patient: frequency, characteristics, and evaluation of factors influencing outcome. Rev Infect Dis. 1989;11:379–390. doi: 10.1093/clinids/11.3.379. [DOI] [PubMed] [Google Scholar]
- 16.Wey SB, Mori M, Pfaller MA, Woolson RF, Wenzel RP. Risk factors for hospital-acquired candidemia. A matched case-control study. Arch Intern Med. 1989;149:2349–2353. [PubMed] [Google Scholar]
- 17.Moors MA, Stull TL, Blank KJ, Buckley HR, Mosser DM. A role for complement receptor-like molecules in iron acquisition by Candida albicans. J Exp Med. 1992;175:1643–1651. doi: 10.1084/jem.175.6.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weissman Z, Shemer R, Kornitzer D. Deletion of the copper transporter CaCCC2 reveals two distinct pathways for iron acquisition in Candida albicans. Mol Microbiol. 2002;44:1551–1560. doi: 10.1046/j.1365-2958.2002.02976.x. [DOI] [PubMed] [Google Scholar]
- 19.Santos R, Buisson N, Knight S, Dancis A, Camadro JM, Lesuisse E. Haemin uptake and use as an iron source by Candida albicans: role of CaHMX1-encoded haem oxygenase. Microbiology. 2003;149:579–588. doi: 10.1099/mic.0.26108-0. [DOI] [PubMed] [Google Scholar]
- 20•.Knight SA, Vilaire G, Lesuisse E, Dancis A. Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect Immun. 2005;73:5482–5492. doi: 10.1128/IAI.73.9.5482-5492.2005. This study establishes a role for the conserved reductive iron uptake pathway in C. albicans utilization of transferrin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo G, Samaranayake LP, Yau JY. Candida species exhibit differential in vitro hemolytic activities. J Clin Microbiol. 2001;39:2971–2974. doi: 10.1128/JCM.39.8.2971-2974.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manns JM, Mosser DM, Buckley HR. Production of a hemolytic factor by Candida albicans. Infect Immun. 1994;62:5154–5156. doi: 10.1128/iai.62.11.5154-5156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe T, Takano M, Murakami M, Tanaka H, Matsuhisa A, Nakao N, Mikami T, Suzuki M, Matsumoto T. Characterization of a haemolytic factor from Candida albicans. Microbiology. 1999;145 (Pt 3):689–694. doi: 10.1099/13500872-145-3-689. [DOI] [PubMed] [Google Scholar]
- 24•.Weissman Z, Kornitzer D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol. 2004;53:1209–1220. doi: 10.1111/j.1365-2958.2004.04199.x. This work establishes that Rbt5-hemoglobin complexes are internalized by endocytosis. [DOI] [PubMed] [Google Scholar]
- 25.Weissman Z, Shemer R, Conibear E, Kornitzer D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol. 2008;69:201–217. doi: 10.1111/j.1365-2958.2008.06277.x. [DOI] [PubMed] [Google Scholar]
- 26.Pendrak ML, Chao MP, Yan SS, Roberts DD. Heme oxygenase in Candida albicans is regulated by hemoglobin and is necessary for metabolism of exogenous heme and hemoglobin to alpha-biliverdin. J Biol Chem. 2004;279:3426–3433. doi: 10.1074/jbc.M311550200. [DOI] [PubMed] [Google Scholar]
- 27.Miethke M. Molecular strategies of microbial iron assimilation: from high-affinity complexes to cofactor assembly systems. Metallomics. 2013;5:15–28. doi: 10.1039/c2mt20193c. [DOI] [PubMed] [Google Scholar]
- 28.Kosman DJ. Molecular mechanisms of iron uptake in fungi. Mol Microbiol. 2003;47:1185–1197. doi: 10.1046/j.1365-2958.2003.03368.x. [DOI] [PubMed] [Google Scholar]
- 29.Hammacott JE, Williams PH, Cashmore AM. Candida albicans CFL1 encodes a functional ferric reductase activity that can rescue a Saccharomyces cerevisiae fre1 mutant. Microbiology. 2000;146 (Pt 4):869–876. doi: 10.1099/00221287-146-4-869. [DOI] [PubMed] [Google Scholar]
- 30.Knight SA, Lesuisse E, Stearman R, Klausner RD, Dancis A. Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology. 2002;148:29–40. doi: 10.1099/00221287-148-1-29. [DOI] [PubMed] [Google Scholar]
- 31••.Ziegler L, Terzulli A, Gaur R, McCarthy R, Kosman DJ. Functional characterization of the ferroxidase, permease high-affinity iron transport complex from Candida albicans. Mol Microbiol. 2011;81:473–485. doi: 10.1111/j.1365-2958.2011.07704.x. This biochemical and cell biological analysis of C. albicans Fet34 establishes its functional equivalence to S. cerevisiae Fet3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almeida RS, Wilson D, Hube B. Candida albicans iron acquisition within the host. FEMS Yeast Res. 2009;9:1000–1012. doi: 10.1111/j.1567-1364.2009.00570.x. [DOI] [PubMed] [Google Scholar]
- 33.Cheng X, Xu N, Yu Q, Ding X, Qian K, Zhao Q, Wang Y, Zhang B, Xing L, Li M. Novel insight into the expression and function of the multicopper oxidases in Candida albicans. Microbiology. 2013;159:1044–1055. doi: 10.1099/mic.0.065268-0. [DOI] [PubMed] [Google Scholar]
- 34.Ramanan N, Wang Y. A high-affinity iron permease essential for Candida albicans virulence. Science. 2000;288:1062–1064. doi: 10.1126/science.288.5468.1062. [DOI] [PubMed] [Google Scholar]
- 35.Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Biol. 2001;33:940–959. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- 36.Larson JA, Howie HL, So M. Neisseria meningitidis accelerates ferritin degradation in host epithelial cells to yield an essential iron source. Mol Microbiol. 2004;53:807–820. doi: 10.1111/j.1365-2958.2004.04169.x. [DOI] [PubMed] [Google Scholar]
- 37••.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217. doi: 10.1371/journal.ppat.1000217. This study describes the unexpected discovery that Als3 is the cell surface receptor for ferritin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Daniels KJ, Oh SH, Green CB, Yeater KM, Soll DR, Hoyer LL. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology. 2006;152:2287–2299. doi: 10.1099/mic.0.28959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJ, Hoyer LL. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- 41.Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE, Jr, Filler SG. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cleary IA, Reinhard SM, Miller CL, Murdoch C, Thornhill MH, Lazzell AL, Monteagudo C, Thomas DP, Saville SP. Candida albicans adhesin Als3p is dispensable for virulence in the mouse model of disseminated candidiasis. Microbiology. 2011;157:1806–1815. doi: 10.1099/mic.0.046326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heymann P, Gerads M, Schaller M, Dromer F, Winkelmann G, Ernst JF. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun. 2002;70:5246–5255. doi: 10.1128/IAI.70.9.5246-5255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haas H. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl Microbiol Biotechnol. 2003;62:316–330. doi: 10.1007/s00253-003-1335-2. [DOI] [PubMed] [Google Scholar]
- 45.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holzberg M, Artis WM. Hydroxamate siderophore production by opportunistic and systemic fungal pathogens. Infect Immun. 1983;40:1134–1139. doi: 10.1128/iai.40.3.1134-1139.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ismail A, Bedell GW, Lupan DM. Siderophore production by the pathogenic yeast, Candida albicans. Biochem Biophys Res Commun. 1985;130:885–891. doi: 10.1016/0006-291x(85)90499-1. [DOI] [PubMed] [Google Scholar]
- 49.Heymann P, Ernst JF, Winkelmann G. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2000;186:221–227. doi: 10.1111/j.1574-6968.2000.tb09108.x. [DOI] [PubMed] [Google Scholar]
- 50••.Hu CJ, Bai C, Zheng XD, Wang YM, Wang Y. Characterization and functional analysis of the siderophore-iron transporter CaArn1p in Candida albicans. J Biol Chem. 2002;277:30598–30605. doi: 10.1074/jbc.M204545200. This report characterizes the C. albicans siderophore transporter, Arn1. [DOI] [PubMed] [Google Scholar]
- 51•.Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol. 2009;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. This review nicely summarizes the data indicating a role for host sequestration of manganese and zinc in antimicrobial defense. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••.Citiulo F, Jacobsen ID, Miramon P, Schild L, Brunke S, Zipfel P, Brock M, Hube B, Wilson D. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog. 2012;8:e1002777. doi: 10.1371/journal.ppat.1002777. This study offers compelling evidence that C. albicans Pra1 serves as a “zincophore” that contributes to pathogenesis in an oral epithelial infection model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soloviev DA, Fonzi WA, Sentandreu R, Pluskota E, Forsyth CB, Yadav S, Plow EF. Identification of pH-regulated antigen 1 released from Candida albicans as the major ligand for leukocyte integrin alphaMbeta2. J Immunol. 2007;178:2038–2046. doi: 10.4049/jimmunol.178.4.2038. [DOI] [PubMed] [Google Scholar]
- 54.Zipfel PF, Skerka C, Kupka D, Luo S. Immune escape of the human facultative pathogenic yeast Candida albicans: the many faces of the Candida Pra1 protein. Int J Med Microbiol. 2011;301:423–430. doi: 10.1016/j.ijmm.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 55.Wilson M. Cambridge University Press, editor. Microbial Inhabitants of Humans. 2005. The gastrointestinal tract and its indigenous microbiota; pp. 251–317. vol University of Cambridge Press. [Google Scholar]
- 56.Pierre JL, Fontecave M, Crichton RR. Chemistry for an essential biological process: the reduction of ferric iron. Biometals. 2002;15:341–346. doi: 10.1023/a:1020259021641. [DOI] [PubMed] [Google Scholar]
- 57.Voisard C, Wang J, McEvoy JL, Xu P, Leong SA. urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis, encodes a protein similar to the erythroid transcription factor GATA-1. Mol Cell Biol. 1993;13:7091–7100. doi: 10.1128/mcb.13.11.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haas H, Angermayr K, Stoffler G. Molecular analysis of a Penicillium chrysogenum GATA factor encoding gene (sreP) exhibiting significant homology to the Ustilago maydis urbs1 gene. Gene. 1997;184:33–37. doi: 10.1016/s0378-1119(96)00570-7. [DOI] [PubMed] [Google Scholar]
- 59.Pelletier B, Beaudoin J, Mukai Y, Labbe S. Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J Biol Chem. 2002;277:22950–22958. doi: 10.1074/jbc.M202682200. [DOI] [PubMed] [Google Scholar]
- 60.Zhou LW, Haas H, Marzluf GA. Isolation and characterization of a new gene, sre, which encodes a GATA-type regulatory protein that controls iron transport in Neurospora crassa. Mol Gen Genet. 1998;259:532–540. doi: 10.1007/s004380050845. [DOI] [PubMed] [Google Scholar]
- 61.Chao LY, Marletta MA, Rine J. Sre1, an iron-modulated GATA DNA-binding protein of iron-uptake genes in the fungal pathogen Histoplasma capsulatum. Biochemistry. 2008;47:7274–7283. doi: 10.1021/bi800066s. [DOI] [PubMed] [Google Scholar]
- 62.Haas H, Zadra I, Stoffler G, Angermayr K. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J Biol Chem. 1999;274:4613–4619. doi: 10.1074/jbc.274.8.4613. [DOI] [PubMed] [Google Scholar]
- 63.Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miele R, Barra D, Bonaccorsi di Patti MC. A GATA-type transcription factor regulates expression of the high-affinity iron uptake system in the methylotrophic yeast Pichia pastoris. Arch Biochem Biophys. 2007;465:172–179. doi: 10.1016/j.abb.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 65.Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, Newport G, Agabian N. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol. 2004;53:1451–1469. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- 66.Pelletier B, Mercier A, Durand M, Peter C, Jbel M, Beaudoin J, Labbe S. Expression of Candida albicans Sfu1 in fission yeast complements the loss of the iron-regulatory transcription factor Fep1 and requires Tup co-repressors. Yeast. 2007;24:883–900. doi: 10.1002/yea.1539. [DOI] [PubMed] [Google Scholar]
- 67••.Chen C, Pande K, French SD, Tuch BB, Noble SM. An Iron Homeostasis Regulatory Circuit with Reciprocal Roles in Candida albicans Commensalism and Pathogenesis. Cell Host Microbe. 2011;10:118–135. doi: 10.1016/j.chom.2011.07.005. This study defines the relationship between Sfu1, Sef1, and Hap43 in regulating C. albicans iron homeostasis and exposes inverse roles of Sfu1 and Sef1 in promoting commensalism and pathogenesis, respectively. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68•.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol. 2004;54:1335–1351. doi: 10.1111/j.1365-2958.2004.04350.x. This study reveals the role of Rim101 in regulating iron uptake genes under alkaline pH conditions. [DOI] [PubMed] [Google Scholar]
- 69.Ramon AM, Porta A, Fonzi WA. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol. 1999;181:7524–7530. doi: 10.1128/jb.181.24.7524-7530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis D, Wilson RB, Mitchell AP. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 2000;68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, Nantel A, Mitchell AP, Filler SG. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol. 2008;10:2180–2196. doi: 10.1111/j.1462-5822.2008.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Hsu PC, Yang CY, Lan CY. Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot Cell. 2011;10:207–225. doi: 10.1128/EC.00158-10. One of two recent studies examining the role of the CBP component Hap43 in C. albicans iron homeostasis and virulence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74•.Singh RP, Prasad HK, Sinha I, Agarwal N, Natarajan K. Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J Biol Chem. 2011;286:25154–25170. doi: 10.1074/jbc.M111.233569. One of two recent studies examining the role of the CBP component Hap43 in C. albicans iron homeostasis and virulence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, White R, Kronstad JW. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jochl C, Moussa TA, Wang S, Gsaller F, Blatzer M, et al. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77•.Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. This comprehensive phenotypic analysis of a large library of transcription factor mutants includes a comparison of iron homeostasis mediators in C. albicans and S. cerevisiae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baek YU, Li M, Davis DA. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot Cell. 2008;7:1168–1179. doi: 10.1128/EC.00108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79•.Chen C, Noble SM. Post-transcriptional Regulation of the Sef1 Transcription Factor Controls the Virulence of Candida albicans in its Mammalian Host. PLoS Pathog. 2012;8:e1002956. doi: 10.1371/journal.ppat.1002956. This study reveals that Sfu1 negatively regulates Sef1 protein stability and nuclear localization under iron-replete conditions, whereas the protein kinase Ssn3 has opposite effects under iron-depleted conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80••.Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013 doi: 10.1038/ng.2710. This study describes the identification of a commensalism-specific cell type that is optimized for the conditions encountered within the mammalian GI tract. [DOI] [PMC free article] [PubMed] [Google Scholar]