Abstract
Background
Breast tumors have been described by molecular subtypes characterized by pervasively different gene expression profiles. The subtypes are associated with different clinical parameters and origin of precursor cells. However, the biological pathways and chromosomal aberrations that differ between the subgroups are less well characterized. The molecular subtypes are associated with different risk of metastatic recurrence of the disease. Nevertheless, the performance of these overall patterns to predict outcome is far from optimal, suggesting that biological mechanisms that extend beyond the subgroups impact metastasis.
Results
We have scrutinized publicly available gene expression datasets and identified molecular subtypes in 1,394 breast tumors with outcome data. By analysis of chromosomal regions and pathways using “Gene set enrichment analysis” followed by a meta-analysis, we identified comprehensive mechanistic differences between the subgroups. Furthermore, the same approach was used to investigate mechanisms related to metastasis within the subgroups. A striking finding is that the molecular subtypes account for the majority of biological mechanisms associated with metastasis. However, some mechanisms, aside from the subtypes, were identified in a training set of 1,239 tumors and confirmed by survival analysis in two independent validation datasets from the same type of platform and consisting of very comparable node-negative patients that did not receive adjuvant medical therapy. The results show that high expression of 5q14 genes and low levels of TNFR2 pathway genes were associated with poor survival in basal-like cancers. Furthermore, low expression of 5q33 genes and interleukin-12 pathway genes were associated with poor outcome exclusively in ERBB2-like tumors.
Conclusion
The identified regions, genes, and pathways may be potential drug targets in future individualized treatment strategies.
Keywords: breast cancer, metastasis, gene expression, microarray, pathway analysis, molecular subtypes
Background
Breast cancer is the most common cancer among women. The local disease is not lethal, but its spread to distant organs is fatal. The risk of metastasis is evaluated by clinical and pathological criteria, but the performance of these methods is far from optimal. Gene expression profiling of tumors has been used for the supervised classification of cancer outcomes in several studies with promising results for the improvement of risk prediction.1–4 Despite these promising clinical results, limited insights into its biological mechanisms has been obtained from the large amount of gene expression data available. A very different approach was used in early studies of global gene expression of breast cancer, where unsupervised hierarchical clustering was used to identify 4–5 molecular subtypes.5–8 These subtypes arise from at least two cell types: basal cells and luminal epithelial cells. Luminal tumors are mainly estrogen receptor (ER)-positive and cluster in two distinct groups (termed luminal A and luminal B) that appear to result in good and poor prognosis, respectively. Basal-like tumors are ER-negative and are characterized by poor prognosis. A distinct group that mainly consists of ER-negative tumors is characterized by the amplification of ERBB2. Finally, a normal-like profile constitutes good prognosis. These subtypes are biologically very meaningful, but their prediction of metastasis is not optimal.
In recent studies, unsupervised and supervised methods have been used to differentiate the molecular subtypes of breast cancer into further subgroups with different outcomes. The discriminating genes have subsequently been related to biological function (for example, immune response among ER-negative tumors).9,10 These studies have identified very broad mechanisms, but they have not pointed at specific biochemical pathways. A very different strategy would be to analyze predefined gene sets representing biological mechanisms, such as the pathways for their association to metastasis. We have previously used this strategy for the analysis of the overall pathways involved in the metastasis of breast cancer.11 Furthermore, we have applied this approach to gene sets representing genomic regions to identify somatic mutations at the deoxyribonucleic acid (DNA)-level that are involved in metastasis.12 However, the analysis of pathways and genomic aberrations has not been applied to analyze the various metastatic mechanisms found within the different subtypes of breast cancer.
We hypothesize that several somatic genomic aberrations and biological pathways characterize the molecular subtypes in breast cancer. Despite the association of the subtypes to metastasis, we expect that mechanisms within a given subtype may be involved in the higher risk of metastasis for a fraction of the tumors. Furthermore, we anticipate that deregulation of the pathways is reflected in gene expression patterns of primary tumor biopsies. We also assume that somatic copy number mutations involving chromosomal regions will be reflected in the overall level of gene expression in these regions. Our aim is to elucidate these metastatic mechanisms within the molecular subtypes and, secondly, to determine the mechanisms that differ between the subgroups.
We have used predefined gene sets representing canonical pathways and chromosomal regions to identify metastatic mechanisms within molecular subtypes. This has been accomplished by a global approach; gene set enrichment analysis (GSEA) examining the entire list of genes ranked according to association to metastasis within each molecular subtype. In a single dataset, this analysis will generally not result in significant findings. However, by performing a meta-analysis of several datasets, we have increased the power to identify somatic mutations among the chromosomal regions and pathways involved in the metastasis of fractions of breast tumors within a specific subtype.
Results
Molecular subtypes of 1,439 tumors
Global gene expression data were collected from 1,586 breast tumors with clinical follow up (Table 1). Molecular subtypes were identified by a single sample prediction approach using nearest centroid classification.8 Single-sample predictions resulted in 1,394 samples with assigned subtypes. Survival analysis confirmed the prognostic impact of the subtypes, with luminal A and normal-like tumors having significantly better prognosis than luminal B, ERBB2, and basal-like tumors (Fig. 1).
Table 1.
Datasets and patients included in the study.
| Data set | Country | Platform | #genes | Outcome | #pt with follow up | #sub typed samples |
|---|---|---|---|---|---|---|
| Amsterdam1 | The Nederlands | Agilent | 24451 | Distant metastasis | 295 | 287 |
| Rotterdam2 | The Nederlands | 133A | 22283 | Distant metastasis | 286 | 277 |
| Stockholm32 | Sweden | 133A+B | 44982 | Death from BC | 159 | 152 |
| Uppsala33 | Sweden | 133A+B | 44982 | Death from BC | 236 | 229 |
| Bild34 | US | 95av2 | 12625 | Metastasis | 158 | 153 |
| Hu8 | 3 Agilent | 22K | Relapse | 105 | 105 | |
| Training total | 1239 | 1203 | ||||
| Mainz9 | Germany | 133A | 22283 | Distant metastasis | 200 | 191 |
| TRANSBIG-S3 | UK | 133A | 22283 | Distant metastasis | 147 | 0 |
| Test total | 347 | 191 | ||||
| Total | 1586 | 1394 |
Figure 1.
Kaplan–Meyer plot of the five molecular subtypes in seven datasets, including 1,394 tumor samples. Molecular subtypes were determined by the single-sample prediction method reported by Hu et al.8 Hazard ratios were calculated by fitting a Cox model to the data for comparing each group to basal-like tumors. The P-value was calculated using the log-rank test.
Genomic aberrations and pathways associated to molecular subtypes
To identify the biological characteristics of the molecular subtypes, we divided the 1,394 samples subtyped with a single sample prediction, to a training set of 1,203 tumors corresponding to six entire datasets and a test set of 191 tumors contained in two datasets (Table 1). We applied GSEA analysis to one subtype at a time compared to all other tumors. This analysis was performed with gene sets representing chromosomal region and with pathway gene sets. A meta-analysis was performed to identify gene sets that were concordantly differentially regulated in the training datasets. For chromosomal regions, a large number of somatic mutations were indicated by the analysis (Table 2). The differential expression of several consecutive regions indicated that larger regions are lost or gained. Validation in the test set resulted in 34% of regions being significant (highlighted in bold in Table 2).
Table 2.
Differentially expressed chromosomal regions.
| Basal | ERBB2 | Lum A | Lum B | Normal |
|---|---|---|---|---|
| Up-regulated | ||||
| 1P34, 1P32, 1P22 2P24, 2P21, 2P16 3Q21, 3Q25, 3Q27, 3Q29 6P21, 6Q23 7P15, 7Q32 10P15 11Q22 12P13, 12P12 16Q13, 16Q24 21Q22 XQ26, XQ28 |
2P11, 2Q37 6Q21 9Q34 12Q12 16Q13 17Q11, 17Q12, 17Q21, 17Q23 22Q11, 22Q13 |
1P33, 1P31, 1Q41 3P22, 3P21, 3P14 5Q11, 5Q13 8P22, 8P21 14Q24 10Q23, 10Q25 17P11 |
4P16 6P22 8P11, 8Q22, 8Q24 12Q13, 12Q24 16P13, 16P12, 16P11 17Q21, 17Q22, 17Q23, 17Q24, 17Q25 20P11, 20Q11, 20Q12, 20Q13 |
2Q32 3P22 5Q13, 5Q32 7P15 8P23, 8P21 9Q21 11Q23 15Q11 XQ13 XQ22 |
| Down-regulated | ||||
| 3P22, 3P21, 3P14 4P16 5Q11, 5Q13, 5Q14, 5Q31 9Q32 10Q23, 10Q25 12Q12, 12Q13, 12Q22 14Q21, 14Q23, 14Q24, 14Q32 15Q22 16P13, 16P12 17P11, 17Q21, 17Q24 |
1P21 3P24 8P22, 8P21 9P24 17P11 18Q12, 18Q21 |
2P25, 2P12 3Q27 6P21 8Q22, 8Q24 10P15 16Q13, 16Q22, 16Q24 17Q11, 17Q23, 17Q25 20P13, 20Q11, 20Q13 22Q11, 22Q13 XQ28 |
1P31, 1P22 2Q32 4P15, 4Q21 5P13 6P25, 6Q21, 6Q22, 6Q23 7P15, 7Q31 9P24, 9Q21 11Q22, 11Q23, 11Q24 |
1Q23, 1Q41, 1Q42 2P12 6P22 8P12, 8Q22, 8Q24 10Q21 17Q25 20Q13 |
Note: Gene sets in bold were also significant in test data set (Mainz).
Pathway analysis using predefined canonical pathways also indicated large differences between the subgroups (Table 3). Of these pathways, 43% were also identified by GSEA analysis in the independent Mainz dataset (highlighted in bold in Table 3).
Table 3.
Differentially expressed pathway gene sets.
| Basal | ERBB2 | Lum A | Lum B | Normal |
|---|---|---|---|---|
| Up-regulated | ||||
|
CELL CYCLE(6) DNA_REPLICATION(3) tRNA SYNTHESIS(2) ACTINY TNF APOPTOSIS(2) CELL_ADHESION PROTEASOME ATRBRCA P53 SIGNALING IL12(2) TH1TH2 ST_B_CELL_ ANTIGEN_RECEPTOR ST_T_CELL_SIGNAL_ TRANSDUCTION ANTIGEN_PROCESSING/ PRES.TOLL_LIKE_ RECEPTOR NATURAL_ KILLER_CELL_CYTOTOX PI3K GLYCOSPHINGOLIPID_ BIOSYNTH ST_FAS_SIGNALING |
BIOSYNTHESIS_OF_ STEROIDS(2) PROTEASOME(2) ARGININE/PROLINE_ METABOLISM |
AMINO ACID METABOLISM(4) ST_JNK_MAPK CIRCADIAN_EXERCISE NITROGEN_ METABOLISM(2) BUTANOATE_ METABOLISM(2) PROPANOATE_ METABOLISM ESTROGEN_SIGNALING SPHINGOLIPID_ METABOLISM FATTY_ACID_ METABOLISM AKT IGF1 GLYCOSPHINGOLIPID_ METABOLISM VIP NUCLEAR_RECEPTORS GLEEVEC CARM_ER PDGF |
OXIDATIVE_ PHOSPHORYLATION(3) MPR P53 PROTEASOME(2) FRUCTOSE/MANNOSE_ METABOLISM(2) IGF1(2) PTDIN DNA_REPLICATION(3) CELL_CYCLE TRANSCRIPTION RAS CARM_ER CHREBP EIF4 RACCYCD BIOSYNTHESIS_OF_ STEROIDS CERAMIDE AMINO_ACID_ METABOLISM(2) CREB |
FATTY_ACID_METABOLISM HISTIDINE_METABOLISM MAPK_SIGNALING WNT_SIGNALING(3) FOCAL_ADHESION ECM PROSTAGLANDIN CARDIACEG ALK PDGF PPARA NUCLEAR_RECEPTORS EGF TGF_BETA_SIGNALING COMPLEMENT_CASCADE(2) SMOOTH_MUSCLE_ CONTRACTION RIBOSOMAL_PROTEINS IGF1 SPRY INTRINSIC |
| Down-regulated | ||||
| TEL MTOR(2) IGF1(2) GPCR VIP AMINO ACID METABOLISM(2) ESTROGEN_ SIGNALING NUCLEAR_ RECEPTORS CHREBP CERAMIDE CARM_ER GLYCOSPHINGOLIPID_ METABOLISM ST_JNK_MAPK BAD |
TEL GLEEVEC IGF1 NITROGEN_ METABOLISM(2) |
CELL_CYCLE(6) IL12(2) GLYCOLYSIS PENTOSE_ PHOSPHATE_(2) PROTEASOME ST_B_CELL_ANTIGEN_ RECEPTOR PTDINS DNA_REPLICATION(3) tRNA_SYNTHESIS(2) ST_T_CELL_SIGNAL_ TRANSDUCTION ACTINY CELL_ADHESION P53_SIGNALING NATURAL_KILLER_ CELL_CYTOTOX |
PROSTAGLANDIN_ SYNTHESIS NTHI ACTIN_ CYTOSKELETON GLYCEROLIPID_ METABOLISM IL1R TOLL LYSINE_DEGRADATION MELANOGENESIS FOCAL_ADHESION WNT_SIGNALING ALK CELL_ADHESION(2) CYTOKINE UREA_CYCLE |
CELL_CYCLE(5) DNA_REPLICATION(5) PENTOSE_PHOSPHATE(2) TRNA_BIOSYNTHESIS(2) OXIDATIVE_ PHOSPHORYLATION(3) CITRATE_CYCLE(3) PROTEASOME(3) GLYCOLYSIS P53 BIOSYNTHESIS_OF_ STEROIDS(2) ATRBRCA ST_FAS_SIGNALING GLUTAMATE_METABOLISM SELENOAMINO_ACID_ METABOLISM RACCYCD MPR |
Notes: Gene sets in bold were also significant in test data set (Mainz). The number of redundant pathway gene sets is given in brackets.
Chromosomal regions and pathways involved in metastasis within subgroups
The above analyses indicate that molecular subtypes significantly account for the metastasis of breast cancer, and that the subtypes are associated with very different biological pathways and somatic mutations. To identify the metastatic mechanisms that extend beyond the molecular subtypes, we performed pathway and chromosomal aberration analyses within each subtype, comparing tumors from patients that experienced metastasis to tumors from patients that did not experience metastasis within the follow-up period. The data were again split into a training set including six datasets, and the two most recently published datasets (Mainz and TRANSBIG-S) were kept for validation purposes. The training set was composed of datasets with different clinical characteristics and treatment statuses of the patients and performed with different microarray platforms. In contrast, the test sets (Mainz and TRANSBIG-S) contained data from node-negative patients not receiving adjuvant medical treatment; all analyses were performed with Affymetrix HG133A chips. The GSEA meta-analysis in the training set resulted in the identification of nine significant regions and pathways (Table S1). Using GSEA for the validation of these regions and pathways in the Mainz dataset did not result in any significant gene sets, probably due to the limited sample size when comparing tumors with poor and good outcomes within subtypes (data not shown).
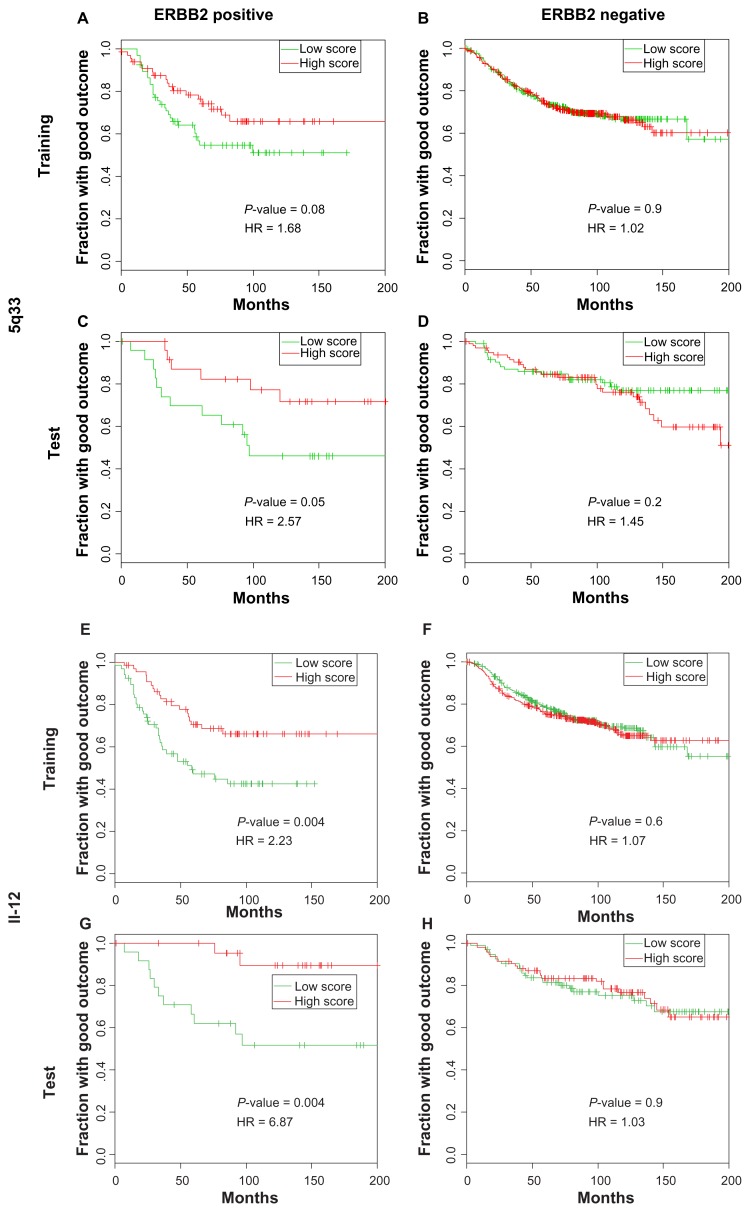
In order to validate the identified regions and pathways with a different method, we investigated whether the overall expression level in each gene set could be used as a prognostic marker in survival analysis. This would also allow us to compare the expression of regions or pathways between single tumors from all datasets. Furthermore, utilizing survival information might help to improve the power of the validation, and combining datasets would also increase the power. Datasets were preprocessed and combined using only genes measured on all chips. The expression level of all genes in each region or pathway identified previously was implemented in the region scores. This method is different from the GSEA meta-analysis described earlier because only genes represented on all platforms were included in the analysis. In 280 basal-like tumors from the training sets, this different approach supported a high level of expression of 5q14 (Fig. 2A) and a low level of expression of tumor necrosis factor receptor (TNFR)2 (Fig. 2E) as predictors of poor outcomes. For ERBB2 tumors, the low expression of 5q33 (borderline significant; Fig. 3A) and interleukin (IL)-12 (Fig. 3E) were associated with poor outcomes, which was in agreement with the GSEA meta-analysis. The remaining five pathways and regions identified by the GSEA meta-analysis were not significant in the survival analysis (Table S1).
Figure 2.
Kaplan–Meyer plots of the 5q14 and TNFR2 groups in basal-like tumors. The training set consisted of 280 basal-like tumors (A and E) and 959 nonbasal-like tumors (B and F). The test set contained 41 basal-like tumors (C and G) and 159 nonbasal-like tumors (D and H). The score is calculated for each sample as the mean standardized expression values for all genes in the gene set The low score group was defined as the third of samples with the lowest score for the indicated gene set, and the high score group was defined as the third of the samples with the highest score. The middle third was excluded from the analysis. The P-values were calculated using the log-rank test.
Abbreviation: TNFR, tumor necrosis factor receptor.
Figure 3.
Kaplan–Meyer plots of 5q33 and IL-12 groups in ERBB2 tumors. The training set consisted of 196 tumors with high ERBB2 expression (A and E) and 1,043 tumors with low ERBB2 expression (B and F). The test set contained 70 tumors with the highest ERBB2 expression (C and G) and 277 tumors with lowest ERBB2 expression (D and H). The score is calculated for each sample as the mean standardized expression values for all genes in the gene set. The low score group was defined as the third of samples with lowest score for the indicated gene set, and the high score group was defined as the third of the samples with the highest score. The middle third was excluded from the analysis. The P-values were calculated using the log-rank test.
Abbreviation: IL, interleukin.
For the validation of metastatic gene sets in independent datasets, two test sets were used. The Mainz data set, where 41 basal-like tumors were identified by single-sample prediction, was used for the validation of the metastatic mechanisms of basal-like tumors. However, only 20 ERBB2 tumors were identified by single-sample prediction in the Mainz dataset. To increase the sample size of ERBB2-positive tumors, the TRANSBIG-S dataset was included. The identification of molecular subtypes failed in this dataset, but ERBB2 status was determined from ERBB2 expression in both datasets. The very unbalanced distribution of ERBB2 and the coregulation of GRB7 located closely to ERBB2 supported this method for the classification of ERBB2 amplification (Fig. S1).
For basal-like tumors, the disadvantage of high 5q14 expression was validated by the survival analysis in the test set (Fig. 2C). To investigate whether this prognostic effect of 5q14 aberration was specific for basal-like tumors, the prognostic performance was also examined in the remaining tumors not belonging to this subgroup. This demonstrated no prognostic impact of 5q14 aberrations in the training set (Fig. 2B) but, surprisingly, a borderline significant opposite effect was found in the test set (Fig. 2D).
The prognostic impact of the TNFR2 pathway identified in the GSEA meta-analysis and in the survival analysis in the training set (Fig. 2E) was also supported by a survival analysis in the test set (Fig. 2G). However, this pathway was also significant in nonbasal-like tumors in the training set, but with smaller separation of the groups (ie, lower hazard ratios [Fig. 2F], and the pathway was only borderline significant among the nonbasal-like tumors in the test set [Fig. 2H]; in addition, a lower hazard ratio was observed among nonbasal-like tumors when compared to the basal-like tumors).
For ERBB2 tumors, the low expression of 5q33 (Fig. 3C) and IL-12 (Fig. 3G) was also associated with poor outcomes in the test set. Interestingly, these mechanisms were specific for tumors with high levels of expression of ERBB2 (Fig. 3B, D, F, and H).
To identify single metastasis candidate genes in the two 5q regions, we compared the expression of metastasizing and non-metastasizing tumors at single gene level. For 5q14, one gene, HAPLN1, was strongly and differentially regulated (Fig. S2), suggesting that this is a plausibly causal metastasis gene. No single gene was strongly and differentially expressed at 5q33 (data not shown).
Discussion
We have conducted a meta-analysis of the tumor gene expression data and identified chromosomal regions and pathways that were strongly associated with the molecular subtypes of breast cancer. Furthermore, some mechanisms were found to be associated with metastasis within the different subgroups. Our results demonstrate that these mechanisms are reflected in the overall gene expression of the chromosomal region and the pathway gene sets, which is in agreement with our hypotheses.
A major finding is the large number of chromosomal regions and pathways associated with the molecular subtypes that were found. This is in agreement with the supposedly different cellular origins of basal-like and luminal-like tumors. The universal nature of the subtypes is supported by several studies9,10 that have used different unsupervised methods to identify the subtypes of breast tumors that resemble the subgroups reported by Sørlie et al.6 ER status, ERBB2 status, and proliferation level are believed to be major characteristics of the molecular subtypes. Our results support the crucial impact of the following mechanisms: 17q amplification in ERBB2-like tumors (Table 2); estrogen signaling in luminal cancers (CARM_ER and ESTROGEN_SIGNALING; Table 3); and elevated cell cycle in basal-like and luminal B tumors. By comparing metastasizing and nonmetastasizing tumors, without stratifying for subtypes, we have previously identified the chromosomal regions and pathways involved in the metastasis of breast cancer.11,12 The identified gene sets strongly overlap with the gene sets that were differentially expressed between molecular subtypes, indicating that the molecular subtypes account for a majority of the metastasis-associated mechanisms. This is strongly supported by the very few metastasis-linked mechanisms we have identified within the molecular subgroups (two regions and two pathways), even though the cut-off for the false discovery rate (FDR) was 0.05 for intra-subgroup analysis and 0.01 for the between-subgroup analysis. However, the considerably smaller sample size in the within-group analysis may bias this conclusion.
Our within-subgroups analysis identified somatic mutations, which were located at different loci on 5q, and which had opposite prognostic effects: high levels of expression of 5q14 were associated with poor outcomes in basal-like cancer, whereas lower levels of expression of 5q33 were associated with poor outcomes among patients with ERBB2-amplified breast tumors. The dual role of 5q aberrations is supported by a few previous publications that used comparative genome hybridization; Friedrich et al13 reported higher frequency of 5q23 losses among cases with distant metastasis and Karlsson et al14 showed that poorer outcomes were associated with 5q31~qter gain for cases with ER-negative tumors. Callaghan et al15 reported a higher frequency of allelic imbalance at 5q21 in patients with lymph node metastasis, indirectly supporting the prognostic significance of this region. However, these studies included few patients and did not subdivide tumors according to basal/ERBB2 status, hampering the conclusion for these aberrations that, according to our results, are specific for basal-like and ERBB2 tumors, respectively.
Our method identifies regions by measuring the differential expression of genes, but it does not (in principle) determine whether the DNA copy number increases in one outcome group or decreases in the opposite group. However, low expression of 5q14 and its surrounding region characterize basal-like tumors according to our analysis, which compared molecular subtypes (Table 2). This is also supported by Adélaïde et al,16 who used array comparative genomic hybridization (aCGH) and reported 5q loss to be frequent in basal-like tumors. This indicates that the loss of 5q14 is common in basal-like tumors, but that patients with basal-like tumors retaining 5q14 have a worse prognosis. This might be explained by the presence of a tumor suppressor gene involved in basal-like cancer development, and another gene promoting metastasis—both of which were located in the region. A similar effect has previously been observed for 16q loss in breast cancer.12,17 We pinpointed one gene, hyaluronan, and proteoglycan link protein 1 (HAPLN1) in the 5q14 region, which had a strong association with the metastasis of basal-like cancers, making this a relevant candidate gene that may be a causal factor in the metastasis of basal-like tumors. HAPLN1 is involved in the stabilization of hyaluronan and vesican. An interaction of hyaluronan and its receptor (CD44) induces signaling events that promote cell growth, migration, and metastasis,18 making genes in this pathway relevant.
ERBB2-like tumors are not characterized by 5q aberrations, as compared to other subtypes (Table 2), but our results indicate that low expression of the 5q33 region is associated with poor outcomes. This may be the result of 5q33 loss in the poor prognosis group, or gain of the region in the good prognosis group. However, this cannot be determined from the present results.
Many pathways have previously been linked to metastasis; however, investigations of the metastatic pathways within different subtypes of breast cancer are sparse, as will be discussed in the following. We identified the high expression of the TNFR2 pathway as an indicator of good prognosis in basal-like cancers. TNFR2 is a receptor with equal affinity for tumor necrosis factor (TNF)a and TNFb. Both of these are produced by activated lymphocytes and can be toxic to many tumor types. Nevertheless, for micropapillary breast carcinomas, overexpression of TNFR2 has been linked with higher incidence of lymph node metastasis, which may conflict with our results.19 However, our finding that the TNFR2 metagene is associated with survival in basal-like and nonbasal-like cancers in both the training set and test set (borderline significance for the nonbasal-like tumors in the test set) strongly indicate the prognostic disadvantage of low TNFR2 pathway expression. According to our results (Table 3), the TNFR2 pathway is not differentially expressed between subgroups. However, the survival analysis within subtypes show smaller separations of high and low TNFR2 score curves (ie, lower hazard ratios for nonbasal-like tumors compared to basal-like tumors; Fig. 2), indicating stronger metastatic prevention by the pathway in basal-like tumors. This is also supported by the GSEA meta-analysis that exclusively identified TNFR2 only among basal-like carcinomas. To our knowledge, no previous studies have described a prognostic influence of TNFR2/TNF signaling among basal-like breast tumors.
The other pathway, IL-12, identified within the ERBB2 subgroup is also associated with good outcomes. This is supported by studies showing that IL-12 activates natural killer cells and enhances their ability to kill tumor cell lines that are treated with herceptin20 However, no benefit was observed by adding IL-12 to the herceptin treatment of breast cancer patients in a small Phase 1 study.21 None of the patients in our analysis received herceptin, indicating that IL-12 signaling has antimetastatic potential, independent of herceptin treatment.
Both IL-12 and TNFR2 signaling are involved in immune activation, suggesting that lymphocyte infiltration is beneficial for patients with ER-negative tumors. This is supported by pathological investigations conducted among subgroups of patients. For instance, Ménard et al22 reported selectively higher survival rates in young breast cancer patients when they had lymphocyte infiltrated tumors, and Aaltomaa et al23 described the benefit of lymphocyte infiltration among patients with rapid proliferating tumors. Several recent publications using gene expression datasets have also identified immune-related gene signatures that distinguish groups with different outcomes. Schmidt et al9 used hierarchical clustering to identify a B-cell signature with a prognostic impact among high-proliferating tumors. Teschendorff et al10 focused their analysis on ER-negative tumors and used an unsupervised clustering technique to identify five subclusters. Among tumors with normal ERBB2 status, they found an immune response gene cluster that was associated with survival. Alexe et al24 also used unsupervised clustering of ERBB2-positive tumors and reported a subtype with upregulated lymphocyte infiltration signal and improved survival. Rody et al25 clustered immune-related genes and identified seven different gene clusters that were shown to represent different immune cell types. A T-cell cluster was associated with survival rates among ER-negative tumors with and without ERBB2 amplification. These studies have used unsupervised clustering techniques and identified subgroups of patients with different rates of survival. Annotation of genes in these patterns showed an overrepresentation of immune system-related genes. The overall conclusion from these studies is that immune response prevents metastasis among patients with ER-negative tumors with or without ERBB2 amplification. However, different mechanisms in tumor cells and immune cells might result in very different outcomes. Elucidation of the most important mechanisms by the global pathway analysis of outcomes within subtypes has not been performed previously. Desmedt et al26 examined a limited set of seven gene modules based on key processes in breast cancer. Modules were defined as groups of genes that are correlated with the expression of seven key candidate genes. One of these key mechanisms, the STAT1 immune response module, was associated with the survival of patients with ER−/ERBB2− as well as ER−/ERBB2+ tumors. In a method that differed greatly from this approach, we analyzed a large collection of knowledge-generated canonical pathways and genome regions to screen for biological processes involved in metastasis within subgroups. In agreement with abovementioned studies, we identified immune related pathways. However, in addition to the previous studies, our results not only identified a broad immune response, but they also pointed directly at specific signaling pathways, suggesting key roles of metastasis prevention for these cytokine pathways. Furthermore, the identified mechanisms are specifically associated to certain subtypes, suggesting different metastasis-related mechanisms depending on ERBB2 status.
Our comparison of molecular subtypes has identified comprehensive differences in pathways and genomic aberrations (Tables 2 and 3). Several of the somatic aberrations are supported by previous studies using aCGH to compare relevant groups. Bergamaschi et al27 examined 89 tumors by gene expression profiling to identify four molecular subtypes; Adélaïde et al16 compared 44 basal and 49 luminal tumors via aCGH, and a recent large study28 performed a meta-analysis of 773 tumors. The results from these studies confirmed many of the regions identified in the present investigation. For basal-like tumors, their results supported the extensive loss of 5q, as well as the loss of 10q23, 12q22, 14q21, 14q23, 15q22, and the gain of 3q25-27, 6p21, 7q32, 10p15, 12p13-12, and 21q22. For luminal A tumors, they also observed 1q41 and 8p22-21 gains and a 16q loss. For luminal B tumors, gains of 8q22-24, 20p11, and 20q13 and a loss of 11q were in agreement with our findings. Finally, 8p22-21 loss in erb2-amplified tumors was also seen by The Cancer Genome Atlas Network.28
Some studies have characterized ER-positive and ER-negative tumors by aCGH (Loo et al29) or by conventional CGH.30 Although this is a more rough differentiation using only two subgroups, many of their results were in agreement with our findings. Loo et al29 identified a gain of 3q25-27 and losses of 3p21, 4p16, 5q, 10q25, 14q32, 16p13, and 17p11 in ER-negative tumors, which is in agreement with our findings for basal-like tumors. Among the ER-negative tumors, they also identified a gain of 17q12 and losses of 8p21 and 17p11, supporting our findings for ERBB2-like tumors. For receptor-positive tumors, a gain of 16p11-13 and a loss of 16q13-24 was in agreement with our results for luminal tumors.
Altogether, many of the regions we have identified are supported by other studies measuring the DNA copy number by more direct techniques, as compared to our indirect expression-based method. This strongly supports our conclusions and also justifies using this approach for mechanisms within subgroups. We identified many regional aberrations differing between subgroups that have not been reported before. This may be explained by our different approach (including using gene expression data instead of directly measuring DNA copy numbers, for example, by aCGH). This has allowed us to considerably increase the sample size because only few small aCGH datasets with relevant information are available.
We have refined the data included in our analysis to obtain rates of distant metastasis or death from breast cancer as the outcome, but for one dataset (Hu), only time to relapse was available. However, this only introduces a minor bias because the majority of the typically observed relapse events included metastasis, and because only one of the six training datasets had this outcome definition. Another potential bias is the inclusion of patients that have received adjuvant treatment, which will bias the conclusion about metastasis biology because some patients will change their outcome status because of the treatment. However, the fraction of patients that respond to treatment and experience good outcomes as a consequence of treatment will typically be below 10% (adjuvant online, http://www.adjuvantonline.com), meaning that this will also be a minor bias. A varying fraction of the tumors in the individual datasets have disseminated cells to the lymph nodes. The classification of lymph node-positive patients without recurrence as nonmetastasis may be controversial. This may bias the results towards the metastatic mechanisms following primary spread to the lymph node. We accepted these biases in the study design to obtain a broad representation of tumor subtypes; the exclusion of lymph node-positive and treated patients would reduce the number of ERBB2-positive and basal-like tumors markedly, resulting in a lack of power in these groups.
Combining the datasets by standardization, as we have done for the survival analysis, assumes that there are comparable characteristics in the included datasets. The varying fraction of node-positive tumors violates this assumption. However, this results in a more conservative test of gene sets in the survival analysis. This may explain why only four of the nine gene sets identified in the meta-analysis were significant in the survival analysis of the training set. Furthermore, these four gene sets are all significant in the test set, even though the sample size is considerably smaller. This is in agreement with the better separation of survival curves and the higher hazard ratios in the test set (Figs. 2C, G, 3C, and G). This may be explained by the very homogeneous test set, which included only node-negative samples from untreated patients performed with the same microarray platform.
Conclusion
By performing a pathway meta-analysis, we have demonstrated that the previously described molecular subtypes account for the majority of metastatic mechanisms in breast cancer. However, by performing the analysis within these subgroups we have identified two different regions on chromosome 5q that impact metastasis among patients with basal-like and ERBB2-amplified tumors, respectively. We suggest one gene, HAPLN1 at 5q14, which may potentially be a metastasis gene for basal-like tumors. The impact of these regions needs to be validated in future large studies using more direct measures of the DNA copy number (for example, aCGH). Furthermore, the role of HAPLN1 and other potential driver genes needs to be addressed in further functional studies. We have also identified two pathways, TNFR2 and IL-12, which are involved in the metastasis of basal-like and ERBB2-amplified breast cancer, respectively. The molecular subgroups already form the basis for different treatments (such as trastuzumab, which is used against ERBB2-amplified tumors); however, future treatment protocols will aim to establish even more individualized strategies. Our study has identified genomic regions, single genes, and pathways that may be potential targets for future drug design for certain subgroups of patients.
Methods
Data sets
Eight publicly available datasets examining gene expression at the ribonucleic acid (RNA) level in primary tumors were included in the analysis. These studies were performed with different platforms, different populations, and so on, as depicted in Table 1. The outcome used is distant metastasis or death from breast cancer, which is nearly always caused by distant metastasis. Only one data set (Hu) included local and regional recurrences. However, nonmetastatic relapse constitutes a minority of clinical cohorts. For the TRANSBIG dataset, samples from Sweden were removed to avoid sample overlap with the Uppsala and Stockholm datasets. The resulting dataset is termed TRANSBIG-S.
The normalizations performed in the studies were retained because the authors found these methods optimal for the datasets, and because the pathway analysis was performed separately in each dataset.
Molecular subtypes
To identify the molecular subtypes, a single sample predictor was applied as described.8 Prior to this, data were preprocessed within each dataset as follows. First, probe sets with maximal expression values were selected whenever more probe sets recognized the same gene using the “collapse to gene symbol” function in GSEA. Data were then column standardized for each sample by subtracting the mean expression of all genes in that sample from each genes expression value, and dividing by the standard deviation for that sample. Next, row median centering was performed within each dataset by subtracting the median expression for a gene across samples from all expression values for that gene. Pearson’s correlation coefficient between each sample and each of the five centroids (defined by Hu et al8) were calculated, and the sample was assigned the subtype with highest correlation coefficient. If the correlation coefficient was below 0.1 for any of the centroids, the sample was not assigned a subtype. Using this method, the samples were forced into the centroids defined by Hu et al.8
GSEA analysis of pathways and genome regions associated with molecular subtypes
To analyze genome regions and pathways that were differentially expressed between the subtypes, we compared one subtype at a time with all other tumors. Only the seven datasets with successfully identified molecular subtypes were included in the analysis. For this analysis, we used original data (ie, not standardized). GSEA version 2.031 was used with 639 curated gene sets representing individual pathways. These pathway gene sets are adopted from KEGG (www.genome.ad.jp/KEGG), GenMapp (http://www.genmapp.org), Biocarta (www.biocarta.com), and so on, and gathered in the Molecular Signature Database implemented in GSEA. Furthermore, we applied the analysis to positional gene sets delimited by cytobands downloaded from the Molecular Signature Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp).
The GSEA program ranks genes according to a signal-to-noise value:
| (1) |
where X is the mean and s is the standard deviation for the two classes A and B (one subtype and the remaining tumors, respectively). When several probes recognized the same gene, the probe with the maximum expression value was extracted using the “collapse to gene set” function. Gene sets represented by less than 15 genes in a dataset were excluded. The output from GSEA is an enrichment score, describing the imbalance in the distribution of ranks of gene expression in each gene set between the compared groups. The enrichment score is normalized according to the size of the gene sets. Then, the gene sets were ranked according to the normalized enrichment score, with gene sets upregulated in the subgroup of interest on the top and downregulated gene sets at the bottom.
GSEA meta-analysis
The ranked lists of gene sets for each analysis generated by GSEA from the seven datasets were integrated so that only gene sets represented in the output from all datasets were included. The initial 639 pathway gene sets were reduced to 347 gene sets passing the threshold (at least 15 genes in gene sets) in all datasets. For the analysis of chromosomal regions, 386 chromosomal gene sets from the Molecular Signature Database were reduced to 188 gene sets.
For each dataset, individual gene sets were assigned a ranking value from 1 to the maximum number of gene sets, according to the ranking performed by GSEA. The mean ranking value for each gene set was calculated across the datasets and, finally, the gene sets were ranked according to this value.
Our null hypothesis is that the expressions of genes in the pathway gene sets are unrelated to the molecular subtype. This means that the ranking value for a given gene set in a given dataset is expected to be a random value between 1 and the maximum number of gene sets analyzed. To simulate the distribution of the mean ranking values across the six test datasets assuming the null hypothesis, a random drawing of the six ranking values was performed 106 times, and the mean value was calculated each time. A null distribution of the mean ranking values was generated from these results. To test the significance for a given gene set, the observed mean ranking value was compared to the null distribution. To fulfill the null hypothesis, an observed mean ranking value should be within 95% of the interval of the null distribution. Correction for multiple testing was performed by calculating the FDR, controlling the expected proportion of incorrectly rejected null hypotheses. These calculations were performed in the R environment (http://www.bioconductor.org). Gene sets with FDR values below 0.01 were considered significant. Validation of the identified gene sets in the test set (Mainz data set) was performed by GSEA analysis using a nominal P-value.
Mechanisms associated with metastasis among each molecular subtype
To identify the chromosomal regions and pathways involved in metastasis within the molecular subtypes, GSEA analysis was performed as described above. Once again, genes were ranked according to the signal-to-noise value:
| (2) |
except that the compared groups were now tumors from patients that developed metastasis (class A) and tumors from patients that did not develop metastasis (class B). This analysis was performed within each subtype in each dataset, and the meta-analysis was applied to identify gene sets concordantly deregulated in metastasizing tumors across datasets for each molecular subtype. Again, 347 pathway gene sets and 188 positional gene sets passed the inclusion threshold. FDR values below 0.05 were considered significant.
Survival analysis
The above GSEA analyses were performed independently in each data set. To get a measure of the overall expression level for all genes in an identified region or pathway for each tumor—and in order to be able to compare these values across datasets—we calculated the region scores as follows. We started out with normalized data. First, probe sets with maximal expression values were selected whenever additional probe sets recognized the same gene using the “collapse to gene symbol” function in GSEA. Then, probe sets recognizing the same genes from the three different platforms were extracted using the gene symbol as an identifier, resulting in 6,654 annotated genes. Data were then row standardized within each dataset by subtracting the mean expression from each expression value for each gene, and dividing by the standard deviation for that gene. Finally, the standardized datasets were combined to a training set consisting of 1,239 tumors and a test set containing 347 tumors. The combination of row standardized datasets assumes that the sample characteristics (such as the clinical parameters) are comparable between the datasets. This introduced a bias in the training set because the dataset contained a variable fraction of lymph node-positive samples and patients receiving adjuvant treatment. However, we retained the node-positive samples to obtain a reasonable number of ERBB2 and basal-like tumors. Patients with these tumors are more likely to have lymph node metastasis and are more likely to receive adjuvant medical treatment.
For the chromosomal regions and pathways, we calculated a score for each sample as the mean standardized expression values for all genes in the gene set. The score is thus a continuous variable representing the overall expression level in the region or pathway. The region score is related to the DNA copy number. However, using gene expression data alone, it is not possible to define a cut-off region in the score between different DNA copy numbers.
The association of region and pathway scores to survival was analyzed by Kaplan–Meyer plots and a log-rank test using surv and pchisq packages, respectively, in R. A high score (region score or pathway score) subset was defined as the third of samples having the highest score, and a low score subset included the third with the lowest score values. The middle third was excluded from the analysis. Hazard ratios were calculated by fitting a Cox model to the data, assuming a constant hazard ratio over time. The P-values were calculated using a log-rank test, with the null hypothesis postulating that there were no differences in survival between the groups.
Validation of regions and pathways involved in metastasis
Validation of prognostic gene sets was performed in the latest published datasets (Mainz, TRANSBIG-S). The patients in these datasets are all node-negative and have not received adjuvant medical treatment; both studies were performed with Affymetrix HG133A chips minimizing bias from the treatment effect, clinical differences, and platform differences. For the validation of metastatic mechanisms in basal-like tumors, 41 basal-like tumors from the Mainz data were used. TRANSBIG-S data where the identification of molecular subtypes failed were excluded for that analysis. For ERBB2-amplified tumors, the number in the Mainz dataset was too small, and the TRANSBIG-S dataset was included, resulting in 347 samples. Instead of using single-sample prediction, we classified tumor ERBB2 status as 20% of tumors with the highest ERBB2 expression. The cut-off was set to 20% of tumors, because this resembles the frequency of ERBB2 amplification in breast cancer.
The region and pathway scores were calculated as described for the training set described above, and once again, the extreme groups (one-third of the highest scores and one-third of the lowest scores) were compared by survival analysis.
Supplementary Materials
Table S1.
GSEA meta-analysis within subtypes and survival analysis in training sets.
| Direction in metastasis | Region or pathway | Mean ranking value | Max ranking value | P-value | fdr | Survival analysis P-value |
|---|---|---|---|---|---|---|
| Basal | ||||||
| Up | 5q14 | 26.8 | 188 | 0.0004 | 0.05 | 0.007 |
| 8q24 | 28.2 | 188 | 0.0005 | 0.05 | 0.17 | |
| Down | TNFR2 | 290 | 347 | 0.0002 | 0.04 | 0.009 |
| ERBB2 | ||||||
| Up | 16Q22 | 27.4 | 188 | 0.0004 | 0.05 | 0.15 |
| Xq22 | 28.4 | 188 | 0.0005 | 0.05 | 0.98 | |
| Fructose_mannose | 32.5 | 347 | 0.00004 | 0.01 | 0.32 | |
| Down | 5q33 | 165 | 188 | 0.0002 | 0.04 | 0.08 |
| IL-12 | 303 | 347 | 0.0002 | 0.04 | 0.004 | |
| GPCRDB | 313 | 347 | 0.00006 | 0.02 | 0.15 | |
Distribution of ERBB2 and GRB7 expression in test set.
Notes: For validation of metastatic mechanisms in ERBB2 positive tumors, a test set including TRANSBIC-S and Mainz was used resulting in a total of 347 samples. Instead of using single sample prediction, we classified tumor ERBB2 status as 20% of tumors with highest ERBB2 expression.
5q14, 321 basale.
Footnotes
Author Contributions
Conceived and designed the experiments: MT, TAK. Analyzed the data: MT, QT. Wrote the first draft of the manuscript: MT. Contributed to the writing of the manuscript: MT, MB, QT, TAK. Agreed with manuscript results and conclusions: MT, MB, QT, TAK. Jointly developed the structure and arguments for the paper: MT, MB, QT, TAK. Made critical revisions and approved final version: MT, MB, QT, TAK. All authors reviewed and approved of the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
Funding
Author(s) disclose no funding sources.
References
- 1.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 3.Desmedt C, Piette F, Loi S, et al. TRANSBIG Consortium. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 4.Thomassen M, Tan Q, Eiriksdottir F, Bak M, Cold S, Kruse TA. Prediction of metastasis from low-malignant breast cancer by gene expression profiling. Int J Cancer. 2007;120(5):1070–5. doi: 10.1002/ijc.22449. [DOI] [PubMed] [Google Scholar]
- 5.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt M, Böhm D, von Törne C, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68(13):5405–13. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 10.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8(8):R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies metastatic pathways and transcription factors in breast cancer. BMC Cancer. 2008;8:394. doi: 10.1186/1471-2407-8-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res Treat. 2009;113(2):239–49. doi: 10.1007/s10549-008-9927-2. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich K, Weber T, Scheithauer J, et al. Chromosomal genotype in breast cancer progression: comparison of primary and secondary manifestations. Cell Oncol. 2008;30(1):39–50. doi: 10.1155/2008/209142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson E, Danielsson A, Delle U, Olsson B, Karlsson P, Helou K. Chromosomal changes associated with clinical outcome in lymph node-negative breast cancer. Cancer Genet Cytogenet. 2007;172(2):139–46. doi: 10.1016/j.cancergencyto.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 15.Callaghan KA, Becker TE, Ellsworth DL, Hooke JA, Ellsworth RE, Shriver CD. Genomic instability and the development of metastatic lymph node tumors. Ann Surg Oncol. 2007;14(11):3125–32. doi: 10.1245/s10434-007-9504-7. [DOI] [PubMed] [Google Scholar]
- 16.Adélaïde J, Finetti P, Bekhouche I, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67(24):11565–75. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 17.Hansen LL, Yilmaz M, Overgaard J, Andersen J, Kruse TA. Allelic loss of 16q23.2–24.2 is an independent marker of good prognosis in primary breast cancer. Cancer Res. 1998;58(10):2166–9. [PubMed] [Google Scholar]
- 18.Götte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66(21):10233–7. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 19.Cui LF, Guo XJ, Wei J, et al. Overexpression of TNF-alpha and TNFRII in invasive micropapillary carcinoma of the breast: clinicopathological correlations. Histopathology. 2008;53(4):381–8. doi: 10.1111/j.1365-2559.2008.03128.x. [DOI] [PubMed] [Google Scholar]
- 20.Parihar R, Dierksheide J, Hu Y, Carson WE. IL-12 enhances the natural killer cell cytokine response to Ab-coated tumor cells. J Clin Invest. 2002;110(7):983–92. doi: 10.1172/JCI15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parihar R, Nadella P, Lewis A, et al. A phase I study of interleukin 12 with trastuzumab in patients with human epidermal growth factor receptor-2-overexpressing malignancies: analysis of sustained interferon gamma production in a subset of patients. Clin Cancer Res. 2004;10(15):5027–37. doi: 10.1158/1078-0432.CCR-04-0265. [DOI] [PubMed] [Google Scholar]
- 22.Ménard S, Tomasic G, Casalini P, et al. Lymphoid infiltration as a prognostic variable for early-onset breast carcinomas. Clin Cancer Res. 1997;3(5):817–9. [PubMed] [Google Scholar]
- 23.Aaltomaa S, Lipponen P, Eskelinen M, et al. Lymphocyte infiltrates as a prognostic variable in female breast cancer. Eur J Cancer. 1992;28A(4–5):859–64. doi: 10.1016/0959-8049(92)90134-n. [DOI] [PubMed] [Google Scholar]
- 24.Alexe G, Dalgin GS, Scanfeld D, et al. High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Cancer Res. 2007;67(22):10669–76. doi: 10.1158/0008-5472.CAN-07-0539. [DOI] [PubMed] [Google Scholar]
- 25.Rody A, Holtrich U, Pusztai L, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11(2):R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desmedt C, Haibe-Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14(16):5158–65. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 27.Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45(11):1033–40. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loo LW, Grove DI, Williams EM, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64(23):8541–9. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- 30.Cingoz S, Altungoz O, Canda T, Saydam S, Aksakoglu G, Sakizli M. DNA copy number changes detected by comparative genomic hybridization and their association with clinicopathologic parameters in breast tumors. Cancer Genet Cytogenet. 2003;145(2):108–14. doi: 10.1016/s0165-4608(03)00094-3. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calza S, Hall P, Auer G, et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 2006;8(4):R34. doi: 10.1186/bcr1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
GSEA meta-analysis within subtypes and survival analysis in training sets.
| Direction in metastasis | Region or pathway | Mean ranking value | Max ranking value | P-value | fdr | Survival analysis P-value |
|---|---|---|---|---|---|---|
| Basal | ||||||
| Up | 5q14 | 26.8 | 188 | 0.0004 | 0.05 | 0.007 |
| 8q24 | 28.2 | 188 | 0.0005 | 0.05 | 0.17 | |
| Down | TNFR2 | 290 | 347 | 0.0002 | 0.04 | 0.009 |
| ERBB2 | ||||||
| Up | 16Q22 | 27.4 | 188 | 0.0004 | 0.05 | 0.15 |
| Xq22 | 28.4 | 188 | 0.0005 | 0.05 | 0.98 | |
| Fructose_mannose | 32.5 | 347 | 0.00004 | 0.01 | 0.32 | |
| Down | 5q33 | 165 | 188 | 0.0002 | 0.04 | 0.08 |
| IL-12 | 303 | 347 | 0.0002 | 0.04 | 0.004 | |
| GPCRDB | 313 | 347 | 0.00006 | 0.02 | 0.15 | |
Distribution of ERBB2 and GRB7 expression in test set.
Notes: For validation of metastatic mechanisms in ERBB2 positive tumors, a test set including TRANSBIC-S and Mainz was used resulting in a total of 347 samples. Instead of using single sample prediction, we classified tumor ERBB2 status as 20% of tumors with highest ERBB2 expression.
5q14, 321 basale.