SUMMARY
Neural progenitor cells (NPCs) are multipotent cells that can self-renew and differentiate into neurons and glial cells. However, mechanisms that control their fate decisions are poorly understood. Here, we show that Smek1, a regulatory subunit of the serine/threonine protein phosphatase PP4, promotes neuronal differentiation and suppresses proliferative capacity of NPCs. We identify the cell polarity protein Par3, a negative regulator of neuronal differentiation, as a novel Smek1 substrate and demonstrate that Smek1 suppresses its activity. We also show that Smek1, which is predominantly nuclear in NPCs, is excluded from nucleus during mitosis, allowing it to interact with cortical/cytoplasmic Par3 and mediate its dephosphorylation by the catalytic subunit PP4c. These results identify the PP4/Smek1 complex as a key regulator of neurogenesis.
INTRODUCTION
Neural stem and progenitor cells located in the ventricular zone (VZ) of the embryonic neocortex are mitotically active, self-renewing cells with the potential to produce differentiated cell types (Temple, 2001). During cortical development, postmitotic neurons generated from NPCs migrate radially out of the VZ and form the cortical plate (CP) in an “inside-out pattern,” eventually establishing a six-layered cortex (Kriegstein et al., 2006). The timing of neuronal differentiation determines the size of the progenitor pool, the final number of neurons, and cortical thickness. However, the molecular mechanisms that control the switch from proliferation to neuronal differentiation of NPCs remain incompletely understood.
Studies of Drosophila neuroblasts show that the serine/threonine protein phosphatase 2A (PP2A) inhibits self-renewal and promotes neuronal differentiation by regulating the phosphorylation status of cell fate determinants, including Numb (Wang et al., 2009). Bazooka, a key component of the Par protein complex, is a well-characterized PP2A substrate in Drosophila neuroblasts (Krahn et al., 2009; Ogawa et al., 2009). PP2A antagonizes phosphorylation of Bazooka by Par1 kinase to control its subcellular localization. In mammals, a protein called Partitioning-defective 3 (Par3), the ortholog of Bazooka, accumulates at the tip of a growing axon in neurons and controls axon specification (Shi et al., 2003). Recently, it has been shown that Par3, which is enriched in the apical domain of NPCs of the VZ (Imai et al., 2006), critically regulates proliferation versus differentiation during cortical development (Bultje et al., 2009; Costa et al., 2008).
PP4, which belongs to the PP2A family, is a protein complex comprised of a catalytic subunit PP4c plus regulatory subunits (Gingras et al., 2005). Smek (also termed PP4R3) has been identified as a PP4 regulatory subunit and implicated in activities as diverse as regulation of MEK (Mendoza et al., 2005), insulin/IGF-1 signaling (Wolff et al., 2006), H2AX phosphorylation (Chowdhury et al., 2008), and histone H3 and H4 acetylation (Lyu et al., 2011). A recent study reported that Falafel (Flfl), the Drosophila homolog of Smek, mediates localization of the adaptor protein Miranda and the cell fate determinant Prospero in neuroblasts (Sousa-Nunes et al., 2009). However, the direct substrate of Smek remains unclear. Here we identify Par3 as a direct substrate of the PP4/Smek1 complex in NPCs and report a novel role for Smek1 in regulating neuronal differentiation.
RESULTS
Smek1 is required for neuronal differentiation of NPCs
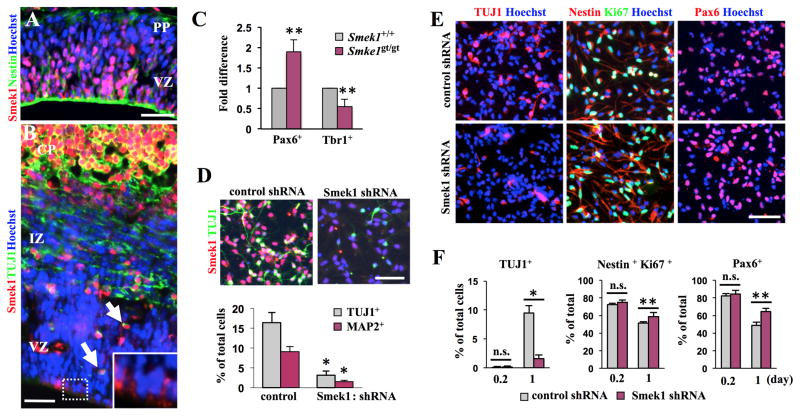
During mouse cortical development, Smek1 is expressed in a distinct temporal and spatial pattern. At E11.5, we observed that Smek1 protein is expressed in most NPCs at the apical side of the forebrain VZ (Figure 1A and S1A). At E14.5, Smek1 protein was detectable primarily in CP neurons (Figure 1B and S1B), while weak Smek1 expression was seen in some NPCs undergoing mitosis at the ventricle surface (Figure 1B, boxes). Interestingly, VZ neurons that migrate to the CP also expressed Smek1 protein (Figure 1B, arrows). In postnatal forebrain, Smek1 protein expression remained detectable in cortical layers I-IV (Figure S1C). Moreover, E14 cortices of Smek1-depleted mice (Smek1gt/gt) exhibited an increase in the number of Pax6-positive cells (an NPC marker) and a decrease in the number of Tbr1-positive cells (a marker of cortical neurons) as compared to E14 cortices of wild-type (Smek1+/+) mice (Figure 1C and S1D).
Figure 1. Smek1 regulates neuronal differentiation in the early phase of NPC differentiation.
(A, B) Smek1 expression in mouse forebrain cortex. Coronal sections from forebrain cortex of E11.5 (A) and E14.5 (B) mouse embryos were immunostained with Smek1 and Nestin (an NPC marker) or TUJ1 (a marker of immature neurons) antibodies. Hoechst dye served as a counterstain. Smek1 proteins are detected mainly in Nestin-positive cells of the VZ of E11.5 cortex (A) and in TUJ1-positive cells of the CP at E14.5 (B). Notably, Smek1 is also observed in cells at the ventricular surface (box) and in TUJ1-positive cells (arrows) of the VZ of E14.5 cortex. Scale bar, 100 μm. PP, preplate; VZ, ventricular zone; IZ, intermediate zone; CP, cortical plate.
(C) Quantitative analysis of Pax6 and Tbr1-positive cells in E14.5 Smek+/+ and Smekgt/gt forebrain cortices. Fold difference was evaluated by normalization of the average number of cells positive for the indicated antibodies per coronal sections against wild-type mice. Error bars represent the mean ± S.D. of three independent experiments. **p < 0.05.
(D) Smek1 depletion inhibits neuronal differentiation. NPCs expressing Smek1 shRNA or control shRNA under control of doxycycline-inducible promoter were cultured in differentiating condition medium containing doxycycline for 6 days and then subjected to immunostaining for TUJ1, MAP2 (a marker of mature neurons), or Smek1. Percentages of TUJ1- and MAP2-positive cells per total cells are shown in the bottom panel. Scale bar, 100 μm.
(E) Smek1 depletion decreases neuronal differentiation. Two days prior to differentiation, expression of Smek1 shRNA or control shRNA was induced by addition of doxycycline. Cells were then cultured under differentiating conditions for indicated times and then immunostained with TUJ1, Nestin, Pax6, or Ki67 antibodies. Scale bar, 100 μm.
(F) Percentages of neuronal cells or undifferentiated NPCs were evaluated by counting TUJ1-positive cells, Nestin/Ki67 double-positive cells, or Pax6-positive cells per total cells. Error bars represent the mean ± S.D. of four independent experiments; each assay was performed in duplicate. *p < 0.001; **p < 0.05; n.s., not significant.
To assess Smek1’ function in neurogenesis, we employed an in vitro culture system using NPCs isolated from the E11.5 mouse forebrain neocortex. NPCs transduced with lentivirus expressing shRNA against Smek1 or control shRNA under control of a doxycycline-inducible promoter (Figure S1E) were cultured in medium containing doxycycline for 6 days under differentiating conditions and then assessed for neurogenesis using TUJ1 (a marker of immature neurons) or MAP2 (a marker of mature neurons). The number of TUJ1- or MAP2-positive cells significantly decreased in Smek1 knockdown cultures compared to cultures expressing control shRNA (Figure 1D), indicating a neuronal differentiation defect. A decrease in number of neurons can be caused by a defect in NPC proliferation or neuronal apoptotic cell death. While no significant difference in the number of apoptotic cell death (as determined by TUNEL staining) was observed between control and Smek1 knockdown cells cultured under differentiation condition (data not shown), Smek1 knockdown NPCs grown under proliferation conditions underwent hyperproliferation (Figure S1F and G). We then asked whether Smek1 regulated the transition of NPCs from proliferative to differentiation states by knocking down Smek1 in NPCs prior to placing them in differentiating culture conditions. Western blotting of cells expressing Smek1 shRNA showed decreased levels of TUJ1 protein relative to controls by day 1 of culture (Figure S1H). At this time point, we found that the percentage of undifferentiated NPCs expressing both Nestin (an NPC marker) and Ki67 (a marker of proliferation) or Pax6 increased in cultures expressing Smek1 shRNA compared to control cultures, while the percentage of TUJ1-positive cells significantly decreased (Figure 1E and F). These findings suggest that Smek1 is required for neuronal differentiation and suppression of NPC proliferative capacity at an early phase of differentiation.
Smek1 recruits PP4c to promote neuronal differentiation
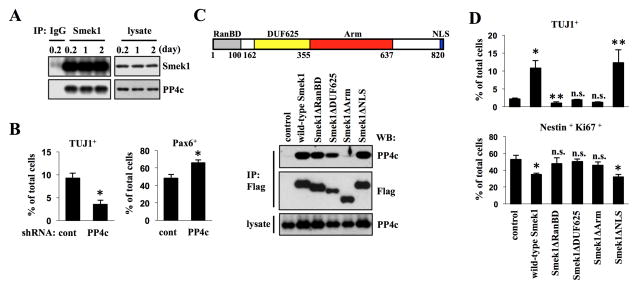
To determine Smek1 as a regulatory subunit of PP4 in neurogenesis, we asked whether Smek1 binds to the catalytic subunit PP4c in NPCs using co-immunoprecipitation. Western blot analysis revealed PP4c in Smek1 but not control immunoprecipitates, indicating that Smek1 physically interacts with PP4c. Such interactions did not change during differentiation (Figure 2A). To examine whether PP4c functions in neurogenesis, NPCs were exposed to lentivirus expressing PP4c or control shRNA and cultured as described in Figure 1D. PP4c knockdown led to changes similar to those accompanying Smek1 knockdown: relative to control cultures TUJ1 expression and the number of TUJ1-positive neurons decreased while Pax6-positive NPCs increased (Figure 2B and S2A and B). We next mapped Smek1 domains required for PP4c interaction. Smek contains four conserved domains: an N-terminal Ran-binding domain (RanBD), a domain of unknown function 625 (DUF625), an armadillo (Arm) repeat region, and a C-terminal nuclear localization sequence (NLS). We constructed a series of Flag-tagged deletion mutants, including Smek1ΔRanBD (lacking amino acid (aa) 2–100), ΔDUF625 (lacking aa 162-355), ΔArm (lacking 350-653), and ΔNLS (lacking aa 809-820) (Figure 2C, top) and introduced them or a wild-type construct into NPCs. PP4c was not be detected in anti-Flag immunoprecipitates from NPCs expressing Flag-Smek1ΔArm (Figure 2C, bottom) but was detected in cells expressing wild-type or other deletion mutants, suggesting that PP4c/Smek1 complex formation requires the Arm repeats. We also found that, while expression of wild-type Smek1 or corresponding ΔNLS mutant in cultures lacking endogenous Smek1 rescued the neuronal differentiation defect, the other mutants did not (Figure 2D and S2C and D). These results indicate that Smek1 regulates neuronal differentiation via its Arm repeats region through PP4c and suggest that both RanBD and DUF625 domains also participate in neurogenesis.
Figure 2. PP4c is required for neuronal differentiation.
(A) Binding of Smek1 to PP4c in differentiating NPCs was confirmed by immunoprecipitation followed by Western blotting using Smek1 or PP4c antibodies.
(B) PP4C knockdown in NPCs blocks their neuronal differentiation. NPCs transduced with lentiviruses expressing PP4c shRNA or control shRNA under control of a doxycycline-inducible promoter were cultured, and then subjected to immunostaining with TUJ1 or Pax6 antibodies. Percentages of neuronal or undifferentiated cells were determined. Error bars represent the mean ± S.D. of three independent experiments. *p < 0.001.
(C) Schematic representation of Smek1 structure (top) showing the Ran-binding domain (RanBD, gray), the domain of unknown function 625 (DUF625, yellow), the armadillo (Arm) repeats region (red), and the nuclear localization sequence (NLS, blue). Numbers indicate amino acids. Western analysis of anti-Flag immunoprecipitates from NPCs transduced with lentiviruses expressing Flag-tagged wild-type or mutant forms of Smek1 (bottom). A Smek1 mutant lacking the Arm repeats region does not bind PP4c.
(D) Neurogenesis defects seen in Smek1 knockdown NPCs are rescued by re-expression of wild-type or ΔNLS mutant Smek1 but not by Smek1ΔRanBD, ΔDUF625, and ΔArm mutants. Smek1 expression was knocked down by shRNA targeting 3′-UTR of Smek1 gene. Smek1 depleted NPCs were then transduced with wild-type Smek1 or mutants and cultured under differentiation conditions. At day 1, cultures were subjected to immunostaining with TUJ1 (upper), or Nestin and Ki67 (lower) antibodies. Percentages of neuronal or undifferentiated cells were determined. Error bars represent the mean ± S.D. of four independent experiments. *p < 0.001; **p < 0.05; n.s., not significant.
Smek1 binds to and mediates Par3 dephosphorylation
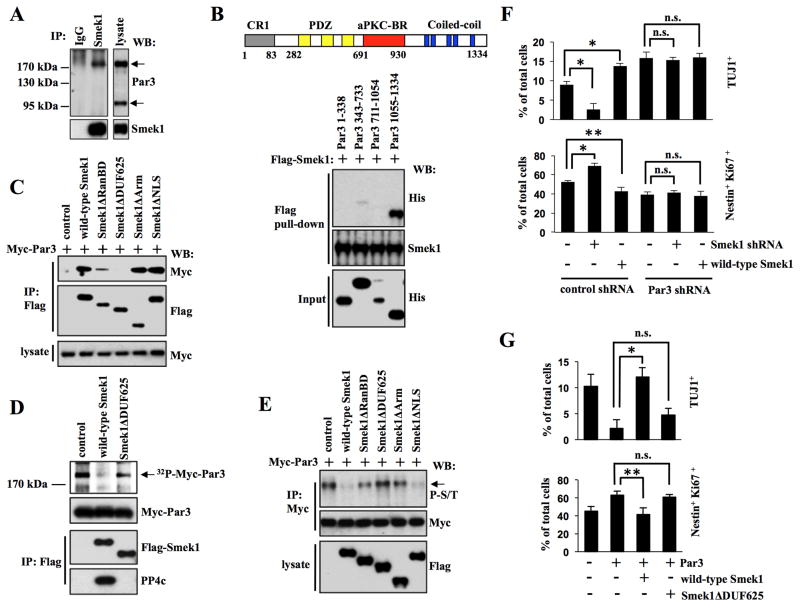
To identify PP4 substrates regulated by Smek1 in NPCs, we employed affinity purification to purify proteins interacting with Smek1. Mass spectrometry analysis identified potential Smek1-binding proteins, including Par3, Kinesin-like protein, coiled-coil domain-containing protein 30 (CCDC 30), heat shock protein 90 (HSP90), PKC lambda, and HDAC1 Figure S3A. Among these, Par3, an intrinsic regulator of neurogenesis, is a particularly attractive candidate (Bultje et al., 2009; Costa et al., 2008). Using an antibody that detects the major isoforms (180, 150, and 100 kDa) of Par3, Western blot analysis revealed the predominant expression of two isoforms, 180 and 100 kDa forms, in NPCs, and that only the 180 kDa Par3 was detectable in Smek1 immunoprecipitates (Figure 3A). To determine whether Smek1/Par3 binding was direct, we performed an in vitro pull-down assay using purified Flag-Smek1 and His-fused Par3 fragments, the latter containing the CR1 domain (aa 1-338), the PDZ domain (aa 343-733), the aPKC-BR domain (aa 711-1054), or the C-terminal coiled-coil region (aa 1055-1334) (Figure S3B). Western blot analysis revealed that Flag-Smek1 pulled down only the Par3 coiled-coil region (Figure 3B), indicating direct binding through that region. Moreover, Par3 was detected in Flag immunoprecipitates derived from NPCs transduced with lentivirus expressing Myc-Par3 plus lentivirus expressing Flag-Smek1 wild-type or Smek1ΔRanBD, Smek1ΔArm, or Smek1ΔNLS constructs but not from NPCs expressing Smek1ΔDUF625 (Figure 3C). This result indicates that Smek1 DUF625 domain is required for Smek1/Par3 interaction.
Figure 3. Smek1 interacts with Par3 and inhibits its function in neuronal differentiation.
(A) Smek1 binds to Par3. NPC lysates were immunoprecipitated with IgG or Smek1 antibodies, and subjected to Western blotting with indicated antibodies.
(B) Smek1 recognizes the Par3 coiled-coil region (amino acids 1055–1334). Upper, schematic structure of Par3 structure containing CR1 (gray, conserved region 1), PDZ (yellow, PSD-95/Dgl/ZO-1), aPKC-BR (red, aPKC-binding region), and coiled-coil (blue, Coiled-coil region) domains. Lower, Flag-Smek1 protein-immobilized beads were incubated with purified His-tagged Par3 protein fragments, and bound proteins were analyzed by Western blotting using anti-His or -Flag antibodies.
(C) The Smek1 DUF625 domain is required for Par3 binding. NPCs were transduced with lentiviruses expressing wild-type Smek1 or Smek1 mutants together with lentivirus expressing Myc-Par3. Anti-Flag immunoprecipitates were analyzed by Western blotting using anti-Myc and -Flag antibodies.
(D) Par3 is dephosphorylated by the Smek1/PP4c complex. 293T cells transfected with wild-type Flag-Smek1, Flag-Smek1ΔDUF625, or a control construct. The Smek1/PP4c-containing protein complex was isolated by anti-Flag immunoprecipiation. Myc-Par3 was 32P-labeled using an NPS lysate and purified by anti-Myc immunoprecipitation. 32P-labeled Myc-tagged Par3 was subjected to a dephosphorylation assay in the presence of a Smek1/PP4c complex. Anti-Flag immunoprecipitates were also subjected to Western blotting with anti-Myc, -Flag, and -PP4c antibodies.
(E) Western blot analysis of anti-Myc immunoprecipitates from NPCs described in C. Western blotting with an anti-phospho-serine/threonine antibody shows that both wild-type Smek1 and the Smek1ΔNLS mutant decrease Par3 phosphorylation.
(F) Smek1 acts upstream of Par3 in neurogenesis. NPCs expressing Smek1 shRNA, wild-type Smek1, Par3 shRNA, or control shRNA were cultured for 1 day under differentiating conditions and then immunostained with TUJ1 or Nestin plus Ki67 antibodies. Percentages of neurons or undifferentiated NPCs were determined by comparing TUJ1-positive or Nestin/Ki67 double-positive cells to total cells, respectively. Error bars represent the mean ± S.D. of three independent experiments. *p < 0.001; **p < 0.05; n.s., not significant.
(G) Smek1 suppresses Par3 regulation of neuronal differentiation. NPCs transduced with lentivirus expressing Par3, wild-type Smek1, Smek1ΔDUF625 mutant, or a control construct were cultured for 1 day under differentiating conditions and then immunostained with TUJ1 or Nestin plus Ki67 antibodies. Percentages of neurons and NPCs were evaluated as described in F. *p < 0.001; **p < 0.05; n.s., not significant.
To assess potential dephosphorylation of Par3 by Smek1, we phosphorylayed Myc-Par3 protein in vitro by incubating it with an NPC lysate and 32P-ATP and then treated it with a complex containing Flag-Smek1 proteins (Figure 3D). 32P-labeling of Par3 was significantly decreased when Par3 protein was incubated with a complex containing wild-type Flag-Smek1 protein and PP4c (Figure 3D). By contrast, treatment with a Flag-Smek1ΔArm protein complex lacking PP4c binding significantly reduced Par3 dephosphorylation. Moreover, Western blot analysis of Par3 immunoprecipitates with an anti-phospho-serine/threonine antibody confirmed that Smek1 and PP4c regulate Par3 phosphorylation through serine/threonine residues (Figure S3C). Since the DUF625 and Arm repeats regions of Smek1 are required for binding to Par3 and PP4c respectively, we examined the Par3 phosphorylation state in NPCs expressing Flag-tagged wild-type or mutant Smek1 together with Myc-tagged Par3. Western blot analysis of Myc-Par3 immunoprecipitates using anti-phospho-serine/threonine antibody showed that overexpression of wild-type Smek1 or Smek1ΔNLS significantly decreased Par3 phosphorylation levels compared to controls, whereas overexpression of Smek1ΔRanBD, ΔDUF625, or ΔArm did not (Figure 3E). These results suggest that, in addition to the DUF625 and Arm, the RanBD domain of Smek1 participates in regulation of Par3 phosphorylation at serine/threonine residues.
Smek1 negatively regulates Par3 in neurogenesis
Next we asked whether Par3 is required for Smek1-mediated neurogenesis. To this end, we assessed the effect of Smek1 loss-or gain-of function on neuronal differentiation in the presence or absence of Par3. NPCs expressing either Smek1 shRNA or wild-type Smek1 were transduced with lentivirus expressing Par3 or control shRNA (Figure S3D). At day 1 after differentiation, in the presence of Par3, knockdown of Smek1 led to a decrease in the number of TUJ1-positive neurons and an increase in the number of Nestin/Ki67 double-positive NPCs, while overexpression of Smek1 had the opposite effect (Figure 3F and S3E). In the absence of Par3 by using shRNA, the number of TUJ1-positve cell was increased and the number of Nestin/Ki67 double positive NPCs was decreased. However, in these cultures knockdown or overexpression of Smek1 did not significantly alter the number of neurons or undifferentiated NPCs. In addition, we also observed increased expression of mRNAs encoding the Notch targets Hes1 and Hes5 in cells expressing Smek1 shRNA compared to control cells (Figure S3F). Moreover, analysis of Notch reporter gene activity revealed that wild-type Smek1 inhibited Notch signaling activity induced by Par3 overexpression, while Smek1ΔDUF625 did not (Figure S3G). Given that Par3 activates Notch signaling (Bultje et al., 2009), these results suggest that Smek1 acts upstream of Par3 to negatively regulate its activity in neurogenesis.
Par3 loss of function promotes neuronal differentiation (Costa et al., 2008), consistent with the effect seen following Smek1 overexpression (Figure 3F). To confirm that Smek1 promotes neurogenesis by suppressing Par3 function, we transduced NPCs with lentiviruses expressing Par3 alone or Par3 together with wild-type Smek1 or Smek1ΔDUF625, cultured them under differentiation conditions, and then neuronal differentiation was quantified by determining the percentage of TUJ1-positive and Nestin/Ki67 double-positive cells one day later. Par3 overexpression decreased the number of TUJ1-positive neurons and increased the number of Nestin/Ki67-positive undifferentiated NPCs compared with control cells (Figure 3G and S3H). As expected, wild-type Smek1 negated the effect of Par3 overexpression, as determined by comparing the percentage of TUJ1-positive and Nestin/Ki67 double-positive cells in cultures expressing both Smek1 and Par3 to cultures expressing Par3 alone. In comparison with wild-type Smek1, no significant change was seen in cultures transduced with Smek1ΔDUF625, which cannot bind Par3. These experiments further confirm that Smek1 negatively regulates Par3 in NPC differentiation.
Dynamic changes in Smek1 subcellular localization facilitate targeting of PP4 to Par3
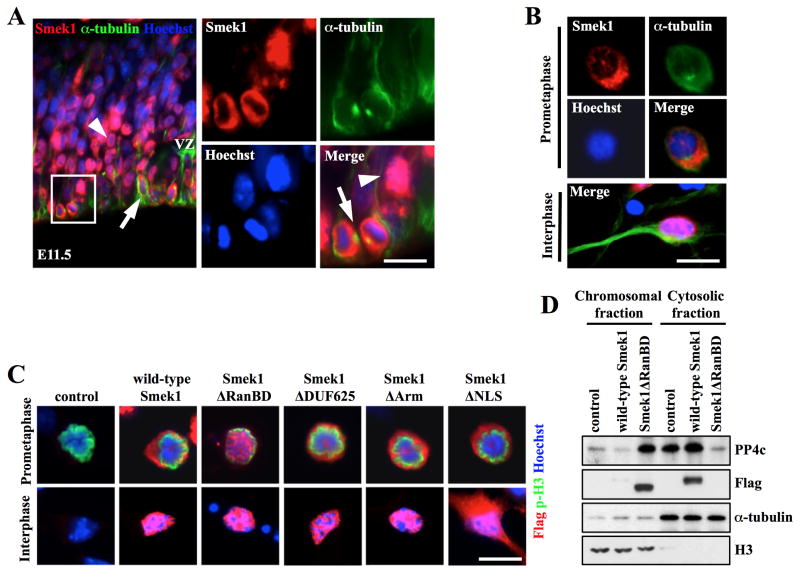
Par3 localizes to the apical cortex of NPCs (Bultje et al., 2009), while Smek1 is predominantly nuclear (Figure 1A and C). To determine if changes in Smek1 subcellular localization occur in NPCs during neurogenesis, coronal sections from E11.5 forebrain were immunostained with anti-Smek1 and -α-tubulin (a cytoplasmic marker) antibodies. Smek1 co-localized with α-tubulin in cells on the ventricular surface (Figure 4A, arrows), indicating a cytoplasmic/cortical localization in mitotic NPCs. In mitotic cells, Par3 showed a similar localization (Figure S4A and B). Moreover, immunostaining of NPC cultures with anti-Smek1 and -α-tubulin antibodies showed that Smek1 undergoes dynamic changes in subcellular localization during mitosis. While Smek1 was nuclear in interphase and prophase cells, it showed a cytoplasmic/cortical localization from prometaphase to anaphase (Figure 4B and S4C). Metaphase and anaphase cells also showed Smek1 enrichment at spindle microtubules.
Figure 4. Smek1 regulates subcellular localization of PP4c but not Par3.
(A, B) Subcellular localization of Smek1 protein in cells in the VZ of E11.5 forebrain cortex (A) and in in vitro cultures (B). Mitoses were determined based on position of microtubules labeled with anti-α-tubulin antibody or chromosomes labeled with Hoechst dye. Smek1 fluorescence is nuclear in interphase cells (arrowheads in A and bottom panel in B) but cytoplasmic in mitoses (arrows in A and top panels in B). Scale bar, 10 μm.
(C) Subcellular localization of Smek1 in mitotic NPC. NPCs were transduced with Flag-tagged wild-type or mutant forms of Smek1. Immunostaining with anti-Flag and anti-phospho-H3 antibodies revealed subcellular localization of wild-type and mutant Smek1 in prometaphase (top panels) and interphase cells (bottom panels). Cells undergoing mitosis are marked by phospho-histone H3. DNA is visualized by Hoechst staining. Scale bar, 10 μm.
(D) Smek1 regulates PP4c subcellular localization. Smek1 knockdown NPCs were transduced with Flag-wild-type Smek1, Flag-Smek1ΔRanBD, or a control construct and arrested in mitosis by nocodazole treatment. Chromosomal and cytosolic proteins were fractionated and subjected to Western blotting with anti-PP4c and –Flag antibodies. α-tubulin and histone H3 expression serve to confirm the purity of cytosolic and chromosomal fractions, respectively.
The RanBD motif of the Dictyostelium discoideum Smek homolog is reportedly critical for its cytoplasmic/cortical localization (Mendoza et al., 2005). To test whether this was the case for mammalian Smek1, Smek1-depleted NPCs were transduced with constructs encoding Flag-tagged wild-type Smek1 or its deletion mutants and immunostained with anti-Flag and anti-phospho-histone H3 (a marker of mitosis and chromatin condensation). Consistent with results reported in Dictyostelium discoideum, the Smek1ΔRanBD mutant failed to localize to the cytoplasm/spindle during mitosis but rather localized in the nucleus and remained there in interphase (Figure 4C). The subcellular localization of other mutants tested resembled that of wild-type Smek1, with the exception of Smek1ΔNLS, which was expressed in both the nucleus and cytoplasm of interphase cells. Smek1ΔRanBD contains domains that can bind Par3 and PP4c, as shown by immunoprecipitation (Figure 2C and 3C). We thus asked whether ectopic expression of Smek1ΔRanBD promoted mislocalization of Par3 and PP4c during mitosis. When we expressed Smek1ΔRanBD ectopically in Smek1-depleted NPCs, cytoplasmic/cortical Par3 remained unchanged while Smek1ΔRanBD was nuclear (Figure S4D). In addition, no difference in localization of Par3 between Smek1-depleted and wild-type Smek1 re-expressing cells was observed, suggesting that Smek1 does not alter Par3 localization. To evaluate PP4c subcellular localization, chromosome-associated and cytosolic protein fractions were isolated from M phase-synchronized NPCs and compared by Western analysis using indicated antibodies (Figure 4D). Interestingly, PP4c protein levels increased in the chromosomal fraction from cells expressing Flag-Smek1ΔRanBD compared to control cells or wild-type Flag-Smek1, while in the cytosolic fraction the level of PP4c protein decreased, indicating altered localization of cytoplasmic PP4c to the nucleus. Taken together, these results demonstrate that PP4c subcellular localization depends on Smek1 localization during mitosis and suggest that cytoplasmic/cortical localization of Smek1 targets PP4 to Par3.
DISCUSSION
Neural stem and progenitor cells have been suggested as potential therapeutics for neurodegenerative disorders. However, understanding molecular and cellular mechanisms underlying their differentiation is a prerequisite to manipulating stem cell behavior. We show that Smek1, an evolutionarily conserved regulatory subunit of PP4, regulates neuronal differentiation and reveal an unreported function of PP4 in mammalian neurogenesis. Moreover, identification of Par3 as a novel Smek1-interacting protein and characterization of its conserved domains reveals a molecular mechanism by which Smek1 targets PP4 to Par3 during mitosis and negatively regulates Par3 function in neurogenesis.
In this study we identify Par3 as a PP4 substrate. We propose that Smek1, through its DUF625 domain, binds directly to the Par3 C-terminus. In NPCs Par3 is primarily cytoplasmic in interphase and mitosis. Thus, nuclear export of Smek1 to the cytoplasm is required for its interaction with Par3. We show dynamic changes in Smek1 subcellular localization in NPCs. While Smek1 localizes exclusively to the nucleus in interphase, during mitosis it becomes cytoplasmic. The RanBD of several proteins reportedly recognizes GTP-bound Ran (RanGTP), which directs assembly of spindle microtubules allowing chromosomal segregation and cytokinesis in mitosis (Carazo-Salas et al., 2001). Smek1 enrichment at spindle microtubules in metaphase and anaphase cells suggests that its RanBD may function in a RanGTP-dependent pathway during mitosis. Notably, nuclear export of Smek1 to the cytoplasm was observed from prometaphase cells when microtubules invade the nuclear space, and deletion of the Smek1 RanBD abolished this effect, as seen by nuclear localization. Thus our data suggest that Smek1 subcellular localization is regulated through the RanBD and that this activity may depend on microtubule dynamics functioning in a Ran-dependent pathway.
Most Smek homologs physically interact with the catalytic subunit PP4c (Gingras et al., 2005; Chowdhury et al., 2008), suggesting that the PP4 complex is evolutionarily conserved. We show that PP4c recognizes the Arm repeats region of Smek1 and its subcellular localization depends on Smek1 localization. Thus, nuclear export of Smek1 during mitosis facilitates dephosphorylation of Par3. This idea is supported by our observation that, while expression of Smek1 induced Par3 dephosphorylation in NPCs, expression of Smek1 mutants lacking RanBD and Arm repeats region did not. Interestingly, studies of Drosophila neuroblasts previously revealed that cell fate specification is tightly linked with phosphorylation status of bazooka protein (Betschinger et al., 2003; Krahn et al., 2009). However, it is now yet clear whether Par3 dephosphorylation directly regulates NPC neurogenesis. Although we could not identify specific phosphorylation sites targeted by PP4, our data defines three conserved domains of Smek1, namely RanBD, DUF625, and Arm repeats, necessary to target PP4 to its substrate Par3 and provides insight into the molecular mechanism by Smek1 to regulate PP4 function in NPCs.
We here show that Smek1 suppresses Par3, a negative regulator of neuronal differentiation. Par3 acts upstream of Notch signaling (Bultje et al., 2009), which critically regulates cell fate decision of NPCs in cortical development (Gaiano and Fishell, 2002). Notch gain-of-function activity inhibits neuronal differentiation (Nye et al., 1994), an effect similar to Smek1 loss-of-function. Moreover, Smek1 inhibits Par3-induced Notch reporter gene activity. Although it remains unclear whether Smek1 inhibits Par3’s ability to activate Notch signaling during mitosis, ensuring a neuronal fate, our data demonstrate Smek1 as a negative regulator of Par3 in regulating neuronal differentiation and suggest a novel role for PP4 in mammalian neurogenesis.
EXPERIMENTAL PROCEDURES
Neural Progenitor Cell culture
NPCs were cultured as described (Lyu et al., 2008). Briefly, neocortices were mechanically dissociated into single cells using a flame-polished Pasteur pipet. Dissociated cells were seeded onto polyornithine (15 μg/ml; Sigma)- and fibronectin (2 μg/ml; Invitrogen)-coated dishes in DMEM/F12 medium containing B27 supplement (Gibco) and fibroblast growth factor 2 (FGF2, 20 ng/ml). To induce differentiation of NPCs, FGF2 was withdrawn from the medium. Expression of cDNAs or shRNAs in NPCs was transduced by lentiviral infection (Supplemental information) and via doxycycline (500 ng/ml; Sigma) treatment. For synchronization, cells were grown in the presence of Aphidicholin (500 nM; Sigma) for 18 hours, washed and further incubated with Nocodazole (100 ng/ml; Sigma) for 6hrs. Cells were then placed in fresh medium for 30 min before fixation and fractionation. For immunoprecipitation and Western blotting, 2 – 4 × 106 cells were seeded onto coated 10 cm dishes and cultured in the presence or absence of FGF2. For immunostaining, 5 × 104 cells were plated onto polyornithine- and fibronectin-coated cover slips.
Immunoprecipitation, Western Blotting and Immunostaining
For immunoprecipitation, Samples containing 300–500μg of protein were incubated with a specific antibody for 2 hrs at 4°C and then with Protein A/G agarose beads (Pierce) overnight. Immunoprecipitates or pull-down samples were eluted using SDS sample buffer and separated using 6, 8, or 10% SDS-PAGE. After blocking, blots were incubated with primary antibody and then with a peroxidase-conjugated secondary antibody. The bound secondary antibody was detected using enhanced chemiluminescence (ECL) reagent (Santa Cruz Biotechnology). For immunostaining, cells grown on cover slips were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Fixed cells and brain sections were incubated with blocking solution containing 2% BSA for 1 h and incubated with antibodies. Cells were incubated with secondary antibodies at room temperature for 1 h and counterstained with Hoechst dye. Images were obtained using a fluorescence microscope with an AxioCam camera (Zeiss) or with a confocal microscope (LSM5 PASCAL, Zeiss). The percentage of antibody-labeled cells was evaluated by quantifying a minimum of 800 cells in 8 randomly chosen microscopic fields. The values obtained from at least three independent experiments were averaged and present as mean ± S.D. The two-tailed Student’s t-test was used for comparison of two experimental groups.
In vitro dephosphorylation assay
Lysates from 293T cells transiently transfected with Myc-Par3 were immunoprecipitated with anti-Myc antibody. Beads were washed and incubated with NPC lysates made with kinase reaction buffer (50mM HEPES, pH 7.5, 20mM MgCl2, 4mM MnCl2, and 20 μCi of 32P-labeled γ-ATP) for 30 min at 20°C. Subsequently, beads were washed and incubated with Flag immunoprecipitates from 293T cells transfected with plasmids expressing wild-type Flag-Smek1, Flag-Smek1 ΔDUF625, or Flag-GFP as a control in phosphatase assay buffer (50mM Hepes, pH7.5, 150mM NaCl, 1mM MnCl2, 3mM DTT, 0.1% Nonidet P-40) for 30 min at 30°C. Reactions were stopped by addition of SDS sample buffer, and samples were analyzed by 6% SDS-PAGE. Dried gels were monitored by autoradiography.
Supplementary Material
Acknowledgments
We thank the USC Transgenic Core Facility for generating mutant mice. This research is funded by a NIH grant to W.L (5R01NS067213) and an NRF grant (NRF-2011-35B-E00015) to J.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, Dykxhoorn DM, Weinstock DM, Pfeifer GP, Lieberman J. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol Cell. 2008;31:33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MR, Wen G, Lepier A, Schroeder T, Gotz M. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development. 2008;135:11–22. doi: 10.1242/dev.009951. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Fishell G. The role of notch in promoting glial and neural stem cell fates. Ann Rev Neurosci. 2002;25:471–490. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Caballero M, Zarske M, Sanchez A, Hazbun TR, Fields S, Sonenberg N, Hafen E, Raught B, Aebersold R. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- Imai F, Hirai S, Akimoto K, Koyama H, Miyata T, Ogawa M, Noguchi S, Sasaoka T, Noda T, Ohno S. Inactivation of aPKClambda results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development. 2006;133:1735–1744. doi: 10.1242/dev.02330. [DOI] [PubMed] [Google Scholar]
- Krahn MP, Egger-Adam D, Wodarz A. PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev Cell. 2009;16:901–908. doi: 10.1016/j.devcel.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Lyu J, Jho EH, Lu W. Smek promotes histone deacetylation to suppress transcription of Wnt target gene brachyury in pluripotent embryonic stem cells. Cell Res. 2011;21:911–921. doi: 10.1038/cr.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J, Yamamoto V, Lu W. Cleavage of the Wnt receptor Ryk regulates neuronal differentiation during cortical neurogenesis. Dev Cell. 2008;15:773–780. doi: 10.1016/j.devcel.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Du F, Iranfar N, Tang N, Ma H, Loomis WF, Firtel RA. Loss of SMEK, a novel, conserved protein, suppresses MEK1 null cell polarity, chemotaxis, and gene expression defects. Mol Cell Biol. 2005;25:7839–7853. doi: 10.1128/MCB.25.17.7839-7853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ohta N, Moon W, Matsuzaki F. Protein phosphatase 2A negatively regulates aPKC signaling by modulating phosphorylation of Par-6 in Drosophila neuroblast asymmetric divisions. J Cell Sci. 2009;122:3242–3249. doi: 10.1242/jcs.050955. [DOI] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN. Hipocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R, Chia W, Somers WG. Protein phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions. Genes Dev. 2009;23:359–372. doi: 10.1101/gad.1723609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- Wang C, Chang KC, Somers G, Virshup D, Ang BT, Tang C, Yu F, Wang H. Protein phosphatase 2A regulates self-renewal of Drosophila neural stem cells. Development. 2009;136:2287–2296. doi: 10.1242/dev.035758. [DOI] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.