Abstract
NMDA receptors are a family of glutamate-gated ion channels that regulate various CNS functions such as synaptic plasticity and learning. However hypo-or hyper-activation of NMDA receptors is critically involved in many neurological and psychiatric conditions such as pain, stroke, epilepsy, neurodegeneration, schizophrenia, and depression. Thus, it is important to identify mechanisms (such as by targeted ubiquitination) that regulate the levels of individual subtypes of NMDA receptors. In this study, we used a series of tagged, carboxy terminal constructs of GluN2D to identify associating proteins from rat brain. Of seven different GluN2D C-terminal fragments used as bait, only the construct containing amino acids 983-1097 associated with an E3 ligase, Nedd4. A direct interaction between GluN2D and Nedd4 was confirmed both in vivo and in vitro. This association is mediated by an interaction between GluN2D's C-terminal PPXY motif and the 2nd and 3rd WW domains of Nedd4. Of the four GluN2 subunits, Nedd4 directly interacted with GluN2D and also weakly with GluN2A. Nedd4 coexpression with GluN2D enhances GluN2D ubiquitination and reduces GluN1/GluN2D NMDA receptor responses. These results identify Nedd4 as a novel binding partner for GluN2D and suggest a mechanism for the regulation of NMDA receptors that contains GluN2D subunit through ubiquitination-dependent downregulation.
1. Introduction
NMDA receptors belong to the superfamily of ionotropic glutamate receptors and are well recognized for their essential role in various forms of synaptic plasticity and learning and memory (Watkins and Evans, 1981; Traynelis et al., 2010). NMDA receptor activation is critical for initiating long-term potentiation and long-term depression (Collingridge, 1987; Malenka and Bear, 2004) and experience-dependent synapse formation/elimination during development (Smith et al., 2009), as well as modulating neuroprotection and excitotoxicity (Hardingham, 2006).
NMDA receptors are heterotetrameric complexes of subunits from three classes: GluN1, GluN2 and GluN3 with most NMDA receptors being composed of two GluN1 and two GluN2 subunits (Traynelis et al., 2010). There are four subtypes of GluN2 subunits, GluN2A-GluN2D. GluN2D-containing receptors differ significantly from other NMDA receptors in their deactivation times (Misra et al., 2000), L-glutamate and glycine affinity (Ikeda et al., 1992; Buller et al., 1995), open probability (Traynelis et al., 2010), CNS distribution, and Mg++ sensitivity (Monyer et al., 1994). They are also developmentally down regulated with protein expression levels reduced by approximately 50-60% by the time of synaptogenesis (Wenzel et al., 1996) with an even greater degree of mRNA down-regulation at this time (Watanabe et al., 1992; Wenzel et al., 1996). Hence NMDA receptors containing the GluN2D subunit are likely to have a specific role in NMDA receptor-dependent synapse formation/elimination that occurs during development and have additional roles in the adult brain.
GluN2D-containing NMDA receptors may also have a special role in neuropathological conditions. Tissue plasminogen activator (TPA) – enhanced stroke damage in the cerebral cortex has been found to be dependent specifically upon GluN2D subunits (Baron et al., 2010; Jullienne et al., 2011). GluN2D may also contribute to white matter injury (Micu et al., 2006) and Creutzfeldt-Jakob disease (Khosravani et al., 2008). Thus, it is important to identify systems that selectively regulate GluN2D containing NMDA receptor subpopulations.
In an effort to determine such mechanisms that regulate GluN2D subunits, we used a proteomic approach to identify proteins that associate with the GluN2D intracellular C-terminal tail. Although the four GluN2 sequences display high homology overall, the intracellular GluN2D C-terminal sequence displays very little homology to other glutamate receptor subunits (Ikeda et al., 1992). This sequence of 460 amino acids is highly enriched in proline residues in patterns reflecting a variety of potential protein-protein interaction motifs. These domains are likely to associate with proteins for downstream signaling of NMDA receptor activity as well as to recruit proteins for the regulation of NMDA receptor activity or degradation.
One such regulatory mechanism is the selective ubiquitination of specific proteins by individual members of the large family of E3-ubiquitin ligases (Yi and Ehlers, 2007). This provides a mechanism for targeting selected proteins for intracellular trafficking and degradation by the ubiquitin-proteosomal system (UPS), the lysosome, or the autophagosome (Clague and Urbe, 2010). In this report we identify and characterize the interaction between the E3 ligase Nedd4 (Neural precursor cell-expressed developmentally down-regulated) (Rotin and Kumar, 2009) and the GluN2D subunit. Nedd4 belongs to the HECT domain-containing family of E-3 ubiquitin ligase proteins (Rotin and Kumar, 2009). As the name reflects, Nedd4 was initially identified as a developmentally down-regulated gene from mouse neural precursor cells isolated from the neural tube (Kumar et al., 1992). Nedd4 is a 105kDa protein with an N-terminal C2 domain, three WW domains (WW1, WW2 and WW3) in the middle region and a C-terminal with a ~ 350 amino acid long HECT domain (Fig. 1E) (Kumar et al., 1997).
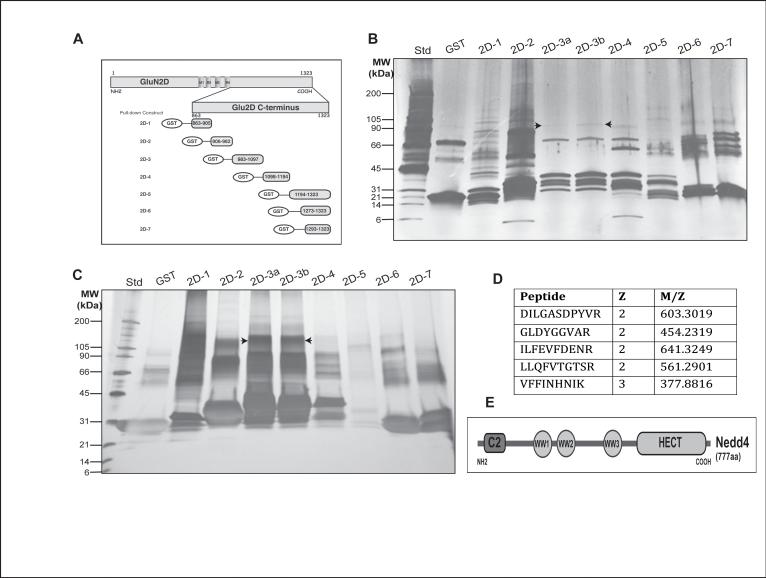
Figure 1.
Identification of Nedd4-1 as a novel GluN2D-interacting protein. Seven different GST-tagged fragments of the GluN2D C-terminal region (A) were used to isolate associating proteins from 10-day-old rat brain. (B and C) Silver-stained SDS-PAGE gels of GluN2D-associated protein complexes. (B) Pull-down was performed in the presence of 1% CHAPS or (C) after the removal of CHAPS by overnight dialysis. (D) Gel bands were cut-out and analyzed by mass spectrometry which identified 5 Nedd4 peptides. (E) Nedd4 modular domains consist of a calcium-sensitive C2 domain, 3 WW domains and a HECT domain.
2. Materials and Methods
2.1. Antibodies and Reagents
Affinity-purified mouse monoclonal antibody against epitope tag HA1.1 was purchased from Covance (Madison, WI). Affinity-purified rabbit antibody against green fluorescent protein (GFP) was purchased from Invitrogen (Carlsbad, CA). Mouse monoclonal and rabbit polyclonal antibodies directed against Myc epitope were purchased from Sigma (St. Louis, MO). A mouse monoclonal antibody against ubiquitin was from BD Bioscience (San Jose, CA). Secondary donkey anti-rabbit horseradish peroxide (HRP) and donkey-anti-mouse HRP were obtained from Jackson ImmunoResearch Laboratories Inc (West Grove, PA). The Gal 4 yeast two-hybrid system was purchased from BD Bioscience (San Jose, CA). The YPD agar was obtained from Bio 101 (La Jolla, CA). Bacto agar and tryptone were from BD Bioscience (San Jose, CA). Glutathione-agarose was acquired from GE healthcare (Piscataway, NJ). The Silver Snap Stain Kit II was from Thermo Fisher Scientific (Rockford, IL). The quick-coupled transcription and translation kit was from Promega (Madison, WI). Protease inhibitor cocktail were obtained from Sigma (St. Louis, MO). MG-132 was from Calbiochem (San Diego, CA) and 35S–methionine was purchased from MP Biomedicals.
2.2. Construction of Recombinant cDNA Constructs
A total of seven overlapping and non-overlapping fragments of GluN2D cytoplasmic region were constructed by PCR amplifying the desired region from GluN2D pBS (kindly provided by Dr. Peter Seeburg, Max Planck Institute, Heidelberg, Germany) and sub-cloned in frame with the coding sequence of GST-tag of pGEX4T-1 vector (obtained from GE Healthcare, Piscataway, NJ). The GluN2D C-terminal fragment constructs were designed to minimize cutting in regions where peptide residues show a greater number of intra-protein contacts (using the CONpro bioinformatics algorithm (Pollastri et al., 2001). In retrospect, this approach prevented cutting within the Nedd4 binding region.
For direct yeast two-hybrid evaluation of protein-protein interactions, the sequence encoding the complete cytoplasmic region of GluN2D (GluN2D-ct; amino acid 841-1302) was amplified by a PCR reaction from full-length GluN2D pBS and sub-cloned into the multiple cloning site (MCS) of pGBKT7 yeast expression vector in frame with GAL4 DNA-binding domain using the restriction enzyme sites NdeI and EcoRI. Full-length sequence of Nedd4 was PCR-amplified from mouse Nedd4 cDNA construct (Magnifico et al., 2003) (plasmid 11426 from Addgene, Boston, MA) and subcloned into the MCS region of pGADT7 GAL4 DNA-activation domain using the restriction enzyme sites NdeI and EcoRI. The Nedd4 construct in pGADT7 yeast vector was used as a template to generate point mutations by using the Quick-change site-directed mutagenesis kit (Stratagene, La Jolla, CA) to generate phenylalanine in place of trytophan in WW domains at position 167 (WW1-W167F), position 323 (WW2-W323F) and position 378 (WW3-W378F). The GluN2D construct in pGBKT7 yeast vector was also used as a template to generate GluN2D C-terminal region carrying mutations in the PPSY motif (PPSY into AASF). Truncated GluN2D C-terminal constructs were generated by inserting stop codons resulting in fragments GluN2D-ctΔ940 and GluN2D-ctΔ1296 using the Quick-change site-directed mutagenesis kit. The yeast two-hybrid vector controls (GAL4-AD SV40 Large T-antigen and GAL4-BD p53) were purchased from BD Bioscience (San Jose, CA).
For in vitro transcription-coupled translation synthesis, various GluN2s C-terminal constructs were made by amplifying the C-terminal nucleotide coding sequence from GluN2A, GluN2B, GluN2C and GluN2D pBS (kindly provided by Dr. Peter Seeburg, Max Planck Institute, Heidelberg, Germany) and subcloning them inframe into the multiple cloning site of pSPUTK vector. For mammalian expression cDNA constructs, GFP-Nedd4 was constructed by amplifying full-length sequence of Nedd4 from Nedd4 cDNA and subcloning it into pEGFP-C1 mammalian expression vector (BD Bioscience) using the restriction sites HindIII and KpnI so that the nucleotide sequence of Nedd4 comes inframe with the GFP nucleotide sequence. HA-tagged ubiquitin in a mammalian vector was obtained from Addgene (Boston, MA). GST-labeled Nedd4 full-length, Nedd4-NH2 terminal, Nedd4 C-terminal, WW1, WW2 and WW3 were made by amplifying the corresponding nucleotide sequence from the Nedd4 cDNA template and subcloning the segment into pGEX4T-1 vector using the restriction enzymes BamHI and EcoRI. All constructs and mutations have been verified at University of Nebraska Medical Center Sequencing facility and are freely available upon request.
2.3. Glutathione S-transferase (GST) Pull-down Assay
Seven different GST-tagged fragments of the GluN2D cytoplasmic region were obtained as purified proteins by first transforming the pGEX4T-1 plasmid GluN2D C-terminal constructs into E.coli BL21 cells (Invitrogen, Carlsbad, CA) and then inducing expression by adding IPTG for 3 hours at 22°C with constant shaking at 300 rpm. Cells were lysed and the GST-tagged fragments were purified by using Glutathione resin. Equal amounts of each of the seven different GST-labeled GluN2D fragments of were used as a bait to pull-down proteins from 10 day-old as well as adult rat brain. In brief, six 10-day old rat pups and 6 adult rats were halothane anaesthetized and their brains quickly removed and submerged into ice-cold lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EGTA and 1% CHAPS detergent) supplemented with protease and phosphates inhibitor cocktails (Sigma). Brains were homogenized by a series of 10 strokes in a Potter-Elvehjem tissue homogenizer followed by 10 seconds of sonication at a setting of 5. After rotating at 4°C 30 minutes, the lysate was cleared from insoluble material by centrifugation at 20,800 × g for 30 minutes. Half of the cleared brain lysate was subjected to detergent removal by performing dialysis at 4°C overnight using the Slide-A-Lyser dialysis cassette (Pierce, Rockford, IL). Equal amounts of brain lysate with and without the presence of CHAPS detergent were independently incubated with the seven GST-tag GluN2D cytoplasmic fragments and the mixture was rotated at 4°C for 2-4 hours. The Glutathione resin was washed with the lysis buffer for 5 times and the bound proteins were eluted with 4M MgCl2.
2.4. One Dimensional SDS Gel-electrophoresis and Silver Staining
In order to remove the presence of high amount of salt from the elution buffer, two methods were followed. In the first method, commercially available Ziptip-C18 (Millipore) was used and the proteins bound to the C18 column were eluted by washing the tip in 2X SDS sample buffer (0.125 M Tris pH 6.8, 4% SDS, 20% glycerol, 0.2 M DTT and 0.02% bromophenol blue). In an alternate procedure, salt was removed by precipitating the eluted proteins using ammonium acetate/10% methanol. The methanol residue was removed and the precipitated proteins were lyophilized to dryness using a SpeedVac concentrator (Thermo Electron, San Jose, CA) then the eluted proteins were again solubilized by using 2X SDS sample buffer. The bound proteins present in the SDS sample buffer were subjected to one-dimensional SDS electrophoresis using the 4-20% gradient Tris-HCl gels (Bio-Rad, Hercules, CA).
Soon after the completion of gel-electrophoresis, silver staining was performed on the gels by using the Silver Snap Stain (Thermo Fisher Scientific, Rockford, IL) according to the manufacture instructions. Silver stained gels were visualized on a light box; the interested bands were excised by a sharp razor blade and subsequently subjected to destaining. Gel slices were kept at 4°C in 10% methanol until further analyzed by mass spectrometry.
2.5. Nano-LC-ESI-Qq-TOF Tandem Mass Spectrometry Analysis
The gel pieces were recovered from the methanol storage solution and resuspended in 25 mM ammonium bicarbonate. The pieces in the mixture were reduced by incubating for 45 minutes at 57°C with 2.1 mM dithiothreitol, in order to reduce the side chains: these side chains were then alkylated with 4.2 mM iodoacetamide for 1 hour in the dark at 21°C. The mixture was digested for 12 hours at 37°C by adding trypsin (12 ng/L), in 25 mM ammonium bicarbonate. The peptides were separated using a 75 μm × 15 cm reverse phase C18 column (LC Packings, Sunnyvale, CA) at a flow rate of 350 nl/min containing 50% acetonitrile and 5% formic acid on an Agilent 1100 series HPLC system equipped with autosampler (Agilent technologies, Palo, Alto, CA). The LC eluent was coupled to a micro-ion spray source attached to a QSTAR Pulsar mass spectrometer (Applied Biosystem, Foster City, CA). Peptides were analyzed in positive ion mode and the MS survey scan were acquired for 1 second, followed by three second MS/MS scans on the two most intense multiply charged ions. A dynamic exclusion window was used to prevent re-sequencing of peptides for sixty seconds.
2.6. Interpretation of MS Spectra
MS/MS spectra were interpreted using Protein Prospector (http://prospector.ucsf.edu), In summary, MS/MS centric peak list were generated using the Macot.d11 script and was searched against the entire Uniprot Muss musculus database. Only proteins with at least one peptide with a Protein Prospector peptide score ≥ 25 and a peptide expectation value of ≤ 0.01 were considered to be positively identified. Protein accession numbers were mapped to its corresponding UniGene entries and if they matched to the same UniGene entry, they were condensed to single protein for identification purpose.
2.7. Direct Yeast Two-hybrid Protein Interaction Assay
In order to test the direct interaction between the GluN2D C-terminal region and full-length Nedd4, a direct yeast two-hybrid assay was performed. In brief, the Saccharomyces cerevisiae strain AH109 was cultured and maintained on YPD agar plates. The two testing cDNAs were transformed into yeast using a lithium acetate procedure as described in the instruction manual for the MATCHMAKER yeast two-hybrid kit (BD Bioscience). The co-transformed yeast were streaked on plates lacking tryptophan and leucine (+His plates) and incubated at 30°C for 3 days. After 3 days, an average of 4-5 colonies were selected, suspended in water and optical density was measured at 600 nm. Equivalent amounts of yeast were then grown on selective plates lacking tryptophan, leucine and histidine (-His plates) as well as on +His plates. The plates were again incubated at 30°C and the growth was analyzed after 3 days. All experiments were replicated 2 to 6 times; the number of experiments is indicated in the corresponding figure legends.
2.8. Cell Culture
Human embryonic kidney 293T (HEK-293T) cell line was obtained from ATCC and cultured in Dulbecco's modified eagle's medium (DMEM, Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Invitrogen) and penicillin/streptomycin (Invitrogen). Cells were maintained in a humidified incubator in 5% CO2 at 37°C. The medium was changed every 2 days and cells were passaged every 4 days. Cells from passages 5-15 were used for all the experimental studies. HEK-293T cells were transfected with a calcium phosphate method when they reached 40-60% confluency in 60 mm plates. For transfection, a maximum of 10 μg of cDNA constructs was mixed with 250 μl mixture of water with 25 μl of 2.5 M CaCl2. This mixture of cDNA, water and CaCl2 was added drop-wise into a separate tube containing 250 μl of 2X HEPES buffered saline solution (pH 7.08) while vortexing. The calcium phosphate precipitate was allowed to form by incubating the transfection mixture for 30 minutes at room temperature and then added to cells. The medium was changed after 6-8 hrs of transfection and cells were harvested after 36-48 hours.
2.9. In vitro Glutathione S-transferase (GST)-[35S] methionine Pull-down Assay
Different forms of GST-tagged Nedd4 were expressed as purified proteins by first transforming E.coli BL21 cells (Invitrogen, Carlsbad, CA) with the corresponding pGEX4T-1 plasmids and then inducing with IPTG for 3 hours at 22°C with constant shaking at 300 rpm. Cells were lysed with the help of sonication and GST-tagged Nedd4 constructs were then purified by using glutathione resin. Equal amounts of different GST-Nedd4 domains or GST alone were used as baits to pull-down [35S]methionine-labeled GluN2 C-terminal regions that were radiolabelled by an in vitro transcription-coupled translation reaction incorporating [35S]methionine according to the manufacturer's protocol (TNT SP6 Coupled Reticulocyte Lysate System, Promega). 10 μl of [35S]methionine-labeled GluN2D C-terminal region was incubated with an equivalent amount of the various GST-Nedd4 constructs in ice-cold buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EGTA and 0.1% Tween-20). After rotating at 4°C for 2 hours, the glutathione resin was washed with the lysis buffer containing 1% NP-40 5 times and the bound proteins were eluted by boiling the samples in 2X SDS loading buffer (0.125 M Tris pH 6.8, 4% SDS, 20% glycerol, 0.2M DTT and 0.02% bromophenol blue) and resolved by SDS-PAGE on a 4-20% gradient gel. Gels were later stained with Coomassie blue, dried and exposed to autoradiographic film.
2.10. Immunoprecipitation and Immunoblotting
For immunoprecipitation experiments using HEK-293T cells, cells were washed twice with ice-cold PBS and harvested in buffer (50 mM Tris-HCl, 150 mM NaCl and 1% NP-40 detergent, pH 7.4) containing protease and phosphatase inhibitors cocktail (Mammalian Protease and Phosphatase Inhibitor cocktail, Sigma) Cells were lysed, briefly sonicated and rotated for 30 minutes at 4°C. The lysate was cleared of insoluble material by centrifugation at 20,800 × g for 30 minutes. Protein concentration of the lysate was determined by Bradford assay (Bio-Rad), and 1 mg protein of the lysate was used for immunoprecipitation. Lysates were then incubated with 5 μg of affinity-purified anti-mouse Myc antibody or rabbit anti-GFP antibody overnight at 4°C followed by 40 μl of Protein A/G agarose (GE Healthcare) for 2 hour at 4°C. Before using, the Protein A/G agarose was washed and blocked with 2% bovine serum albumin (w/v) (BSA, Sigma) so as to block the nonspecific interaction of protein with the beads. Finally, Protein A/G agarose was washed for 3 times in ice-cold lysis buffer without BSA using low speed centrifugation (1000 × g, 5 minutes) at 4°C and the proteins were denatured and eluted by boiling the samples in SDS-PAGE loading buffer. Proteins were separated on 4-20% gradient SDS-PAGE gels, transferred to PVDF membrane and blocked in PBS containing 0.05% Tween-20 (w/v) and 1% (w/v) polyvinylpyrolidone (PVP) overnight at 4°C. The next day, membranes were incubated with primary antibody in blocking solution for 90 minutes, followed by 3 washes with PBS-T (0.1% Tween-20, w/v). HRP-conjugated secondary antibody (1: 25000 dilution) was then added to the membrane for 1 hour and the immunoblot was developed by enhanced chemiluminescence (GE Healthcare).
2.11. Ubiquitination Assay
To determine ubiquitination, HEK-293T cells were washed twice with ice-cold PBS and harvested in buffer (50 mM Tris-HCl, 150 mM NaCl and 1% NP-40 detergent and 1% SDS, pH 7.4) containing a cocktail of protease and phosphatase inhibitors. Cells were lysed, briefly sonicated and rotated for 30 minutes at 4°C. The lysate was cleared of insoluble material by centrifugation at 20,800 × g for 30 minutes. For each ubiquitination assay, 1mg of lysate protein was diluted 10-fold in lysis buffer without SDS so that that the final concentration of SDS was 0.1 %. Lysates were then incubated with 5 μg of affinity-purified anti-mouse Myc antibody overnight at 4°C followed by 40 μl of Protein A/G agarose (GE healthcare) for 2 hour at 4°C. Before using, the Protein A/G agarose was washed and blocked with 2% bovine serum albumin (w/v) (BSA, Sigma) so as to block the nonspecific interaction of protein with the beads. Finally, Protein A/G agarose was washed 5 times in ice-cold lysis buffer containing 0.1 % SDS using low speed centrifugation (1000 × g, 5 minutes) at 4°C and the proteins were denatured and eluted out by boiling the samples in 2 × SDS loading buffer. Proteins were separated on 4-12% gradient SDS-PAGE gels, transferred to PVDF membrane and blocked in PBS containing 0.05% Tween-20 (w/v) and 1% (w/v) polyvinylpyrolidone (PVP) overnight at 4°C. The next day, membranes were incubated with primary antibody in blocking solution for 90 minutes, followed by 3 washes with PBS-T (0.1% Tween-20, w/v) at room temperature. HRP-conjugated donkey anti-mouse secondary antibody (1:25000) was added to the membrane and incubated at room temperature for 1 hour. Membrane was then washed with PBS-T (0.1% Tween-20, w/v) and the immunoblot was later developed with enhanced chemiluminescence (GE Healthcare).
2.12. Oocyte Electrophysiology
Oocytes were removed and isolated from mature female Xenopus laevis (Xenopus One, Ann Arbor, MI, USA) according to the procedures approved by the University of Nebraska Medical Center's Institutional Animal Care and Use Committee in compliance with National Institute of Health guidelines. GluN1a, GluN2D, Nedd4 and Nedd4-C744E RNAs were prepared using mMessage mMachine (Ambion/Invitrogen). A total of 50 nl of the RNA mixture containing constant amount of GluN1a and GluN2D with or without Nedd4 or catalytically inactive mutant Nedd4-C744E were injected into the oocyte cytoplasm. Oocytes were then incubated in ND-96 solution at 17°C prior to electrophysiological assay. Electrophysiological responses were measured using a standard two-microelectrode voltage clamp (model OC-725B; Warner Instruments, Hamden, CT). Buffer containing 116 mM NaCl, 2 mM KCl, 0.3 mM BaCl2, and 5 mM HEPES, pH 7.4 was used for the recording. Response magnitude was determined by the steady plateau response elicited by bath application of 10 μM L-glutamate plus 10 μM glycine and held at a membrane potential of -60 mV. Responses were digitized for quantification (Digidata 1440A and pClamp-10; Molecular Devices, Sunnyvale, CA).
3. Results
3.1. Identification of Nedd4 as a GluN2D-interacting protein
To identify potential GluN2D-ubiquitin ligases, we performed a mass spectrometry-based identification of proteins that associate with GST-GluN2D C-terminal region fragments. Seven GST-GluN2D C-terminal fragments were used as baits to pull-down interacting proteins from 10-day old rat brain lysate (Fig. 1A). The associating proteins were separated on an SDS-PAGE gel and silver-stained. Individual gel bands were excised, trypsin digested, and subjected to LC-MS/MS-based identification. As shown in Fig. 1B and 1C, the different GST-constructs pulled-down distinct sets of proteins, revealing many interacting proteins. Most of these bands were not pulled-down with GST alone and were specific to the fragment being used, signifying the specific association of proteins with each GluN2D C-terminal fragment. Two different pull-downs with fragment 3 show the same protein pattern.
Among the >100 proteins identified only one E3 ubiquitin ligase was identified, Nedd4 (neural precursor cell-expressed developmentally down-regulated, UniProt accession P46935). This protein was identified in pull-down experiments in either the presence or absence of CHAPS and from a total of 5 different Nedd4 peptides (Fig. 1D). In each case, the Nedd4 peptide was isolated only by GluN2D-ct fragment #3. As Nedd4 did not associate with the GST alone construct, or with the other GluN2D C-terminal constructs, Nedd4 appears to specifically associate with fragment #3's GluN2D -amino acids 983-1097.
3.2. GluN2D mediates direct interaction with Nedd4 through its C-terminal region
In order to test whether the GluN2D C-terminal region could potentially mediate a direct association with Nedd4, we performed a direct yeast two-hybrid analysis using the C-terminal region of GluN2D and the full-length coding region of Nedd4. As shown in Figure 2A, yeast growth was observed after streaking the nonselective plate (+His), suggesting the successful cotransformation of both the GluN2D C-terminal region and the full-length Nedd4. After transferring the cotransformed yeast to selective plates (-His), growth still persisted, suggesting a direct interaction between the GluN2D C-terminal region and full-length Nedd4. Control yeast cotransformed with either GluN2D C-terminus and SV40 T antigen or Nedd4 and p53 did not show any growth on selective plates (negative control) whereas cells cotransformed with P53 and SV40 grew on the (-His) plates (positive control).
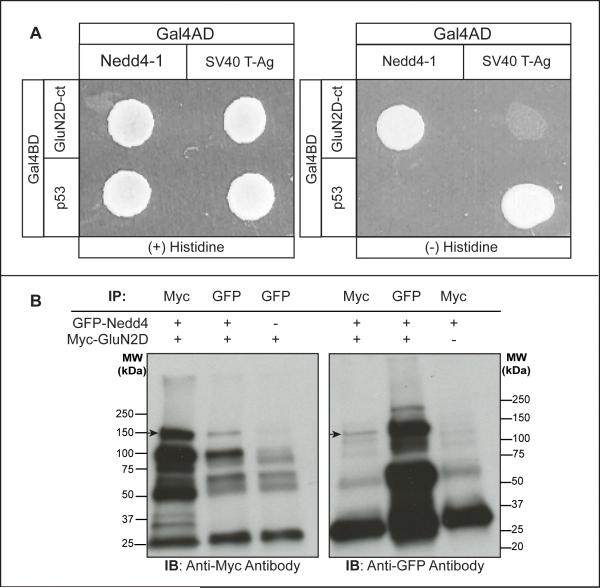
Figure 2.
GluN2D interaction with ubiquitin ligase Nedd4-1. (A) The yeast strain Saccharomyces cerevisae was cotransformed with the Gal4 activation domain fusion constructs of Nedd4-1 or SV40 T Ag (control) together with the Gal4 binding domain fusion constructs of GluN2D C-terminal region or p53 (control). The cotransformed yeast was assayed for its growth on nonselective (+ histidine; left) and selective (histidine; right) media. Nedd4-1 and GluN2D C-terminal constructs required coexpression to allow yeast growth on selective media as did the p53/SV40 T-Ag control (n = 3).
(B) Western blots of full-length GluN2D and Nedd4 immunoprecipitates. Myc-GluN2D and GFP-Nedd4 were co-transfected, or transfected alone, in HEK-293T cells and immunoprecipitated by anti-myc or anti-GFP antibodies as indicated and subjected to Western blot with anti-myc or anti-GFP antibodies. Arrows indicate the corresponding bands at the predicted molecular weight for GluN2D (left panel) and Nedd4 (right panel). Anti-myc antibodies could immunoprecipitate GFP-Nedd4 only if coexpressed with myc-GluN2D and anti-GFP antibodies could immunoprecipitate myc-GluN2D only if coexpressed with GFP-Nedd4 (n = 2).
3.3. NMDA receptor subunit GluN2D and Nedd4 associate in vivo
The above studies show that Nedd4 can directly associate with the GluN2D C-terminal region. To determine if the full-length functional GluN2D subunit interacts and forms a complex with Nedd4 in vivo, we performed co-immunoprecipitation utilizing the HEK-293T cell line. An NH2-terminal Myc-tagged GluN2D was cotransformed with NH2-terminal GFP-tagged Nedd4 in HEK293T cells and coimmunoprecipitation was performed using tag-specific antibodies. After immunoprecipitating Myc-GluN2D and immunoblotting with a rabbit anti-GFP tag antibody, a major band of 130kDa was detected, suggesting that GFP-Nedd4 associated with Myc-GluN2D and was pulled-down with Myc-GluN2D in HEK-293T cells (Fig. 2B). Conversely, Myc-GluN2D was detected as a major band of 150kDa following GFP-Nedd4 immunoprecipitation and anti-myc immunoblotting. The association of GluN2D with Nedd4 was specific, as MycGluN2D was not pulled-down by anti-GFP antibodies when using cells only expressing Myc-GluN2D. Likewise, GFP-Nedd4 was not pulled down by anti-Myc antibodies from cells only expressing GFP-Nedd4. Specific immunoprecipitation of GluN2D and Nedd4 by either protein suggests that GluN2D and Nedd4 associate with each other in a complex in vivo.
3.4. Nedd4 interacts with the C-terminal region of GluN2D through its WW domains
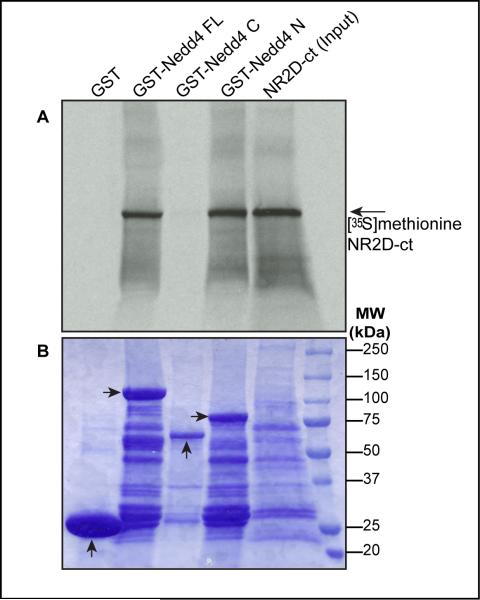
Processed Nedd4 is 777 amino acids long and contains an N-terminal C2 domain, three WW domains in the middle and a C-terminal HECT domain (Fig. 1E). To further define the molecular region of Nedd4 that is essential for its association with the C-terminal region of GluN2D, we developed a radioactive in vitro protein interaction assay using GST-tagged recombinant purified Nedd4 protein and [35S]methionine-labeled GluN2D C-terminal. The full-length coding region of Nedd4 was used as a template to generate GST fusion protein constructs containing the Nedd4 C2 and WW domains (amino acids 52-422), a C-terminal construct containing the Nedd4 HECT domain (amino acids 422-777), and a full-length GST-tagged Nedd4. These constructs, and GST-tag alone, were tested for their ability to pull-down [35S]methionine-labeled C-terminal GluN2D. As shown in Fig. 3, the [35S]methionine-labeled GluN2D C-terminal region was successfully pulled-down by GST-tagged full-length purified Nedd4 protein, further corroborating our previous results that Nedd4 directly associates with the C-terminal region of GluN2D. The GluN2D C-terminal was also specifically pulled-down by the GST-Nedd4 NH2-terminal fragment but not with the GST-Nedd4 C-terminal region. GST alone did not show any association with [35S]methionine-labeled GluN2D (negative control). As the NH2-terminal region of Nedd4 possesses three WW domains, which are small modular protein-protein interaction domains, Nedd4's specific association with the [35S]methionine-labeled C-terminal of GluN2D indicates the potential role of WW domains of Nedd4 in mediating its direct interaction with GluN2D.
Figure 3.
The Nedd4 N-terminal is required for Nedd4 interaction with the C-terminal region of GluN2D. (A) Autoradiograph of GST-Nedd4 pull-downs of [35S]methionine-labeled GluN2D C-terminal protein using GST-Nedd4 constructs containing either the full-length Nedd4 (GST-Nedd4 FL), the C-terminal (GST-Nedd4 C, containing the HECT domain) or the N-terminal (GST-Nedd4 N, containing the C2 domain and 3 WW domains). Following pull-down, proteins were separated by SDS PAGE, stained with Coomassie blue (B), and the dried gel subjected to autoradiography. Arrows in bottom panel indicate the respective GST-constructs (n = 2).
3.5. Both WW2 and WW3 domains of Nedd4 can associate with the C-terminal region of GluN2D
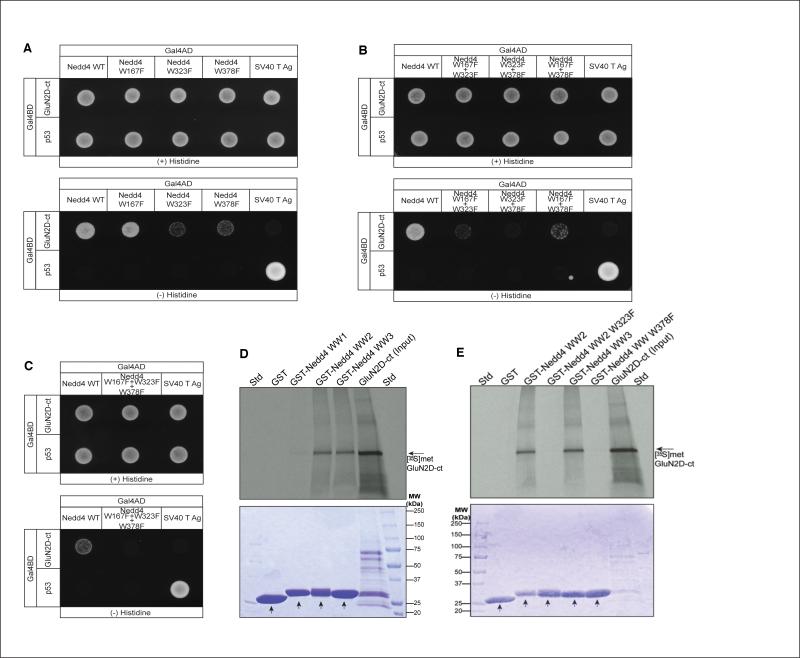
To determine which Nedd4 WW domain might be interacting with GluN2D, we utilized a direct yeast-two hybrid assay in addition to an in vitro GST pull-down approach. In constructs used in the direct yeast two-hybrid assay, we mutated the second conserved tryptophan residue present in one of the three WW domains into a phenylalanine residue, which should eliminate that WW domain's ability to bind (Lu et al., 1999) while leaving the other two WW domains intact. Yeast cotransformed separately with each of the three individually-mutated WW domains and the GluN2D C-terminal showed strong growth only in the case of mutated WW1 (W167F) (Fig. 4A bottom), while both WW2 (W323F) and WW3 (W378F) mutants of Nedd4 showed reduced growth on selective (-His) plates (Fig. 4A bottom). This suggests that both WW2 and WW3 domains of Nedd4 are required for optimal interaction with the C-terminal of GluN2D.
Figure 4.
WW2 and WW3 domains of Nedd4 are essential in mediating Nedd4 association with the C-terminal region of GluN2D. (A) Yeast were cotransformed with the Gal4 activation domain fusion constructs of wild-type as well as full length Nedd4 with and without single point mutants: Nedd4-wt, Nedd4-WW1-W167F, Nedd4-WW2-W323F, Nedd4-WW3-W378F and SV40 TAg (positive control with p53) together with the Gal4 binding domain fusion construct of the GluN2D C-terminal region or p53. Cotransformed yeast were assayed for growth on the selective (-histidine) and nonselective (+histidine) media. Yeast cotransformed with GluN2D C-terminal region and Nedd4 carrying the point mutation in WW2 domain (W323F), or with the WW3 domain mutation (W378F), displayed very weak growth on the selective (-histidine) media as compared to the yeast cotransformed with GluN2D C terminal region and Nedd4 WW1 W167F mutant or wild-type Nedd4. (B) Yeast two-hybrid analysis of GluN2D C-terminal interactions with full length Nedd4 constructs with two mutated WW domains or (C) the all three WW domains mutated. Yeast cotransformed with GluN2D C-terminal region and Nedd4 carrying double point mutation in both WW2 and WW3 domain (W323F + W378F) or the Nedd4 triple WW domain mutant failed to grow on the selective (-histidine) media. Barely detectable growth was observed with cells expressing the WW1 + WW2 or WW1 + WW3 mutations. (D, E) GST-tagged purified proteins of Nedd4 were used to pull down [35S]methionine-labeled GluN2D C-terminal protein which was then separated by SDS PAGE and stained with Coomassie blue (bottom panels, loading controls, arrows indicate the GST construct). The gel was then dried and exposed to autoradiographic film (top panels). In (D), individual Nedd4 WW domains are tested for their abililty to pull-down radiolabelled GluN2D C-terminal. In (E), Nedd4's WW2 and WW3 with and without point mutations are tested for their ability to pull-down radiolablelled GluN2D C-terminal (n = 2).
To further evaluate the role of WW2 and WW3 separately in mediating Nedd4 association with GluN2D, we cotransformed the yeast with GluN2D and Nedd4 carrying the tryptophan mutation in two of its WW domains, leaving only one WW intact (Fig. 4B). Yeast cotransformed with GluN2D and Nedd4 WW2 and WW3 double mutant (W323F and W378F) did not show growth on the selective plates. However, weak growth of yeast was observed in the case of WW1-WW3 (W167F and W378F) and WW1-WW2 (W167F-W323F) double mutants with more growth in the case of the WW1-WW3 double mutant, suggesting that the WW2 domain might display higher affinity toward GluN2D C-terminal region than WW3 (Fig. 4B bottom). As expected, mutating all three WW domains also eliminated growth (Fig. 4C bottom). Thus, both WW2 and WW3 domains of Nedd4 can associate with the C-terminal region of GluN2D but with different affinities. Moreover, Nedd4's WW1 does not interact with GluN2D.
The importance of individual WW domains of Nedd4 in mediating its interaction with GluN2D was also evaluated by the GST pull-down approach (Figs. 4D and 4E). Purified GST-tagged recombinant proteins of each of the three WW domains were produced and used as baits to pull-down the [35S]methionine-labeled GluN2D C-terminal region. As shown in Figure 4D both GST-WW2 and GST-WW3 when tested alone were able to pull-down [35S]methionine-labeled GluN2D. In contrast, neither GST-WW1 nor the GST-tag itself was able to pull down the GluN2D construct. Mutating the second conserved tryptophan residue into phenyalanine in both WW2 and WW3 domains, completely abolished the interaction of these domains with the [35S]methionine-labeled C-terminal region of GluN2D (Fig. 4E). These results further validate that both WW2 and WW3 domains of Nedd4 interact specifically with the NMDA receptor subunit GluN2D.
3.6. The direct physical association of GluN2D with Nedd4 is mediated by the presence of a PPXY motif in the GluN2D C-terminal region
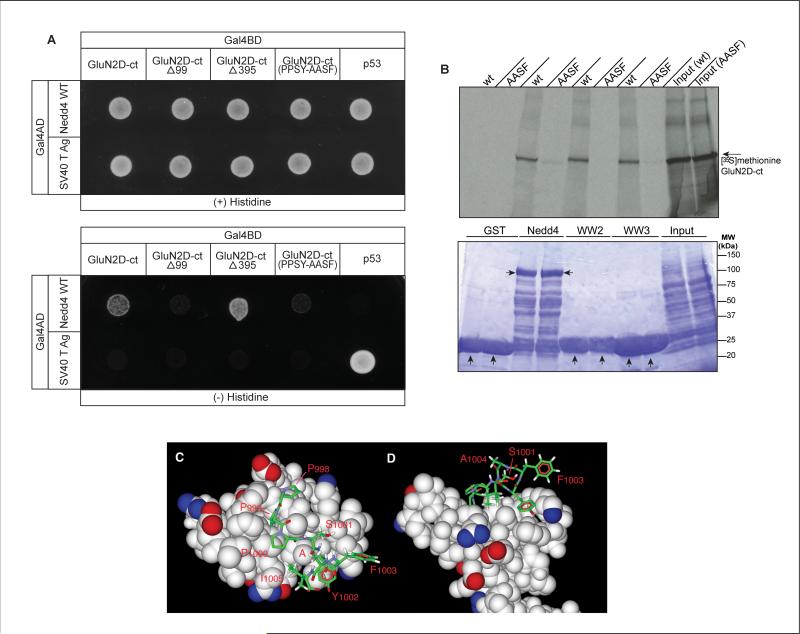
In the mass spectrometry-based identification of GluN2D-associated proteins, the multiple peptides of Nedd4 were only found associated with the third fragment of the GluN2D C-terminal. This suggests that GluN2D harbors a Nedd4-interacting motif in this 115 amino acid segment. Consistent with this region being the site of Nedd4 interaction, we found that truncating the GluN2D C-terminal after 99 amino acids eliminated the vast majority of the Nedd4 interaction but truncation after 395 amino acids did not reduce Nedd4 association in the yeast two-hybrid system (Fig. 5A).
Figure 5.
The PPSY motif of GluN2D interacts with Nedd4. (A) Yeast two-hybrid analysis of truncated and mutated GluN2D C-terminal constructs binding with Nedd4. Nedd4 or the SV40 T-antigen were cotransformed with GluN2D C-terminal constructs with the full length GluN2D C-terminal (GluN2D-ct), the GluN2D C-terminal truncated after 99 amino acids (GluN2D-ctΔ99), or truncated after 395 amino acids (GluN2D-ct Δ395), or the full length GluN2D C-terminal with the triple point mutation of the PPSY motif converted to AASF or p53 (control). Deletion of Nedd4 binding motif PPSY (GluN2D-ctΔ99) or mutation of PPSY motif eliminates GluN2D interaction with the Nedd4. (B) Top: Autoradiograph of [35S]methionine-labeled GluN2D C-terminal protein after pull-down by purified GST-Nedd4, GST-Nedd4 WW2, and GST-Nedd4 WW3. Mutation of the GluN2D's PPSY motif to AASF (PPSY-AASF) prevented pull-down by Nedd4, Nedd4-WW2, and Nedd4-WW3. Bottom panel shows Nedd4 loading. (C, D) -Space-filled model of the Nedd4 WW3 domain structure (1I5H) bound to a stick representation of the PPPSYFAI sequence of GluN2D based upon a homology model of the ENaC peptide PPPNYDSL bound to rat Nedd4 (Kanelis et al., 2001). Images in C are rotated 90° horizontally relative to D; non-conserved amino acid residues S1001, F1003, and A1004 (“A”) have their side-chains directed away from the Nedd4 interface, I1005 can occupy a hydrophobic pocket occupied by a leucine in ENaC. A red star indicates the C-terminal end of the peptide. (n = 3).
The GluN2D C-terminal fragment #3 that pulled-down Nedd4 has a PPXY motif which has been shown to interact with Nedd4 in the case of several proteins such as Notch (Sakata et al., 2004), epithelial Na+ channel (ENAC) (Staub et al., 1996; Fotia et al., 2004), and ErbB4 (Zeng et al., 2009). To test if this PPXY sequence in GluN2D is mediating GluN2D association with Nedd4, we co-transformed yeast with two-hybrid constructs containing the GluN2D C-terminal region carrying a mutated PPXY motif (AASF) and full-length Nedd4 (Fig. 5A). Yeast growth was greatly diminished with the GluN2D construct carrying the mutated motif AAXF, suggesting that the C-terminal region of GluN2D associates with Nedd4 most strongly through its PPXY motif. A similar result was found using the [35S]methionine-labeled GluN2D C-terminal pull-down assay (Fig. 5B). GluN2D association with Nedd4 was greatly reduced by mutating the PPXY motif. Furthermore, the mutation of GluN2D's PPXY motif blocked the binding to both Nedd4's WW2 and WW3 tested individually (Fig. 5B).
Homology modeling of the Nedd4-GluN2D c-terminal interaction using the ENaC peptide interaction with the Nedd4 WW3 domain (Kanelis et al., 2001) is shown in Figs. 5 C and D. In the PPXY binding motif, the GluN2D sequence is PPSY with the serine-1001 side-chain pointing away from the binding interface, consistent with the observation that this residue can be variable. Modeling also suggests that the conserved proline preceeding the PPXY sequence and the side-chain of isoleucine-1005 (substituting for leucine in ENaC) also can weakly contribute to binding near the PPXY binding hydrophobic pocket while side-chains of phenylalanine 1003 and alanine 1004 do not interact with Nedd4.
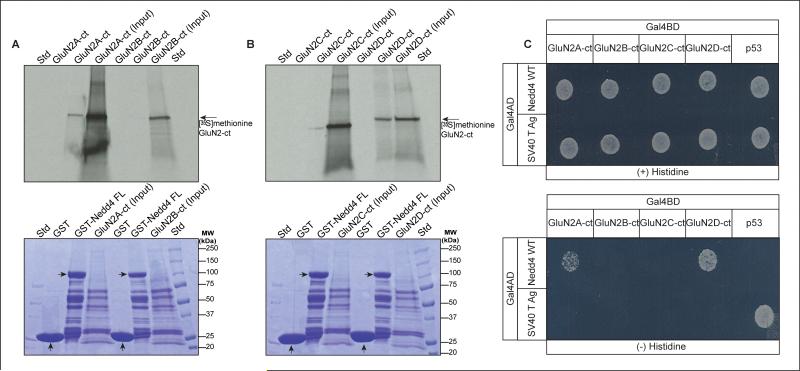
3.7. Nedd4 interacts preferentially with the NMDA receptor GluN2D subunit
To determine the specificity of the Nedd4 interaction among the GluN2 family of subunits, we tested the ability of GST-Nedd4 recombinant protein to pull-down C-terminal domains corresponding to each member of the GluN2 family. As shown in Fig. 6, Nedd4 was selective for the C-terminal constructs of both GluN2A and GluN2D, however the association with GluN2D appeared to be much stronger (Fig. 6B). There was no detectable association with GluN2B (Fig. 6A). There appeared to be weak GluN2C - Nedd4 association seen in the experiment shown in Fig. 6B, but this was not consistent among experiments and not confirmed by the yeast two-hybrid assay (Fig. 6C).
Figure 6.
Nedd4 selectivity among GluN2 receptor subunits. (A, B) Top panel: Autoradiographs of [35S]methionine-labeled GluN2 C-terminal protein after pull-down by purified GST or GST-Nedd4 (see bottom panel for GST or GST-Nedd4 label) and resolving on SDS-PAGE. The bottom panel shows the Coomassie blue staining of the gels used in the top panel, and the expression and loading of the GST or GST-Nedd4 constructs (arrows).
(C) Yeast two-hybrid analysis of Nedd4 binding with the GluN2 C-termini. Yeast were cotransformed with Gal4 activation domain (Gal4AD) and Gal4 binding domain (Gal4BD) fusion constructs as indicated and grown on non-selective (-histidine) and selective (-histidine) media. Yeast cotransformed with GluN2D C-terminal region and Nedd4 shows significant growth on selective media (-histidine) while there was negligible growth when the GluN2B and GluN2C C-terminal regions were cotransformed with Nedd4. Yeast expressing Nedd4 with the GluN2A construct had low levels of growth (n = 3).
3.8. Nedd4 enhances ubiquitination-dependent modification of GluN2D
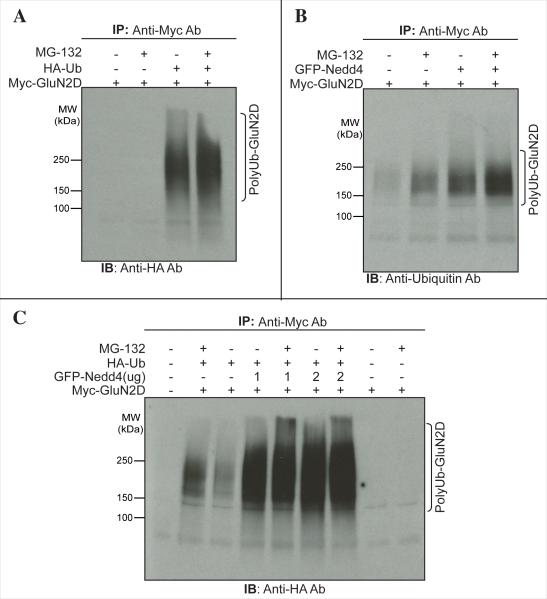
Nedd4 is a member of the HECT domain-containing E-3 ubiquitin ligases that catalyze ATP-dependent transfer of ubiquitin polypeptide from E-2 conjugating enzymes to a lysine residue in the target protein. The process of ubiquitination generally results in protein degradation by either the proteosome, the lysosome, or the autophagosome (Clague and Urbe, 2010). Studies have also shown that ubiquitination is sometimes also necessary for protein trafficking and sorting to different subcellular compartments (Yi and Ehlers, 2007). The C-terminal region of GluN2D possesses multiple lysine residues, but whether it undergoes ubiquitination-dependent post-translational modification has not been previously demonstrated. We sought to determine whether, in an in vivo system, the GluN2D subunit is subject to ubiquitination. The ubiquitination assay was performed using HEK-293T cells expressing Myc-tagged GluN2D along with GluN1 and with or without the HA-tagged ubiquitin and in the presence or absence of the proteosomal inhibitor MG-132. GluN2D was immunoprecipitated and the resulting blot was probed with anti-HA tag antibody. As shown in Figure 7A, a broad smear in the PAGE gel was observed after coexpression with HA-ubiquitin, consistent with polyubiquitination of GluN2D. Inhibition of the proteosome with MG-132 caused a modest enhancement of GluN2D ubiquitination. Thus, GluN2D expressed in HEK-293T cells undergoes ubiquitination. To test if Nedd4 can cause GluN2D ubiquitination, we expressed Myc-GluN2D along with GluN1 (to promote complex assembly and cell surface localization) with or without GFP-tagged Nedd4 and determined the presence of endogenous ubiquitination of the Myc-GluN2D immunoprecipitates. Staining against ubiquitin showed that GluN2D appeared in a smear-like pattern (Fig. 7B), consistent with polyubiquitination. Coexpression of Myc-GluN2D with GFP-Nedd4 greatly enhanced the level of endogenous ubiquitin in the GluN2D immunoprecipitate, suggesting that Nedd4 promotes GluN2D polyubiquitination. Likewise, coexpression of Nedd4 along with HA-tagged ubiquitin enhanced GluN2D polyubiquitination (Fig. 7C).
Figure 7.
The NMDA receptor subunit GluN2D undergoes Nedd4-dependent polyubiquitination. (A) Full-length Myc-tagged GluN2D was cotransfected with HA-tagged ubiquitin (HA-Ub) in HEK-293T cells as indicated. The cells were treated with or without 10 μM proteosomal inhibitor MG-132 for 12 hours and then analyzed for ubiquitination by anti-HA tag Western blotting of anti-myc (GluN2D) immunoprecipitates. Polyubiquitination of GluN2D was detected in a smear-like pattern. (B, C) Myc-tagged GluN2D was cotransfected with GFP-Nedd4 and/or HA-tagged ubiquitin (HA-Ub) in HEK-293T cells and treated with our without MG-132 as indicated. GluN2D was then analyzed for ubiquitination by anti-ubiquitin (B) or anti-HA tag (C) Western blotting of anti-myc (GluN2D) immunoprecipitates. Increased polyubiquitination of GluN2D was detected in the presence of GFP-Nedd4. MG-132 increased both basal and Nedd4enhanced ubiquitination (n = 3).
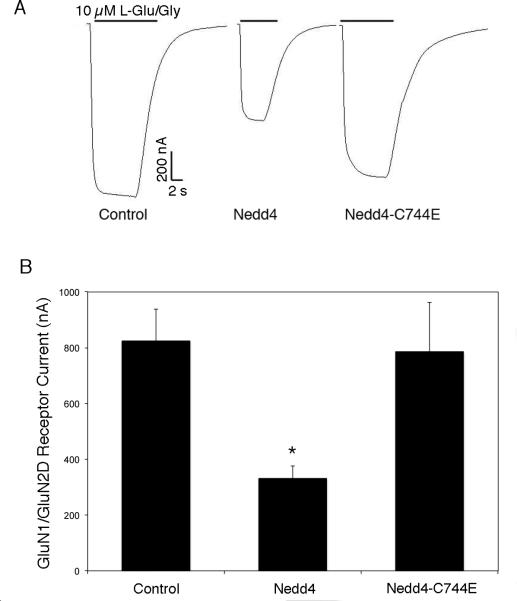
Since polyubiquitination is generally associated with the subsequent internalization or degradation of target protein, it is likely that Nedd4-mediated ubiquitination of GluN2D results in a reduction in the levels of cell surface GluN2D subunits. A frequently used system for studying the effects of Nedd4 on cell surface proteins is the Xenopus oocyte expression system (Kamynina et al., 2001; Fotia et al., 2004; Ekberg et al., 2007). Accordingly, we co-injected cRNA for GluN1 and GluN2D with and without Nedd4 cRNA into Xenopus oocytes, and then performed two-electrode voltage clamp to determine the effect of Nedd4 coexpression on GluN1/GluN2D receptor function (Fig. 8). GluN1/GluN2D receptor responses evoked by 10 μM L-glutamate plus 10 μM glycine were obtained 4 to 6 times for each cell and averaged for a single observation. In the absence of Nedd4 (control), GluN1/GluN2D receptor responses were 826 ± 112 nA (n = 6 cells). Co-expression of GluN1/GluN2D with Nedd4 significantly reduced NMDA receptor response to 333 ± 43 nA (p < 0.05, Student's t-test, one- and two-tailed tests, n = 6). However, GluN1/GluN2D receptor responses in cells co-expressing an equivalent amount of catalytically-inactive Nedd4 (C744E) were not significantly different from control (786 ± 177 nA; n = 6), but were significantly different (p < 0.05) from cells expressing Nedd4.
Figure 8.
Down-regulation of functional, cell surface GluN1/GluN2D receptors expressed in Xenopus oocytes after co-expression of active, but not inactive, Nedd4. (A) Representative current traces recorded by two-electrode voltage clamp of GluN1/GluN2D receptor responses evoked by 10 μM L-glutamate plus 10 μM glycine. NMDA receptors were expressed alone (Control) or with wild type Nedd4 (Nedd4) or with catalytically inactive Nedd4 (Nedd4-C744E). (B) Average GluN1/GluN2D receptor responses current responses (± s.e.m.) in the absence (Control) or presence of active Nedd4 or catalytically inactive Nedd4, n = 6 for each condition. *The average response size in cells containing Nedd4 was statistically different from both the Control and Nedd4-C744E conditions (p < 0.05).
4. Discussion
NMDA receptors containing the GluN2D subunit have quite distinct physiological properties relative to other NMDA receptors. Thus, there are likely to be mechanisms that differentially regulate GluN2D activity and location. The intracellular C-terminal tail of GluN2D contains a proline-rich sequence with several potential protein-protein interaction motifs for small modular domains, such as SH3, WW and EVH. To identify GluN2D binding partners that may regulate GluN2D subunit levels and function, we performed a proteomic characterization of rat brain proteins that can interact with the GluN2D C-terminal, identifying Nedd4 as a GluN2D-interacting protein. Of seven different GluN2D C-terminal pull-down constructs used, only the 115 amino acid fragment corresponding to amino acids 983-1097 associated with Nedd4. The specific sites of Nedd4-GluN2D association were further characterized by three distinct approaches, yeast two-hybrid, in vitro labeled 35S[Met] protein pull-down and in vivo expression followed by co-immunoprecipitation. These studies confirm that full-length GluN2D can associate with Nedd4 in both purified protein preparations or in a cellular, in vivo context. Furthermore, we demonstrate a specific association between the GluN2D PPXY motif and both WW2 and WW3 domains of Nedd4. This association was specific for the GluN2D subunit; no Nedd4 interaction was seen with GluN2B or Glu2C subunits and only a very weak interaction was found for GluN2A subunits. The weak association of GluN2A with Nedd4 may be due to the amino acid sequence SPRY in the GluN2A C-terminal. While this sequence is not the canonical PPXY motif, the WW domains of Nedd4 can associate with a LPXY sequence in the protein Commissureless (Myat et al., 2002).
GluN2D can bind through its PPXY motif to either the WW2 or WW3 domain of Nedd4. A similar redundant interaction is seen with the Saccharomyces cerevisiae protein Rsp5p, a Nedd4 homologue. As seen for the Nedd4/GluN2D interaction, both the 2nd and 3rd WW domains of Rsp5p, and not the WW1 domain, bind the LPKY motif of Mga2p as well as that of Spt23p (Bhattacharya et al., 2008). Rsp5p can also bind simultaneously to both Bsd2 and Tre1 through Rsp5's WW2 and WW3 domains, respectively. In this case, the catalytic HECT domain is positioned such that it ubiquitinates a third protein, Smf1 (Sullivan et al., 2007). Since, there are two GluN2D subunits in the tetrameric GluN1/GluN2D receptor complex, it is possible that one molecule of Nedd4 can bind two GluN2D subunits providing high affinity and spatially restricting the catalytic HECT domain. If both WW2 and WW3 can bind simultaneously in the tetrameric complex, then one might expect Nedd4 to preferentially associate with NMDA receptors containing two GluN2D subunits (GluN1/GluN2D) over receptors with a single GluN2D subunit such as GluN1/GluN2B/GluN2D.
For Nedd4 substrate proteins, such as Notch (Sakata et al., 2004), epithelial Na+ channel (ENAC) (Staub et al., 1996; Fotia et al., 2003), ErbB4 (Zeng et al., 2009), the association of the WW domains of Nedd4 to the target protein's PPXY motif leads to ubiquitination and down-regulation. Consistent with these findings, co-expression of Nedd4 with GluN2D increased the ubiquitination levels of GluN2D and significantly reduced the magnitude of GluN1/GluN2D receptor-mediated currents. This suggests that Nedd4 association reduces GluN2D function, mostly likely by promoting GluN2D ubiquitination and internalization. This is consistent with the observation that coexpression of catalytically-inactive Nedd4 did not reduce GluN1/GluN2D receptor activity.
Relatively little is known about the functional role of Nedd4 in neuronal systems. Nedd4 expression is reduced in the brain after the second postnatal week (Kumar et al., 1997). The diminishing levels of Nedd4 expression during synaptogenesis correlates with GluN2D expression in brain and suggests that these two proteins may play a role in the developmental processes regulated by NMDA receptors during or preceding synaptogenesis. Recently, Nedd4 has been shown to ubiquitinate and down-regulate the GluA1 AMPA receptor subunit (Schwarz et al., 2010; Lin et al., 2011) as well as the glutamate transporters EAAT3 and EAAT4 (Yang et al., 2008). Hence, Nedd4 activation has multiple targets in neurons and synapses where it is localized in postsynaptic spines and dendrites (Lin et al., 2011).
Since Nedd4 is widely expressed in neurons of the CNS (Allen Brain Atlas, Kumar et al., 1997), especially during development when GluN2D expression is higher, it is highly likely that these two proteins are co-expressed in many cells. Thus, given the robust and specific, dual-interaction shown by these proteins, activated Nedd4 probably ubiquitinates GluN2D subunits. Nevertheless, it will be important to characterize these interactions in neuronal cells. Such an association of Nedd4 with GluN2D can account for the observation that GluN2D mRNA levels decrease to a greater extent than corresponding decreases in protein levels during the first few weeks postnatally (Wenzel et al., 1996). If Nedd4 is a major contributor to GluN2D subunit degradation prior to synaptogenesis, then the parallel reduction in Nedd4 and GluN2D mRNA levels in the second week postnatally would increase GluN2D protein turnover time resulting in less down-regulation of GluN2D protein than GluN2D mRNA.
The specific association of Nedd4 with GluN2D also provides a potential mechanism for activity-dependent down regulation of GluN2D-containing NMDA receptors. The C2 domain of Nedd4 is a protein-lipid and protein-protein interaction domain and plays a role in calcium-dependent translocation of Nedd4 to the plasma membrane (Plant et al., 1997; Plant et al., 2000). In addition, the C2 domain of Nedd4 binds to and inhibits the catalytic HECT domain (Wang et al., 2010). The presence of calcium releases this auto-inhibition and thus activates Nedd4. Since GluN1/GluN2D receptors are calcium permeable, their activation may activate and recruit Nedd4 to GluN2D-containing complexes resulting in GluN2D downregulation. Along these lines, AMPA receptor activation leads to Nedd4-dependent ubiquitination of GluA1 subunits in a calcium-dependent manner (Schwarz et al., 2010).
Although Nedd4 is developmentally-downregulated, Nedd4 has been shown to be upregulated in adult cerebral cortex following ischemic brain injury (Sang et al., 2006). This upregulation of Nedd4 and the Nedd4 adapter protein Ndfip1 has been reported to have neuroprotective activity. It is possible that Nedd4 upregulation contributes to neuroprotection through GluN2D subunits downregulation. As mentioned above, there is evidence of a role for GluN2D subunits in initiating cortical neuronal cell death that is enhanced by tissue plasminogen activator following stroke (Baron et al., 2010; Jullienne et al., 2011). Furthermore, we have found that GluN2D knockout mice have significantly reduced ischemic damage in the cerebral cortex following middle cerebral artery occlusion (Monaghan et al., 2010).
Nedd4 provides a highly selective mechanism for the discrete regulation of NMDA receptors that contain GluN2D subunits. As the role of GluN2D in synaptic function and neurological disorders becomes better defined, it will be important to further define the role of Nedd4 in limiting GluN2D function.
Highlights.
Proteomic based identification of Nedd4 as a novel GluN2D-interacting protein in rat brain
Both WW2 and WW3 domains of Nedd4 interact with GluN2D
Presence of PPXY motif on GluN2D C-terminal is essential in mediating GluN2D interaction with Nedd4
Nedd4 acts as a specific ubiquitin ligase for GluN2D
Nedd4 downregulates GluN1/GluN2D NMDA receptor response in Xenopus oocytes
Acknowledgements
The authors thank Drs. David Lynch, Shigetada Nakanishi, and Peter Seeburg for providing NMDA receptor cDNA constructs and the American Heart Association Predoctoral Fellowship (V.G.). We also gratefully acknowledge the UCSF Mass Spectrometry Facility supported by the National Institutes of Health NIGMS 8P41GM103481 (to A.L.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baron A, Montagne A, Casse F, Launay S, Maubert E, Ali C, Vivien D. NR2D-containing NMDA receptors mediate tissue plasminogen activator-promoted neuronal excitotoxicity. Cell Death Differ. 2010;17:860–871. doi: 10.1038/cdd.2009.172. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Zoladek T, Haines DS. WW domains 2 and 3 of Rsp5p play overlapping roles in binding to the LPKY motif of Spt23p and Mga2p. The international journal of biochemistry & cell biology. 2008;40:147–157. doi: 10.1016/j.biocel.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller AL, Larson HC, Morrisett RA, Monaghan DT. Glycine modulates ethanol inhibition of heteromeric N-methyl-D-aspartate receptors expressed in Xenopus oocytes. Mol Pharmacol. 1995;48:717–723. [PubMed] [Google Scholar]
- Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Collingridge G. Synaptic plasticity. The role of NMDA receptors in learning and memory. Nature. 1987;330:604–605. doi: 10.1038/330604a0. [DOI] [PubMed] [Google Scholar]
- Ekberg J, Schuetz F, Boase NA, Conroy SJ, Manning J, Kumar S, Poronnik P, Adams DJ. Regulation of the voltage-gated K(+) channels KCNQ2/3 and KCNQ3/5 by ubiquitination. Novel role for Nedd4-2. J Biol Chem. 2007;282:12135–12142. doi: 10.1074/jbc.M609385200. [DOI] [PubMed] [Google Scholar]
- Fotia AB, Ekberg J, Adams DJ, Cook DI, Poronnik P, Kumar S. Regulation of neuronal voltage-gated sodium channels by the ubiquitin-protein ligases Nedd4 and Nedd4-2. J Biol Chem. 2004;279:28930–28935. doi: 10.1074/jbc.M402820200. [DOI] [PubMed] [Google Scholar]
- Fotia AB, Dinudom A, Shearwin KE, Koch JP, Korbmacher C, Cook DI, Kumar S. The role of individual Nedd4-2 (KIAA0439) WW domains in binding and regulating epithelial sodium channels. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2003;17:70–72. doi: 10.1096/fj.02-0497fje. [DOI] [PubMed] [Google Scholar]
- Hardingham GE. Pro-survival signalling from the NMDA receptor. Biochem Soc Trans. 2006;34:936–938. doi: 10.1042/BST0340936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa M, Mori H, Araki K, Sakimura K, Watanabe M, Inoue Y, Mishina M. Cloning and expression of the epsilon 4 subunit of the NMDA receptor channel. FEBS letters. 1992;313:34–38. doi: 10.1016/0014-5793(92)81178-o. [DOI] [PubMed] [Google Scholar]
- Jullienne A, Montagne A, Orset C, Lesept F, Jane DE, Monaghan DT, Maubert E, Vivien D, Ali C. Selective inhibition of GluN2D-containing N-methyl-D-aspartate receptors prevents tissue plasminogen activator-promoted neurotoxicity both in vitro and in vivo. Molecular neurodegeneration. 2011;6:68. doi: 10.1186/1750-1326-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamynina E, Debonneville C, Bens M, Vandewalle A, Staub O. A novel mouse Nedd4 protein suppresses the activity of the epithelial Na+ channel. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001;15:204–214. doi: 10.1096/fj.00-0191com. [DOI] [PubMed] [Google Scholar]
- Kanelis V, Rotin D, Forman-Kay JD. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nature structural biology. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- Khosravani H, Zhang Y, Tsutsui S, Hameed S, Altier C, Hamid J, Chen L, Villemaire M, Ali Z, Jirik FR, Zamponi GW. Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J Gen Physiol. 2008;131:i5. doi: 10.1085/JGP1316OIA5. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- Kumar S, Harvey KF, Kinoshita M, Copeland NG, Noda M, Jenkins NA. cDNA cloning, expression analysis, and mapping of the mouse Nedd4 gene. Genomics. 1997;40:435–443. doi: 10.1006/geno.1996.4582. [DOI] [PubMed] [Google Scholar]
- Lin A, Hou Q, Jarzylo L, Amato S, Gilbert J, Shang F, Man HY. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J Neurochem. 2011;119:27–39. doi: 10.1111/j.1471-4159.2011.07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine-or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, Lipkowitz S, Weissman AM. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J Biol Chem. 2003;278:43169–43177. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:521. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Micu I, Jiang Q, Coderre E, Ridsdale A, Zhang L, Woulfe J, Yin X, Trapp BD, McRory JE, Rehak R, Zamponi GW, Wang W, Stys PK. NMDA receptors mediate calcium accumulation in myelin during chemical ischaemia. Nature. 2006;439:988–992. doi: 10.1038/nature04474. [DOI] [PubMed] [Google Scholar]
- Misra C, Brickley SG, Wyllie DJ, Cull-Candy SG. Slow deactivation kinetics of NMDA receptors containing NR1 and NR2D subunits in rat cerebellar Purkinje cells. J Physiol. 2000;525(Pt 2):299–305. doi: 10.1111/j.1469-7793.2000.t01-1-00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan DT, Zhao H, Gautam V, Sun H, Mayhan W. Deletion of the NMDA receptor subunit GluN2D leads to reduced cell death in the cerebral cortex in the mouse middle cerebral artery occlusion model. Society for Neuroscience. 2010 (abstract), #873.2. [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Myat A, Henry P, McCabe V, Flintoft L, Rotin D, Tear G. Drosophila Nedd4, a ubiquitin ligase, is recruited by Commissureless to control cell surface levels of the roundabout receptor. Neuron. 2002;35:447–459. doi: 10.1016/s0896-6273(02)00795-x. [DOI] [PubMed] [Google Scholar]
- Plant PJ, Yeger H, Staub O, Howard P, Rotin D. The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J Biol Chem. 1997;272:32329–32336. doi: 10.1074/jbc.272.51.32329. [DOI] [PubMed] [Google Scholar]
- Plant PJ, Lafont F, Lecat S, Verkade P, Simons K, Rotin D. Apical membrane targeting of Nedd4 is mediated by an association of its C2 domain with annexin XIIIb. J Cell Biol. 2000;149:1473–1484. doi: 10.1083/jcb.149.7.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri G, Baldi P, Fariselli P, Casadio R. Improved prediction of the number of residue contacts in proteins by recurrent neural networks. Bioinformatics. 2001;17(Suppl 1):S234–242. doi: 10.1093/bioinformatics/17.suppl_1.s234. [DOI] [PubMed] [Google Scholar]
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nature reviews Molecular cell biology. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Sang Q, Kim MH, Kumar S, Bye N, Morganti-Kossman MC, Gunnersen J, Fuller S, Howitt J, Hyde L, Beissbarth T, Scott HS, Silke J, Tan SS. Nedd4-WW domain-binding protein 5 (Ndfip1) is associated with neuronal survival after acute cortical brain injury. J Neurosci. 2006;26:7234–7244. doi: 10.1523/JNEUROSCI.1398-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz LA, Hall BJ, Patrick GN. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J Neurosci. 2010;30:16718–16729. doi: 10.1523/JNEUROSCI.3686-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GB, Heynen AJ, Bear MF. Bidirectional synaptic mechanisms of ocular dominance plasticity in visual cortex. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2009;364:357–367. doi: 10.1098/rstb.2008.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Lewis MJ, Nikko E, Pelham HR. Multiple interactions drive adaptor-mediated recruitment of the ubiquitin ligase rsp5 to membrane proteins in vivo and in vitro. Mol Biol Cell. 2007;18:2429–2440. doi: 10.1091/mbc.E07-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Peng Q, Lin Q, Childress C, Carey D, Yang W. Calcium activates Nedd4 E3 ubiquitin ligases by releasing the C2 domain-mediated auto-inhibition. J Biol Chem. 2010;285:12279–12288. doi: 10.1074/jbc.M109.086405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M. Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 1992;3:1138–1140. doi: 10.1097/00001756-199212000-00027. [DOI] [PubMed] [Google Scholar]
- Watkins JC, Evans RH. Excitatory amino acid transmitters. Annual review of pharmacology and toxicology. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- Wenzel A, Villa M, Mohler H, Benke D. Developmental and regional expression of NMDA receptor subtypes containing the NR2D subunit in rat brain. J Neurochem. 1996;66:1240–1248. doi: 10.1046/j.1471-4159.1996.66031240.x. [DOI] [PubMed] [Google Scholar]
- Yang L, Wang S, Sung B, Lim G, Mao J. Morphine induces ubiquitin-proteasome activity and glutamate transporter degradation. J Biol Chem. 2008;283:21703–21713. doi: 10.1074/jbc.M800809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol Rev. 2007;59:14–39. doi: 10.1124/pr.59.1.4. [DOI] [PubMed] [Google Scholar]
- Zeng F, Xu J, Harris RC. Nedd4 mediates ErbB4 JM-a/CYT-1 ICD ubiquitination and degradation in MDCK II cells. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:1935–1945. doi: 10.1096/fj.08-121947. [DOI] [PMC free article] [PubMed] [Google Scholar]