Abstract
Epigenetic regulation of gene expression orchestrates dynamic cellular processes that become perturbed in human disease. An understanding of how subversion of chromatin-mediated events leads to pathologies such as cancer and neurodevelopmental syndromes may offer better treatment options for these pathological conditions. Chromodomain Helicase DNA-binding protein 5 (CHD5) is a dosage-sensitive tumor suppressor that is inactivated in human cancers, including neural-associated malignancies such as neuroblastoma and glioma. Here we report a detailed analysis of the temporal and cell type-specific expression pattern of Chd5 in the mammalian brain. By analyzing endogenous Chd5 protein expression during mouse embryogenesis, in the neonate, and in the adult, we found that Chd5 is expressed broadly in multiple brain regions, that Chd5 sub-cellular localization undergoes a switch from the cytoplasm to the nucleus during mid-gestation, and that Chd5 expression is retained at high levels in differentiated neurons of the adult. These findings may have important implications for defining the role of CHD5-mediated chromatin dynamics in the brain and for elucidating how perturbation of these epigenetic processes leads to neuronal malignancies, neurodegenerative diseases, and neurodevelopmental syndromes.
1. Introduction
Chromodomain Helicase DNA-binding protein 5 (CHD5) is a predicted chromatin remodeling protein implicated in human cancer (Bagchi and Mills, 2008b; Bagchi et al., 2007; Fujita et al., 2008; Thompson et al., 2003). As a member of the CHD class of ATP-dependent chromatin remodelers, CHD5 contains functional motifs including chromo, ATP-dependent helicase, and DNA binding domains (Thompson et al., 2003). CHD proteins have been shown to modulate transcriptional activation, repression, and elongation (Murawska and Brehm, 2011). Like its closest family members CHD3 and CHD4, CHD5 has dual plant homeodomains (PHDs) that mediate binding to unmodified histone 3 (H3) (Oliver et al., 2012; Paul et al., 2013), an interaction we showed essential for tumor suppression (Paul et al., 2013). CHD5 maps to 1p36, a region of the genome frequently deleted in diverse human cancers (Aarts et al., 2006; Barbashina et al., 2005; Bello et al., 1994; Bello et al., 1995; Bieche et al., 1993; Bieche et al., 1999; Brodeur et al., 1977; Caron et al., 2001; Cheung et al., 2005; Dong et al., 2004; Dracopoli et al., 1989; Ezaki et al., 1996; Felsberg et al., 2004; Godfried et al., 2002; Hoggard et al., 1995; Kleer et al., 2000; Leister et al., 1990; Melendez et al., 2003; Moley et al., 1992; Mori et al., 2003; Mori et al., 1998; Mulligan et al., 1993; Nagai et al., 1995; Piaskowski et al., 2005; Poetsch et al., 2003; Praml et al., 1995; Wada et al., 1988; White et al., 1995; White et al., 2005; Zhou et al., 2004). In addition to being commonly deleted, CHD5 is silenced by methylation in neuroblastoma (Koyama et al., 2012), lung cancer (Zhao et al., 2011), and colorectal cancer (Mokarram et al., 2009; Mulero-Navarro and Esteller, 2008; Wang et al., 2009); CHD5 is mutated in cancers of the breast (Sjoblom et al., 2006), ovary (Bell D, 2011; Gorringe et al., 2008; Jones et al., 2010), prostate (Berger et al., 2011; Robbins et al., 2011), liver (Li et al., 2011) as well as in squamous cell carcinoma (Agrawal et al., 2011), neuroblastoma (Okawa et al., 2008), glioma (McLendon, 2008), and melanoma (Lang et al., 2011); and CHD5 levels correlate directly with survival following anti-cancer therapy (Garcia et al., 2010b; Koyama et al., 2012; Wong et al., 2011).
While a cursory view of Chd5 expression has been reported in brain (Egan et al., 2013; Garcia et al., 2010a; Potts et al., 2011; Thompson et al., 2003), a detailed characterization of the pattern of Chd5 expression in the brain during development, in the neonate, and in the adult has not, to our knowledge, been reported thus far. Our interest in the function of Chd5 in the brain is three-fold. First, CHD5 is frequently inactivated in neuronal malignancies such as neuroblastoma and glioma. Second, we have defined Chd5 as a regulator of Cdkn2a/b, a locus encoding components of pathways that regulate renewal of neural stem cells (Bagchi et al., 2007; Bruggeman et al., 2005). Third, recent exome sequencing efforts implicate CHD proteins in autism (O’Roak et al., 2012). As a step to define the role that Chd5 plays in brain homeostasis and in neuronal malignancies, we assessed precisely when and where it is expressed in the developing brain.
2. Results
We used immunohistochemistry (IHC) to detect Chd5 protein expression in the brain throughout development, including embryonic, neonatal, and adult stages. After demonstrating that the available antibody was specific for Chd5 (Supplemental Figure 1), we analyzed an extensive developmental series starting at E8.5, throughout gestation, and into postnatal and adult stages of development. We performed co-immunostaining with a panel of neuronal, glial, and progenitor markers in order to define the cell type in which Chd5 was expressed.
2.1 Chd5 expression during embryogenesis and post-natal stages
Chd5 expression was first detectable at embryonic day 10.5 (E10.5) of development (Figure 1A). At this developmental stage, Chd5 was detected in the cytoplasm of cells of the ventral rhombencephalon and the posterior mesencephalon, where it was expressed in Nestin-positive neuronal progenitors (Figure 1B; Supplemental Figure 2). Chd5 was also detected in dorsal root ganglia, a non-CNS neuronal tissue, where it co-localized with NeuN-expressing differentiating neurons (Figure 1B).
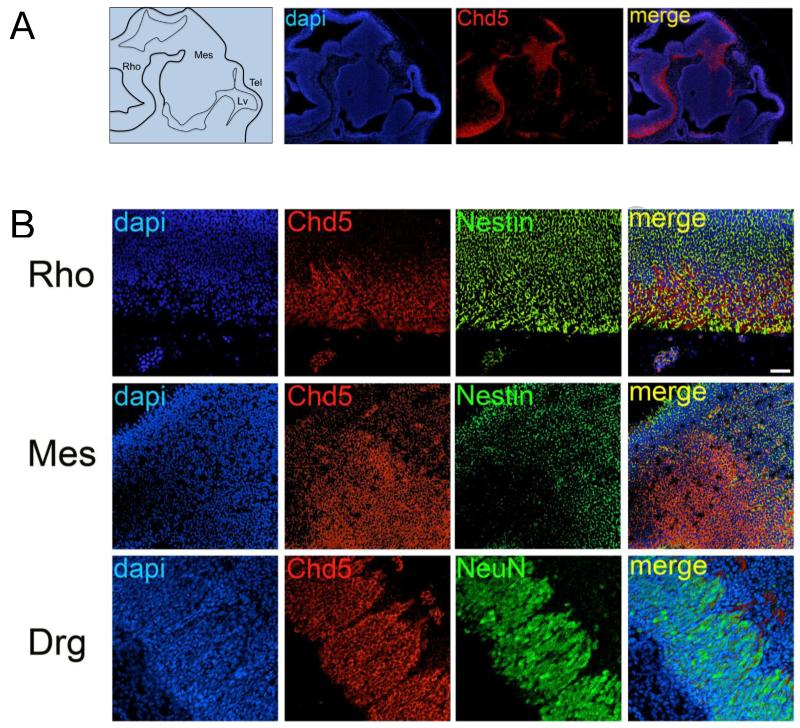
Figure 1. Chd5 expression in brain of E10.5 embryos.
A) Left panel: Schematic of E10.5 brain. Right panels: At embryonic stage E10.5 cytoplasmic Chd5 is detected in a cytoplasmic pattern in the ventral rhombencephalon and posterior mesencephalon. Scale bar, 100μM. Co-localization coefficient for merged panel = −0.277. B) Upper panel: Chd5 is detected in the cytoplasm of Nestin-positive progenitors in the ventral rhombencephalon. Middle panel: Chd5 is detected in the posterior mesencephalon. Lower panel: Chd5 is localized in the cytoplasm of NeuN-positive differentiating neurons in the dorsal root ganglia. Scale bar, 50μM. Rho, rhombencephalon; Mes, mesencephalon; Tel, telencephalon; Lv, lateral ventricle. Co-localization coefficient for merged panels = 0.160 (upper panel), 0.174 (middle panel), 0.441 (lower panel).
In brains of E13.5 embryos, the pattern of Chd5 expression changed significantly; at this stage it was expressed in the mesencephalon, rhombencephalon, and telencephalon (Figure 2A). Chd5 expression in the mesencephalon and rhombencephalon was localized to the nucleus (Figure 2B; Supplemental Figure 3). In these regions Chd5 expression overlapped predominantly with NeuN, a marker of neurons. However, a portion of Chd5-expressing regions also expressed Nestin, a marker of neuronal progenitor cells. In the telencephalon, Chd5 was detected in the cytoplasm in the upper layers of the developing cortex, where newly formed differentiating neurons reside, as seen by the presence of Chd5-positive/NeuN-positive neurons. Whereas Nestin-positive progenitor cells of the telencephalon were located in the lower layers, the majority of Chd5-positive cells were located in the upper layers.
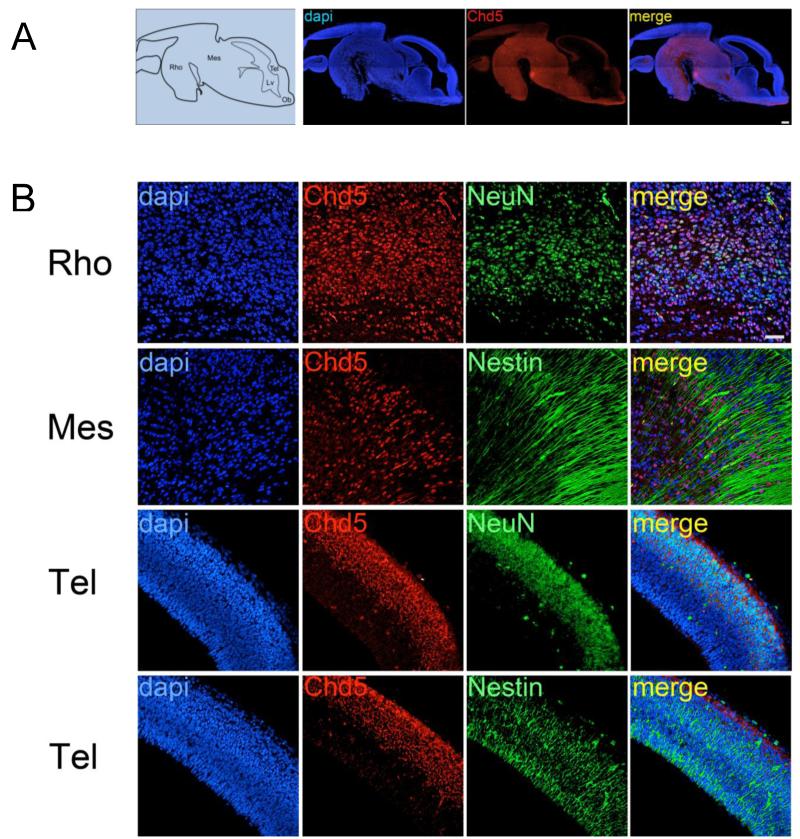
Figure 2. Chd5 expression in brain of E13.5 embryos.
A) Left panel: schematic of E13.5 brain. Right panels: Chd5 is detected in the nucleus throughout the in the rhombencephalon and mesencephalon. Scale bar, 200μM. Co-localization coefficient for merged panel = 0.66. B) Upper panels: Chd5 co-localizes predominantly with NeuN in neurons in the rhombencephalon. Second row panels: In some brain regions, Chd5 is expressed in Nestin-positive progenitors. Third row panels: Chd5 is detected in the cytoplasm of differentiating NeuN-positive neurons located in the dorsal region of the telencephalon. Fourth row panels: Chd5 expression does not co-localize with Nestin-positive progenitors in the ventral region of the telencephalon. Scale bar, 50μM. Rho, rhombencephalon; Mes, mesencephalon; Tel, telencephalon; Lv, lateral ventricle; Ob, olfactory bulb. Co-localization coefficients for merged panels = 0.614 (upper panel), −0.222 (upper middle panel), 0.259 (lower middle panel), 0.263 (lower panel).
At late stages of gestation (E18.5), Chd5 expression was detected in the nucleus in all brain regions analyzed, and appeared to be specific to NeuN-positive differentiated neurons (Figure 3A). This nuclear and neuronal-specific expression pattern of Chd5 observed in brains of E18.5 embryos persisted throughout post-natal stages of development (Figure 3B). To determine whether Chd5 was expressed earlier stages of neurogenesis, we analyzed the pattern of Tuj1 expression, a marker of less differentiated neurons (Figure 3B). This demonstrated that Chd5 is expressed at early stages of neuronal differentiation.
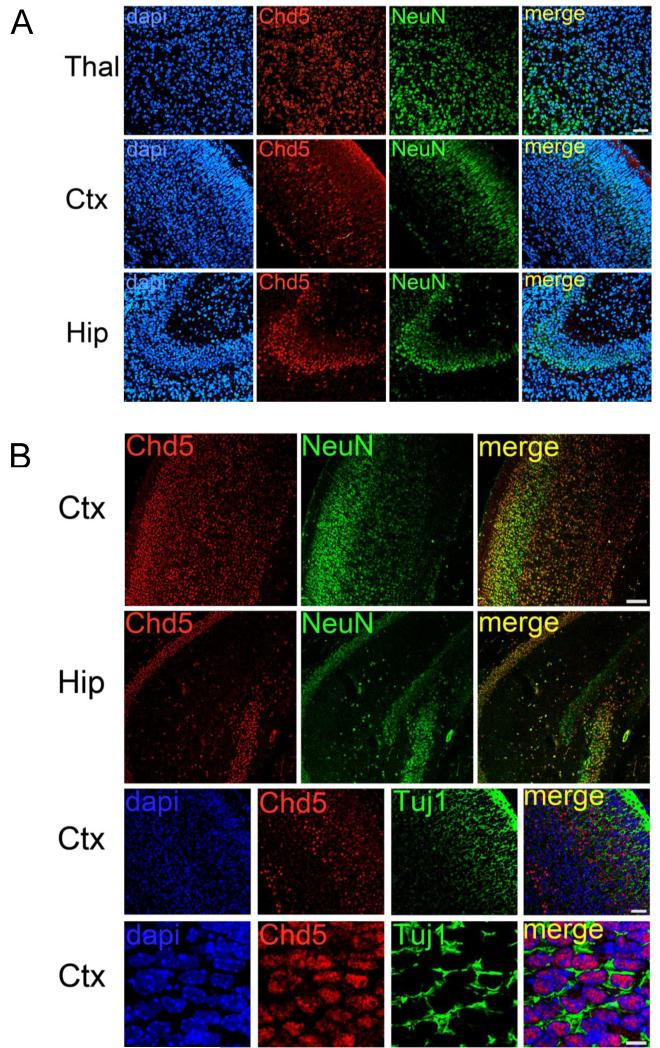
Figure 3. Chd5 expression in brain of E18.5 embryos and P7 neonates.
A) Starting at embryonic stage E18.5, Chd5 is expressed in NeuN-positive neurons throughout the brain. Upper row panels: thalamus. Middle row panels: cortex. Lower row panels: hippocampus. Scale bar, 50μM. Co-localization coefficients for merged panels = 0.641 (upper panel), 0.336 (middle panel), 0.655 (lower panel). B) In P7 neonatal brains, Chd5 is expressed in NeuN-positive neurons. Two upper row panels: Chd5 is expressed in all cortical layers; Chd5 is expressed in the CA1-3 regions of the hippocampus but is not detected in the P7 dentate gyrus. Two lower panels: Chd5 is expressed in Tuj1-positive differentiating neurons. Scale bar, 100μM (two upper rows), 50μM (third row), 10μM (fourth row). Thal, thalamus; Ctx, cortex; Hip, hippocampus. Co-localization coefficients for merged panels = 0.66 (upper panel), 0.62 (upper middle panel), −0.1 (lower middle panel), −0.143 (lower panel).
2.2 Chd5 expression in adult mouse brain
In adult mice, Chd5 was detected in NeuN-positive neurons throughout the brain (Figure 4A, B). In the cortex, Chd5 expression was observed in all cortical layers, being more highly expressed in cortical layers 2 and 3. In the hippocampus, Chd5 was detected robustly in CA1-3 regions, and was detected at lower levels in the dentate gyrus. Chd5 was detected in the caudate-putamen, thalamus, and olfactory bulb, albeit not as strongly as in cortex and hippocampus. In the cerebellum, Chd5 was detected most robustly in Purkinje neurons, as determined by co-localization of Chd5 with Calb1. In addition, Chd5 was detected at relatively lower levels in molecular layer and granule layer neurons of the cerebellum.
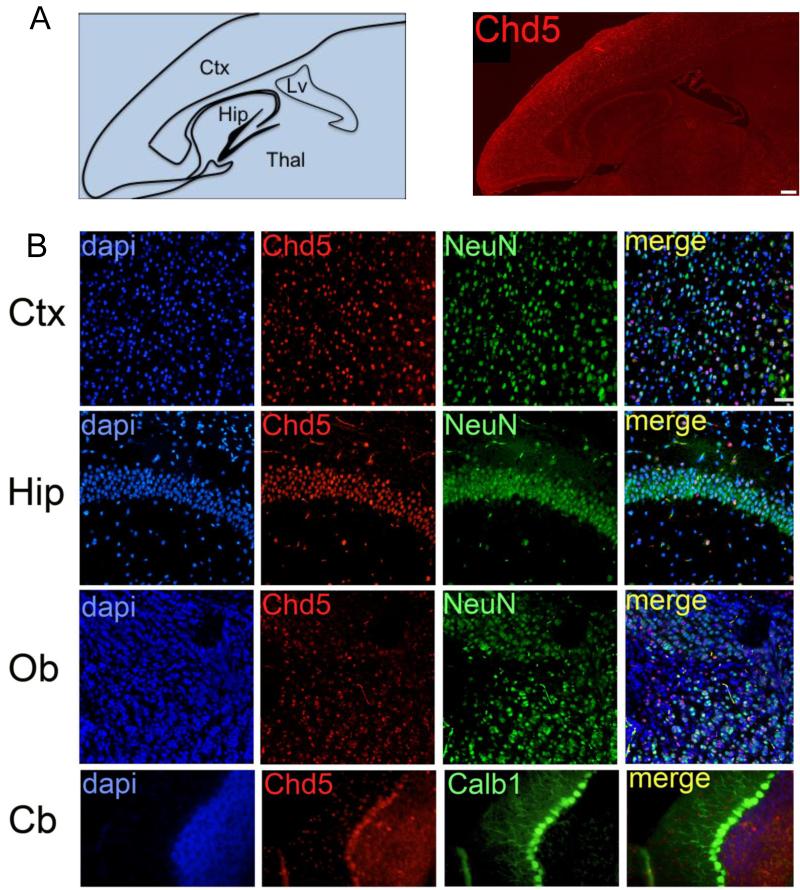
Figure 4. Chd5 expression in adult brain.
A) Left panel: schematic of adult P45 brain. Right panel: Chd5 is expressed throughout the adult brain. Scale bar, 400μM. B) Upper row panels: Chd5 is expressed in NeuN-positive neurons of the cortex. Second row panels: Chd5 is expressed in NeuN-positive neurons in the CA1 region of the hippocampus. Third row panels: Chd5 is expressed in NeuN-positive cells of the olfactory bulb. Fourth row panels: Chd5 is expressed robustly in Calb1-positive Purkinje neurons of the cerebellum, and is expressed at lower levels in molecular layer and granule neurons of the cerebellum. Scale bar, 50μM. Thal, thalamus; Ctx, cortex; Hip, hippocampus; Lv, lateral ventricle; Ob, olfactory bulb; Cb, cerebellum. Co-localization coefficients for merged panels = 0.72 (upper panel), 0.606 (upper middle panel), 0.503 (lower middle panel), 0.063 (lower panel).
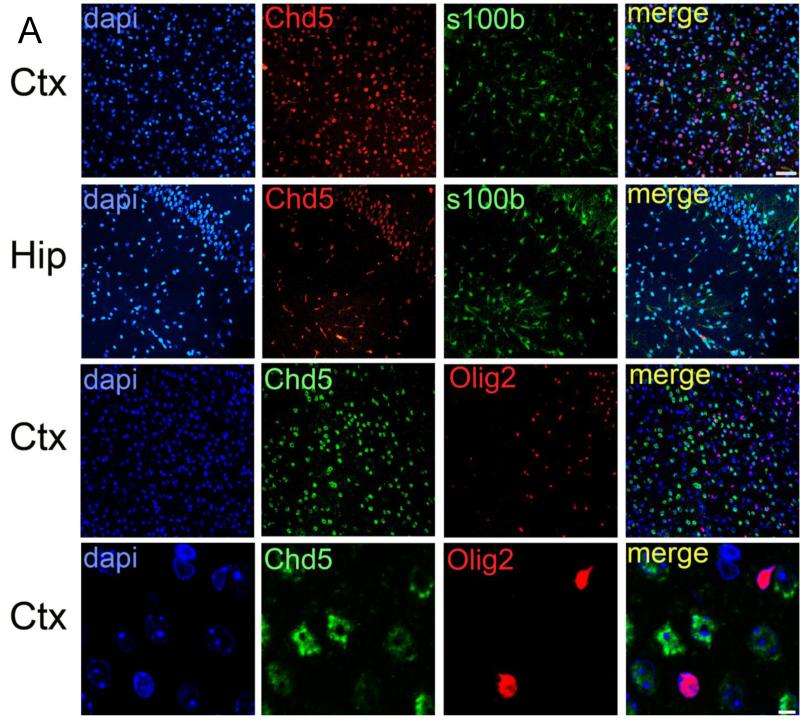
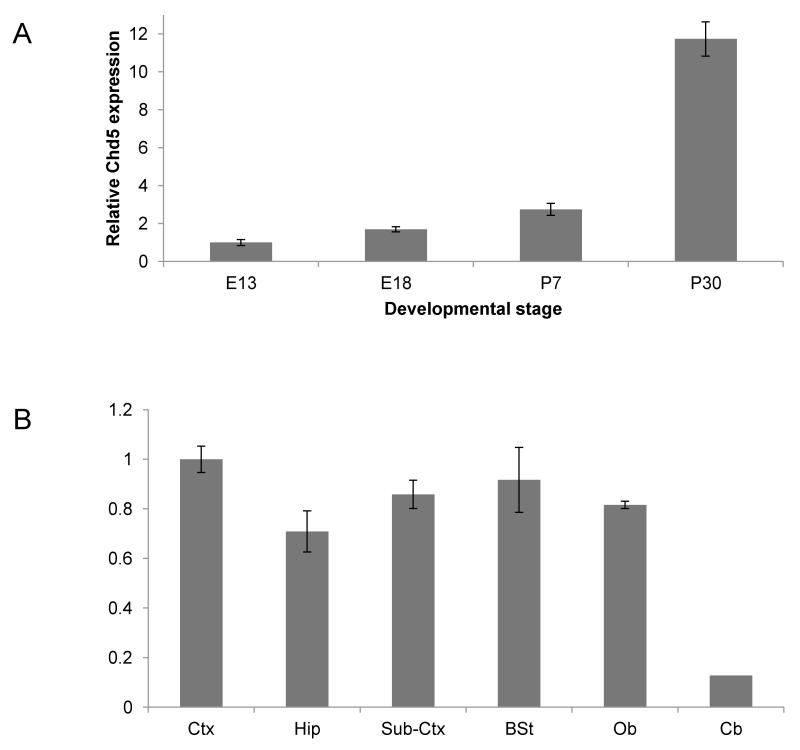
Chd5 expression did not co-localize with S100b or Olig2 in either postnatal or adult stages, indicating that Chd5 expression is not detectable in astrocytes or oligodendrocytes (Figure 5). qPCR analyses indicated that Chd5 transcript is induced as neurogenesis proceeds during development, and that Chd5 is expressed robustly in many regions of the brain (Figure 6).
Figure 5. Chd5 expression is not detectable in astrocytes or oligodendrocytes.
Chd5 is not detected in S100b-positive astrocytes of the cortex (upper panels) or hippocampus (upper middle panels). Chd5 is not detected in Olig2-positive oligodendrocytes of the cortex (two lower panels). Scale bar, 50μM (upper three rows), 5μM (bottom row). Ctx, cortex; Hip, hippocampus. Co-localization coefficients for merged panels = 0.074 (upper panel), 0.081 (upper middle panel), −0.057 (lower middle panel), −0.067 (lower panel).
Figure 6. Developmental and regional expression of Chd5 in the brain.
A) qPCR analysis of Chd5 expression during embryogenesis and after birth. B) qPCR analysis of Chd5 expression in different regions of the brain. Ctx, cortex; Hip, hippocampus, Sub-Ctx, sub-cortex; BSt, brain stem; Ob, olfactory bulb; Cb, cerebellum.
3. Discussion
Up to the present, only a very limited characterization of Chd5 expression in the mammalian brain existed (Egan et al., 2013; Garcia et al., 2010a; Thompson et al., 2003). A more extensive view of the dynamics of Chd5 expression during brain development and maturation is crucial for understanding the role that Chd5 plays in neurogenesis and in brain homeostasis. Moreover, a better understanding of the pattern of Chd5 expression might help to shed light on Chd5’s previously characterized role as a tumor suppressor, and more specifically its involvement in tumorigenesis in the brain.
Our characterization of Chd5 is the first in-depth spatiotemporal report of a member of the Chd family of chromatin remodelers. Although other Chd family members, such as Chd3, Chd4, Chd7, and Chd8 have been reported to be expressed in brain (Batsukh et al., 2010; Feng et al., 2013; Potts et al., 2011), a detailed spatiotemporal characterization such as the one performed here for Chd5 is still lacking. Given the involvement of CHD8 mutations in autism (O’Roak et al., 2012), it is essential to define the dynamics and pattern of CHD expression in the brain to determine how different members of the CHD family interact with one another. These expression data will set the stage for assessing how CHD proteins regulate chromatin-mediated events vital for brain function and for determining how perturbation of these epigenetic events cause brain connectivity to go awry in the disease state.
Our findings presented here show that Chd5 is a neuronal marker in newborn and adult mouse brain, as it is detected throughout the brain in neurons but is not detectable in astrocytes or oligodendrocytes. Chd5’s presence in progenitor cells of the brain during embryogenesis at a stage when neuronal progenitors initiate differentiation and shift from a proliferative into a post-mitotic state is in agreement with Chd5’s functional role as a tumor suppressor and its ability to inhibit cellular proliferation (Bagchi and Mills, 2008a; Bagchi et al., 2007). During neuronal differentiation, anti-proliferative mechanisms are put into action and Chd5 plays a major part in this process, due to its ability to activate the p16/Rb and p19/p53-mediated pathways. Interestingly, it is at this same phase of differentiation that Chd5 expression shifts its localization from the cytoplasm to the nucleus, which is the expected sub-cellular location for this putative chromatin remodeler. Whether this shift is the result of cytoplasmic retention of the protein until a proper developmental cue or the expression of a different isoform remains to be explored.
Chd5 expression is induced in differentiating neuronal progenitors and is sustained in neuronal progeny, indicating a role for Chd5 in neuronal differentiation and/or maturation. We suggest a role for Chd5 in the process of neurogenesis based on the pattern of its expression in differentiating neurons as they shift from a progenitor state into a mature neuron. Presumably such function, when impaired, could set the stage for malignancies of the brain. While we previously implicated CHD5 loss in human glioma, our findings here indicate that Chd5 is not expressed in glial cells, suggesting the possibility that Chd5-mediated chromatin dynamics controls cell fate decisions. Whether compromised Chd5 has a functional consequence in the brain remains to be determined. Interestingly, CHD family members have recently been implicated in neurodevelopmental disorders, such as CHD8 in autism (O’Roak et al., 2011). Taken together, these findings suggest a new role for chromatin remodelers and Chd5 in particular, as key players in the brain.
4. Experimental procedures
4.1. Immunohistochemistry and microscopy
Brains were dissected from euthanized animals, fixed in 4% formaldehyde in PBS (Gibco) for 15 minutes at room temperature, followed by three 5 minutes washes in PBS. Fixed brains were sectioned at 10μm and embedded in paraffin. Sections were deparaffinized, rehydrated, processed in a pressure cooker in Tris-EDTA buffer (pH=9), and blocked in goat serum (Gibco, Life Technologies, USA) at RT for 30 minutes. Sections were incubated with rabbit anti-Chd5 (1:250, Santa Cruz sc-68389), mouse anti-NeuN (1:500, Millipore MAB377), mouse anti-Nestin (1:1000 Abcam ab6142), rabbit anti-Olig2 (1:500, Millipore AB9610), mouse anti-s100b (1:200, Abcam ab4066), mouse anti-Calb1 (1:500 Sigma Aldrich C9848). The secondary antibodies used were goat anti-rabbit Alexa Fluor 594 (1:2000, Molecular Probes A11037), goat anti-mouse Alexa Fluor 488 (1:2000, Molecular Probes A10680). Sections were mounted using Vectashield (Vector, H1000). Slides were visualized and imaged using the Zeiss LSM 710 confocal microscope. Quantification of co-localization coefficients was performed using Velocity 6.3.
Supplementary Material
Supplemental Figure 1. Characterization of the anti-Chd5 antibody
A) IHC of minus primary antibody control does not produce spurious signal in brain. B) IHC using a distinct anti-Chd5 antibody (Paul et al., 2013) produces a similar pattern in brain. C) Incubation with a Chd5 blocking peptide abrogates IHC signal in brain. D) Western analysis in whole brain lysates generates a distinct band for Chd5. Scale bars, 50μM.
Supplemental Figure 2. Cytoplasmic localization of Chd5
Chd5 (red) is expressed in the cytoplasm of E10.5 rhombencephalon. Scale bar, 20μM.
Supplemental Figure 3. Nuclear localization of Chd5 correlates with stage of neurogenesis.
A) Wide-field view of forebrain of E13.5 embryo shows pattern of Chd5 expression (red). Scale bar, 200μM. B) High magnification view of panel A, showing border of intense vs. less intense Chd5 expression. Scale bar, 100μM. C) Similar region as shown panel B, showing expression of Chd5 (red) and NeuN (green). Note that Chd5 expression is nuclear in NeuN-positive differentiated neurons. Scale bar, 50μM.
Highlights.
Chd5 is expressed in many regions throughout the brain.
The subcellular localization of Chd5 switches from cytoplasmic to nuclear during mid-embryogenesis.
Chd5 is expressed robustly in differentiated neurons.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarts M, Dannenberg H, deLeeuw RJ, van Nederveen FH, Verhofstad AA, Lenders JW, Dinjens WN, Speel EJ, Lam WL, de Krijger RR. Microarray-based CGH of sporadic and syndrome-related pheochromocytomas using a 0.1-0.2 Mb bacterial artificial chromosome array spanning chromosome arm 1p. Genes Chromosomes Cancer. 2006;45:83–93. doi: 10.1002/gcc.20268. [DOI] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Research. 2008;68:2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res. 2005;11:1119–1128. [PubMed] [Google Scholar]
- Batsukh T, Pieper L, Koszucka AM, von Velsen N, Hoyer-Fender S, Elbracht M, Bergman JEH, Hoefsloot LH, Pauli S. CHD8 interacts with CHD7, a protein which is mutated in CHARGE syndrome. Human molecular genetics. 2010;19:2858–2866. doi: 10.1093/hmg/ddq189. [DOI] [PubMed] [Google Scholar]
- Bell D. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. e.a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello MJ, de Campos JM, Kusak ME, Vaquero J, Sarasa JL, Pestana A, Rey JA. Molecular analysis of genomic abnormalities in human gliomas. Cancer Genet Cytogenet. 1994;73:122–129. doi: 10.1016/0165-4608(94)90195-3. [DOI] [PubMed] [Google Scholar]
- Bello MJ, Leone PE, Vaquero J, de Campos JM, Kusak ME, Sarasa JL, Pestana A, Rey JA. Allelic loss at 1p and 19q frequently occurs in association and may represent early oncogenic events in oligodendroglial tumors. Int J Cancer. 1995;64:207–210. doi: 10.1002/ijc.2910640311. [DOI] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Champeme M-H, Matifas F, Cropp CS, Callahan R, Lidereau R. Two distinct regions involved in 1p deletion in human primary breast cancer. Cancer Research. 1993;53:1990–1994. [PubMed] [Google Scholar]
- Bieche I, Khodja A, Lidereau R. Deletion mapping of chromosomal region 1p32-pter in primary breast cancer. Genes Chromosomes Cancer. 1999;24:255–263. doi: 10.1002/(sici)1098-2264(199903)24:3<255::aid-gcc11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Sekhon G, Goldstein MN. Chromosomal aberrations in human neuroblastomas. Cancer. 1977;40:2256–2263. doi: 10.1002/1097-0142(197711)40:5<2256::aid-cncr2820400536>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H, Spieker N, Godfried M, Veenstra M, van Sluis P, de Kraker J, Voute P, Versteeg R. Chromosome bands 1p35-36 contain two distinct neuroblastoma tumor suppressor loci, one of which is imprinted. Genes Chromosomes Cancer. 2001;30:168–174. doi: 10.1002/1098-2264(200102)30:2<168::aid-gcc1072>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Cheung TH, Lo KW, Yim SF, Poon CS, Cheung AY, Chung TK, Wong YF. Clinicopathologic significance of loss of heterozygosity on chromosome 1 in cervical cancer. Gynecol Oncol. 2005;96:510–515. doi: 10.1016/j.ygyno.2004.10.035. [DOI] [PubMed] [Google Scholar]
- Dong Z, Pang JS, Ng MH, Poon WS, Zhou L, Ng HK. Identification of two contiguous minimally deleted regions on chromosome 1p36.31-p36.32 in oligodendroglial tumours. Br J Cancer. 2004;91:1105–1111. doi: 10.1038/sj.bjc.6602093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracopoli NC, Harnett P, Bale SJ, Stanger BZ, Tucker MA, Housman DE, Kefford RF. Loss of alleles from the distal short arm of chromosome 1 occurs late in melanoma tumor progression. Proc Natl Acad Sci U S A. 1989;86:4614–4618. doi: 10.1073/pnas.86.12.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O’Connell DJ, Rraklli V, Dolan MJ, Chadderton N, Hansen K, et al. CHD5 Is Required for Neurogenesis and Has a Dual Role in Facilitating Gene Expression and Polycomb Gene Repression. Developmental cell. 2013;26:223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Ezaki T, Yanagisawa A, Ohta K, Aiso S, Watanabe M, Hibi T, Kato Y, Nakajima T, Ariyama T, Inazawa J, et al. Deletion mapping on chromosome 1p in well-differentiated gastric cancer. Br J Cancer. 1996;73:424–428. doi: 10.1038/bjc.1996.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsberg J, Erkwoh A, Sabel MC, Kirsch L, Fimmers R, Blaschke B, Schlegel U, Schramm J, Wiestler OD, Reifenberger G. Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol. 2004;14:121–130. doi: 10.1111/j.1750-3639.2004.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Khan MA, Bellvis P, Zhu Z, Bernhardt O, Herold-Mende C, Liu HK. The Chromatin Remodeler CHD7 Regulates Adult Neurogenesis via Activation of SoxC Transcription Factors. Cell stem cell. 2013;13:62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Fujita T, Igarashi J, Okawa ER, Gotoh T, Manne J, Kolla V, Kim J, Zhao H, Pawel BR, London WB, et al. CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. Journal of the National Cancer Institute. 2008;100:940–949. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia I, Mayol G, Rodriguez E, Sunol M, Gershon TR, Rios J, Cheung NK, Kieran MW, George RE, Perez-Atayde AR, et al. Expression of the neuron-specific protein CHD5 is an independent marker of outcome in neuroblastoma. Molecular cancer. 2010a;9:277. doi: 10.1186/1476-4598-9-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfried MB, Veenstra M, Valent A, Sluis P, Voute PA, Versteeg R, Caron HN. Lack of interstitial chromosome 1p deletions in clinically-detected neuroblastoma. Eur J Cancer. 2002;38:1513–1519. doi: 10.1016/s0959-8049(02)00137-5. [DOI] [PubMed] [Google Scholar]
- Gorringe KL, Choong DY, Williams LH, Ramakrishna M, Sridhar A, Qiu W, Bearfoot JL, Campbell IG. Mutation and methylation analysis of the chromodomain-helicase-DNA binding 5 gene in ovarian cancer. Neoplasia. 2008;10:1253–1258. doi: 10.1593/neo.08718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard N, Brintnell B, Howell A, Weissenbach J, Varley J. Allelic imbalance on chromosome 1 in human breast cancer II microsatellite repeat analysis. Genes, Chromosomes & Cancer. 1995;12:24–31. doi: 10.1002/gcc.2870120105. [DOI] [PubMed] [Google Scholar]
- Jones S, Wang TL, Kurman RJ, Nakayama K, Velculescu VE, Vogelstein B, Kinzler KW, Papadopoulos N, Shih IM. Low-grade serous carcinomas of the ovary contain very few point mutations. J Pathol. 2010 doi: 10.1002/path.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Bryant BR, Giordano TJ, Sobel M, Merino MJ. Genetic Changes in Chromosomes 1p and 17p in Thyroid Cancer Progression. Endocr Pathol. 2000;11:137–143. doi: 10.1385/ep:11:2:137. [DOI] [PubMed] [Google Scholar]
- Koyama H, Zhuang T, Light JE, Kolla V, Higashi M, McGrady PW, London WB, Brodeur GM. Mechanisms of CHD5 Inactivation in Neuroblastomas. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012 doi: 10.1158/1078-0432.CCR-11-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J, Tobias ES, Mackie R. Preliminary evidence for involvement of the tumour suppressor gene CHD5 in a family with cutaneous melanoma. Br J Dermatol. 2011 doi: 10.1111/j.1365-2133.2011.10223.x. [DOI] [PubMed] [Google Scholar]
- Leister I, Weith A, Bruderlein S, Cziepluch C, Kangwanpong D, Schlag P, Schwab M. Human colorectal cancer: high frequency of deletions at chromosome 1p35. Cancer Res. 1990;50:7232–7235. [PubMed] [Google Scholar]
- Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon R. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. e.a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez B, Cuadros M, Robledo M, Rivas C, Fernandez-Piqueras J, Martinez-Delgado B, Benitez J. Coincidental LOH regions in mouse and humans: evidence for novel tumor suppressor loci at 9q22-q34 in non-Hodgkin’s lymphomas. Leuk Res. 2003;27:627–633. doi: 10.1016/s0145-2126(02)00278-3. [DOI] [PubMed] [Google Scholar]
- Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, Nouraie M, Green R, Lee E, Smoot DT, Ashktorab H. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 2009;4:e7012. doi: 10.1371/journal.pone.0007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moley JF, Brother MB, Fong CT, White PS, Baylin SB, Nelkin B, Wells SA, Brodeur GM. Consistent association of 1p loss of heterozygosity with pheochromocytomas from patients with multiple endocrine neoplasia type 2 syndromes. Cancer Res. 1992;52:770–774. [PubMed] [Google Scholar]
- Mori N, Morosetti R, Mizoguchi H, Koeffler HP. Progression of myelodysplastic syndrome: allelic loss on chromosomal arm 1p. Br J Haematol. 2003;122:226–230. doi: 10.1046/j.1365-2141.2003.04434.x. [DOI] [PubMed] [Google Scholar]
- Mori N, Morosetti R, Spira S, Lee S, Ben-Yehuda D, Schiller G, Landolfi R, Mizoguchi H, Koeffler HP. Chromosome band 1p36 contains a putative tumor suppressor gene important in the evolution of chronic myelocytic leukemia. Blood. 1998;92:3405–3409. [PubMed] [Google Scholar]
- Mulero-Navarro S, Esteller M. Chromatin remodeling factor CHD5 is silenced by promoter CpG island hypermethylation in human cancer. Epigenetics-Us. 2008;3:210–215. doi: 10.4161/epi.3.4.6610. [DOI] [PubMed] [Google Scholar]
- Mulligan LM, Gardner E, Smith BA, Mathew CG, Ponder BA. Genetic events in tumour initiation and progression in multiple endocrine neoplasia type 2. Genes Chromosomes Cancer. 1993;6:166–177. doi: 10.1002/gcc.2870060307. [DOI] [PubMed] [Google Scholar]
- Murawska M, Brehm A. CHD chromatin remodelers and the transcription cycle. Transcription. 2011;2:244–253. doi: 10.4161/trns.2.6.17840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Negrini M, Carter SL, Gillum R, Rosenberg AL, Schwartz GF, Croce CM. Detection and cloning of a common region of loss of heterozygosity at chromosome 1p in breast cancer. Cancer Research. 1995;55:1752–1757. [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature genetics. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa ER, Gotoh T, Manne J, Igarashi J, Fujita T, Silverman KA, Xhao H, Mosse YP, White PS, Brodeur GM. Expression and sequence analysis of candidates for the 1p36.31 tumor suppressor gene deleted in neuroblastomas. Oncogene. 2008;27:803–810. doi: 10.1038/sj.onc.1210675. [DOI] [PubMed] [Google Scholar]
- Oliver SS, Musselman CA, Srinivasan R, Svaren JP, Kutateladze TG, Denu JM. Multivalent recognition of histone tails by the PHD fingers of CHD5. Biochemistry. 2012;51:6534–6544. doi: 10.1021/bi3006972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Kuo A, Schalch T, Vogel H, Joshua-Tor L, McCombie WR, Gozani O, Hammell M, Mills AA. Chd5 Requires PHD-Mediated Histone 3 Binding for Tumor Suppression. Cell Rep. 2013 doi: 10.1016/j.celrep.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaskowski S, Rieske P, Szybka M, Wozniak K, Bednarek A, Pluciennik E, Jaskolski D, Sikorska B, Liberski PP. GADD45A and EPB41 as tumor suppressor genes in meningioma pathogenesis. Cancer Genet Cytogenet. 2005;162:63–67. doi: 10.1016/j.cancergencyto.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Poetsch M, Dittberner T, Woenckhaus C. Microsatellite analysis at 1p36.3 in malignant melanoma of the skin: fine mapping in search of a possible tumour suppressor gene region. Melanoma Res. 2003;13:29–33. doi: 10.1097/00008390-200302000-00006. [DOI] [PubMed] [Google Scholar]
- Potts RC, Zhang P, Wurster AL, Precht P, Mughal MR, Wood WH, 3rd, Zhang Y, Becker KG, Mattson MP, Pazin MJ. CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PLoS One. 2011;6:e24515. doi: 10.1371/journal.pone.0024515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praml C, Finke LH, Herfarth C, Schlag P, Schwab M, Amler L. Deletion mapping defines different regions in 1p34.2-pter that may harbor genetic information related to human colorectal cancer. Oncogene. 1995;11:1357–1362. [PubMed] [Google Scholar]
- Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, Beckstrom-Sternberg J, Barrett M, Long J, Chinnaiyan A, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–1011. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- Wada M, Yokota J, Mizoguchi H, Sugimura T, Terada M. Infrequent loss of chromosomal heterozygosity in human stomach cancer. Cancer Res. 1988;48:2988–2992. [PubMed] [Google Scholar]
- Wang X, Lau KK, So LK, Lam YW. CHD5 is down-regulated through promoter hypermethylation in gastric cancer. J Biomed Sci. 2009;16:95. doi: 10.1186/1423-0127-16-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PS, Maris JM, Beltinger C, Sulman E, Marshall HN, Fujimori M, Kaufman BA, Biegel JA, Allen C, Hilliard C, et al. A region of consistent deletion in neuroblastoma maps within human chromosome 1p36.2-36.3. Proc Natl Acad Sci U S A. 1995;92:5520–5524. doi: 10.1073/pnas.92.12.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, Kok M, Winter C, Gregory SG, Hogarty MD, Maris JM, et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- Wong RR, Chan LK, Tsang TP, Lee CW, Cheung TH, Yim SF, Siu NS, Lee SN, Yu MY, Chim SS, et al. CHD5 Downregulation Associated with Poor Prognosis in Epithelial Ovarian Cancer. Gynecol Obstet Invest. 2011;72:203–207. doi: 10.1159/000323883. [DOI] [PubMed] [Google Scholar]
- Zhao R, Yan Q, Lv J, Huang H, Zheng W, Zhang B, Ma W. CHD5, a tumor suppressor that is epigenetically silenced in lung cancer. Lung Cancer. 2011 doi: 10.1016/j.lungcan.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Zhou CZ, Qiu GQ, Zhang F, He L, Peng ZH. Loss of heterozygosity on chromosome 1 in sporadic colorectal carcinoma. World J Gastroenterol. 2004;10:1431–1435. doi: 10.3748/wjg.v10.i10.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Characterization of the anti-Chd5 antibody
A) IHC of minus primary antibody control does not produce spurious signal in brain. B) IHC using a distinct anti-Chd5 antibody (Paul et al., 2013) produces a similar pattern in brain. C) Incubation with a Chd5 blocking peptide abrogates IHC signal in brain. D) Western analysis in whole brain lysates generates a distinct band for Chd5. Scale bars, 50μM.
Supplemental Figure 2. Cytoplasmic localization of Chd5
Chd5 (red) is expressed in the cytoplasm of E10.5 rhombencephalon. Scale bar, 20μM.
Supplemental Figure 3. Nuclear localization of Chd5 correlates with stage of neurogenesis.
A) Wide-field view of forebrain of E13.5 embryo shows pattern of Chd5 expression (red). Scale bar, 200μM. B) High magnification view of panel A, showing border of intense vs. less intense Chd5 expression. Scale bar, 100μM. C) Similar region as shown panel B, showing expression of Chd5 (red) and NeuN (green). Note that Chd5 expression is nuclear in NeuN-positive differentiated neurons. Scale bar, 50μM.