Abstract
Ultraviolet photodissociation (UVPD) mass spectrometry was used to characterize the structures of amphiphilic glycosphingolipids and gangliosides in comparison to collision induced dissociation (CID) and higher energy collision dissociation (HCD) in a high performance Orbitrap mass spectrometer. UVPD produced the widest array of fragment ions diagnostic for both the ceramide base and oligosaccharide moieties. CID and HCD generated mainly glycosidic B/Y and C/Z cleavages of the oligosaccharides moieties and very few informative fragments related to the hydrophobic ceramide base. Several unique cleavages at the sphingoid base and the fatty acid chain occurred upon UVPD, as well as a wider variety of cross ring cleavages (A/X ions), thus affording differentiation of isobaric gangliosides. An LC-UVPD-MS strategy allowed the elucidation of 27 gangliosides among five different classes.
Introduction
Glycosphingolipids are generally considered one of the most complex lipids and contain two hydrophobic chains (ceramide) and a glycan head moiety (oligosaccharide).1,2 The first of the chains is a unique amine-containing lipid referred to as the sphingoid base; the other is a fatty acid tail. The complexity of glycosphingolipids arises from the variability in the potential number and type of saccharides, in addition to the length, position, saturation and configuration of the carbon chains of both the sphingoid base and the fatty acid tail. In particular, gangliosides are sialic acid-containing glycosphingolipids that are found throughout all eukaryotes and some virus and prokaryotic organisms. Gangliosides are ubiquitously distributed throughout many different tissues and biological fluids but are in especially high concentrations in the nervous system where they are localized at the cell membranes and influence cell structure and cell signaling.1 The oligosaccharide portions are known to interact with exogenous compounds such as neighboring cells, extracellular proteins and pathogens. The lipid moieties are inserted into the cell wall and can act as mediators for intercellular and extracellular signaling. The number of oligosaccharides and lengths of the hydrophobic moieties in gangliosides are known to change with brain development and ageing.3 More specifically mutations in the protein N-acetylgalactosaminyltransferase are known to affect the expression of complex gangliosides and have been connected to several diseases such as Alzheimer's disease, Huntington's disease, Parkinson's disease and AIDS related dementia.3–7 Irregularities in ganglioside synthesis have been linked to several different types of cancer, and thus gangliosides are frequently used as diagnostic biomarkers of the stages of cancer.7,8
The growing interest in profiling and quantifying cellular lipids, such as gangliosides, has been fueled by the increasing recognition of the importance of lipids in signaling pathways and their vital architectural role in cell membranes.9 Both of these critical functional features of lipids have also motivated the evaluation of lipid profiles as biomarkers of health and disease status. Advances in the field of mass spectrometry have proven indispensable for the characterization of complex lipids and lipid mixtures, as evidenced by several recent reviews.10– 12 In particular, technological improvements in separation and ion activation methods have been pivotal for accelerating broader and deeper studies of lipid profiles and lipidomics. For example, Reid et al. demonstrated the benefits of high resolution mass spectrometry paired with higher energy collision dissociation (HCD) and collision induced dissociation (CID) for analysis of lipids from cell extracts, thus identifying over 600 different lipids.13,14 Shotgun lipidomics, a strategy which combines direct infusion ESI-MS and multidimensional mass spectrometry to directly analyze lipids from organic extracts without HPLC separation, utilizes neutral loss and precursor ion scans to fingerprint lipids within complex mixtures.15,16 Others have reported the advantages of using nanoChip-LC devices which offer a simplified chromatographic separation mode coupled with highly sensitive nanoESI-mass spectrometry.17–19 Recently a wide variety of ambient ionization methods have been applied for lipidomic problems, thus providing a facile method to profile lipids from surfaces like tissue slices or TLC plates.20–26
Most mass spectrometric methods have utilized CID or more recently HCD to confirm lipid type and structure,13,14,27–33 and a few of the many studies are summarized here as representative examples of the status of the field. One of the first studies reported the elucidation of glycosphingolipids and gangliosides via keV CID for which details about the ceramide portion were revealed in the positive mode and information about the carbohydrate moiety was determined in the negative mode.27 Low energy CID proved effective for differentiation of deprotonated alpha2-3 and alpha2-6 sialylated neolacto-series gangliosides.28 Another low energy CID method, sustained off-resonance irradiation CID, was used to sequence deprotonated sialylated and sulfated glycosphingolipids in an FTICR mass spectrometer.29 A selected reaction monitoring CID method was developed using a triple quadrupole mass spectrometer to map gangliosides in milk based on a characteristic N-acetylneuraminic acid fragment ion of m/z 290.30 Comparison of CID, HCD, and pulsed Q dissociation (PQD) methods on an Orbitrap mass spectrometer showed that HCD provided the most unique and accurate fingerprint fragmentation patterns of sulfoquinovosyldiacylglycerol lipids.14 There remain, however, common shortcomings of CID and HCD arising from a lack of consistent diagnostic fragment ions, thus preventing confident elucidation of such key features as isomerization of unsaturated C-C bonds. Structurally specific features of lipids have been elucidated via MSn methods implemented on traps.31–33 These MSn experiments, although elegant, require larger ion abundances and/or longer scan times due to the multi-stage nature of the ion isolation and activation sequences, thus preventing their ready adaptation for low ion signals in a high throughput mode. Continuation of the development of lipids as biomarkers and elucidating their role in cellular processes will require new MS/MS methods that provide even greater structural specificity and sensitivity, especially for isomer differentiation and quantification.
A number of new activation methods have been developed for or applied to lipid analysis in order to overcome some of the deficiencies of traditional collision-based MS/MS methods (e.g. insufficient fragmentation to pinpoint double bond positions or characterize fatty acid chains). Infrared multiphoton dissociation (IRMPD), electron capture issociation (ECD), and electron detachment dissociation (EDD) have been evaluated for gangliosides, and this comparison showed that ECD allowed identification of both portions of the ceramide moiety as well the presence of an acetyl group for those with an N-acetylated sugar.34 Both ECD and electron transfer dissociation (ETD) methods have been utilized for the analysis of glycerophosphocholine lipids after adduction with metal ions and proved useful for revealing the degree of unsaturation of the fatty acid substituents35,36 but exhibited limited advantages over CID-based methods. Extensive use of electron-based activation methods for many classes of lipids has been impeded by their acidic nature which makes them more readily ionized in the negative mode via deprotonation rather than as the multi-protonated species best suited for ECD and ETD, which is largely why electron-based activation methods have not been extensively adopted by the lipidomics community. An innovative alternative strategy for elucidation of double bond positions in unsaturated lipids, termed ozone electrospray ionization-MS (OzESI-MS)37–39 or ozone induced dissociation-MS (OzID-MS) depending on the mode of implementation,40–42 exploited a unique ozone-induced fragmentation process that led to product ions arising from a specific double bond cleavage. This method was recently adapted for ambient ionization.41 Similarly Cooks et al. demonstrated that low temperature plasma ionization, another type of ambient ionization, produced ozone in situ and induced preferential oxidation and cleavage of lipid double bonds, allowing the assignment of the double bond positions of unsaturated fatty acids.21
Over the past decade, ultraviolet photodissociation (UVPD) has gained general recognition as a versatile ion activation option for the analysis of proteins, nucleic acids and more recently lipids and glycans.43–59 Several methods utilizing UVPD have been developed for lipid analysis. Reilly et al. reported that 157 nm UVPD of leukotrienes produced CID-like fragment ions as well as UVPD-specific diagnostic radical ions that allowed differentiation of isomers.55 More recently Blanksby et al. have demonstrated a new radical directed dissociation (RDD) method that led to intrachain fragmentation of fatty acyl groups. This approach entailed attachment of a 4-iodobenzyl group to the lipid, either by non-covalent adduction or covalent derivatization, which served as a UV chromophore at 266 nm and a radical initiator.56,57 RDD-based methods afforded the differentiation of isobaric lipids and determination of chain branching based on selective C-C cleavages that CID or OzID could not provide alone. We have previously demonstrated the benefits of UVPD (193 nm) for characterization of lipid A structures, and the rich fragmentation patterns generated by UVPD of lipid anions offered a means to elucidate unusual amino acid modifications of the lipid A structures.58,60,61 It is this latter work that motivated the present study which is aimed specifically at the structural characterization of gangliosides.
As shown in this report, UVPD results in the production of several types of diagnostic fragment ions, including glycosidic cross-ring cleavages and C-C and C-N bond cleavages at the sphingosine and fatty acid moieties. These UVPD-specific cleavages enhance the elucidation of the hydrophobic lipid chains and the structures of glycans that have proven to be particular problematic by other MS/MS methods and show considerable promise for isomer differentiation. This UVPD methodology was then utilized in conjunction with HPLC to elucidate a complex ganglioside mixture.
Experimental
Reagents and Solution Preparation
C12 β-D-lactosyl ceramide (LacCer(18:1/12:0)), a mixture of isolated GM3 gangliosides from bovine milk, and a complex mixture of porcine brain gangliosides were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Solutions for direct infusion and HPLC-MS mass spectrometry were prepared by diluting stock solutions to 1 μM in 50/50 methanol/water. All solvents used for solution preparation and LC-MS were LC or HPLC-MS grade and purchased from Sigma Aldrich (Saint Louis, MI).
Mass Spectrometry and Liquid Chromatography
All experiments were performed on an Orbitrap Elite mass spectrometer (Bremen, Germany) modified to perform ultraviolet photodissociation (UVPD) within the instrument's higher collision energy cell in the negative mode using a previously described set up.62 The mass spectrometer was equipped with a Coherent ExiStar XS excimer laser (Santa Clara, CA) producing 193 nm photons at a net energy of 4 mJ/pulse. All ESI mass spectra were acquired in the Orbitrap mass analyzer using a resolution of 240,000 (at m/z 400) and MS/MS spectra, also in the Orbitrap, were collected using a resolution of 15,000 (at m/z 400). All MS/MS spectra were collected using an isolation window of 3 m/z. CID mass spectra were collected using an activation qz value of 0.25 and a normalized collision energy (NCE) set at 39% with a total activation time of 20 ms. HCD experiments were collected using an activation qz value of 0.1 and an activation time of 0.1 ms. UVPD mass spectra were acquired using ten 5 ns pulses at 500 Hz using a qz value of 0.1. All direct infusion experiments were performed using solutions diluted to 1 μM (80:20 MeOH/H2O) and directly infused at 3 μL/min. For all experiments, the ESI source was set to 4 kV and with a sheath gas flow setting of 20 units.
Separation of ganglioside mixtures was undertaken with a Dionex Ultimate 3000 microbore liquid chromatography system (Sunnyvale, CA) equipped with an Agilent Poroshell 120 EC-C18 column (3 mm X 50 mm, 2.7 micron particles). Mobile phase A consisted of 50:50 methanol/water with 0.05% NH4OH, and mobile phase B (MPB) consisted of isopropyl alcohol (IPA) with 0.05% NH4OH. A linear gradient, using a flow rate of 300 μL/min, was employed starting at 15% MPB and ending at 90% MPB over a 20 minute period. The column was then flushed by holding at 90% MPB for 5 minutes before re-equilibrating the column at 15% MPB for 3 minutes. One microgram injections (20 μL volume using approximately 25 μM solutions in 50/50 methanol/water) of ganglioside extracts were injected onto the column and analyzed in triplicate. In all cases (CID, HCD, UVPD), the MS/MS spectra were based on single scans with each scan consisting of five microscans. Data dependent LC-MS was performed using a full mass scan (m/z range of 600-2000) followed by UVPD on the top five peaks from the survey scan event, which equated to about 40 MS/MS events per chromatographic peak.
Workflow for Identification of Gangliosides and Ganglioside Nomenclature
Ganglioside mixtures analyzed via direct infusion MS and LCMS were manually searched in the LIPID Metabolites and Pathways Strategy structure database (LIPID MAPS, www.lipidmaps.org.) Experimental masses were obtained from the m/z values in the ESI mass spectra and matched to all known sphingolipids within the lipid database. Search results were further narrowed by eliminating matches outside a mass error range of 10 ppm, thus reducing the potential for false positives. Final structures were elucidated by assigning the MS/MS fragment ions observed in CID, HCD or UVPD-MS spectra. MS/MS fragment ions were manually assigned based on the nomenclature originally proposed by Domon and Costello27 and later updated by Adams and Ann63 (see Figure 1). Ganglioside structure assignment and labeling followed the nomenclature proposed by the IUPAC-IUB Joint Commission on Biochemical Nomenclature for glycolipids.64
Figure 1.
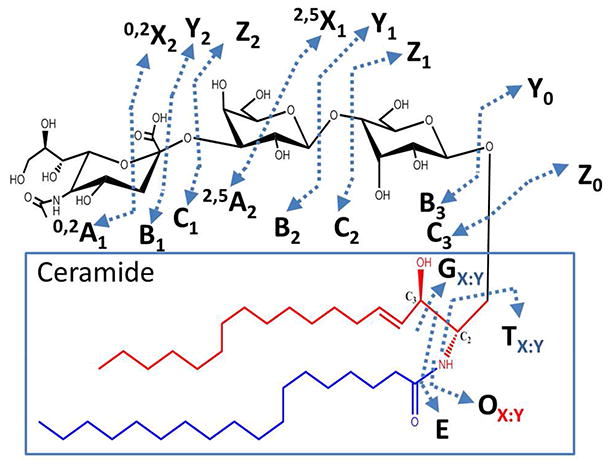
MS/MS nomenclature, adapted to include cleavages and types of fragment ions produced by UVPD of glycosphingolipids. The glycan moiety is designated in black, the sphingoid base carbon tail is designated in red, and the fatty acid tail is designated in blue. The ceramide, composed of the sphingosine and fatty acid chains, is boxed in blue. The UVPD-specific fragment ions are labelled as G, O, and E. G is produced upon cleavage of the bond between C2-C3 in the sphingosine tail. O is created upon loss of the amide-linked fatty acid chain. E is produced upon cleavage of the C2-C3 sphingoid base bond and the amide-linked fatty acid chain. For G, O and T ions, the subscript X designates the total number of carbons in the hydrophobic chain and Y designates the number of unsaturated carbon-carbon bonds in the fragment ion.
Results and Discussion
We report the utilization of CID, HCD, and 193 nm UVPD to characterize gangliosides using an Orbitrap mass spectrometer in the negative mode. Gangliosides are sialic acid-containing glycosphingolipids that can vary in the number of sugars and the lengths of the attached sphingoid and fatty acid groups. CID and HCD result primarily in glycosidic cleavages with little to no cleavages of the ceramide moieties. The analytical merits of UVPD are compared relative to CID and HCD for the structural analysis of gangliosides with respect to the overall diversity of fragment ions for each activation method. CID, HCD and UVPD mass spectra are presented for LacCer(18:1/12:0), GM3 gangliosides from bovine milk, and GD1 and GT1b from porcine brain extracts. Finally the benefits of LC-UVPD-MS are demonstrated for the high throughput analysis of gangliosides from a complex porcine brain extract.
MS/MS of LacCer(18:1/12:0) and Milk Gangliosides
CID, HCD and UVPD mass spectra for singly deprotonated LacCer(18:1/12:0) (see structure in Supplemental Figure 1) are shown in Figure 2. The CID spectrum was dominated by glycosidic cleavages that resulted in Y0, Y1 and Z1 ions. A minor cross ring cleavage product (0,2X1) and a single oligosaccharide terminal B2 fragment ion were also observed, but no cleavages occurred at the hydrophobic ceramide. The corresponding HCD mass spectrum (Figure 2B) extensively mirrored the CID mass spectrum, but also displayed two additional terminal saccharide ions, the B1 and C1 ions. The UVPD mass spectrum is strikingly different, with B/Y and C/Z pairs and several new fragment ion types (labelled G, O and E which are highlighted green in Figure 2C) arising from cleavages of the hydrophobic ceramide moiety (refer to Figure 1). In fact, the most dominant product ion upon UVPD was the G12:0 ion where M corresponds to the specific cleavage of the C2-C3 sphingosine bond and 12:0 represents the carbon chain length and hydrogen saturation of the fatty acid chain. Another informative ceramide cleavage occurred at the C-N amide bond which links the fatty acid chain moiety to the sphingosine moiety and is labelled O18:1 with the numerical subscript conveying the length and saturation of the sphingosine hydrophobic chain. One other prominent UVPD-specific ion (E type ion) evolves from cleavage of both the C2-C3 linkage and the amide-linked fatty acid chain. These new UVPD-specific G, O, and E ions, unlike the conventional B/Y and C/Z glycosidic cleavages observed upon CID and HCD, are highly diagnostic for the ceramide chains. Neither HCD nor CID promoted cleavages at the sphingosine/fatty acid moieties nor provided any information to characterize the glycosphingolipid ceramide structure. The total numbers of carbons in the LacCer(18:1/12:0) ceramide could be elucidated based on the Yo fragment ion upon CID or HCD. However this Y0 fragment ion did not provide specific structural information about the lengths of each of the two constituent hydrocarbon chains. UVPD produced the widest and most diverse array of LacCer(18:1/12:0) fragment ions and allowed the most complete structural analysis of both alkyl chains in the ceramide base and elucidation of the glycan sugars.
Figure 2.
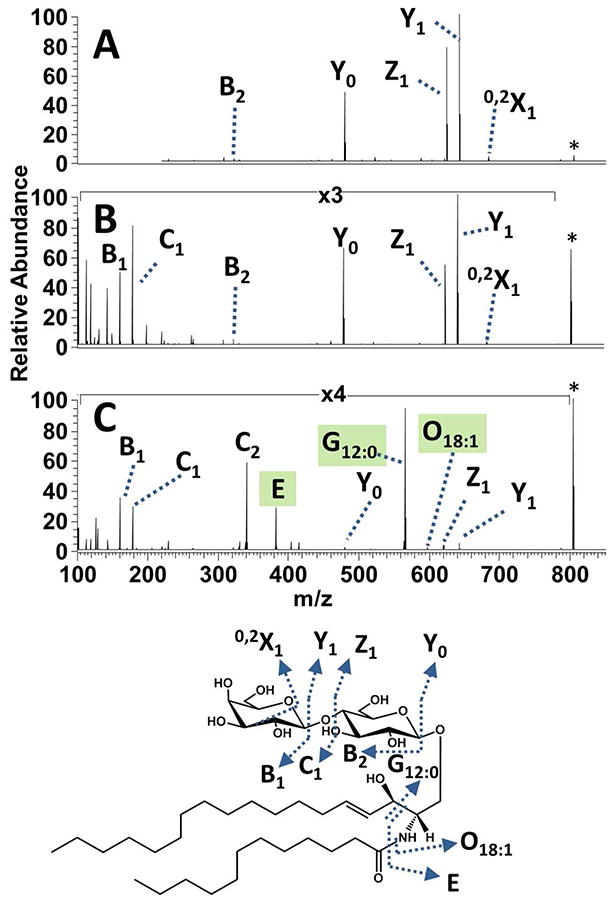
(A) CID, (B) HCD and (C) UVPD mass spectra of the singly deprotonated sphingoglycolipid LacCer(d18:1/12:0). The precursor ion is labelled with an asterisk. A comprehensive fragmentation map is shown below the spectra.
An extract of GM3 gangliosides from bovine milk with various chain lengths and common sugar moieties was analyzed by ESI-MS/MS as described above. The CID spectrum of the singly deprotonated ganglioside GM3(18:1/23:0) in Figure 3A shows dominant glycosidic cleavages leading to Y ions and a few low abundance Z ions. Two low abundance cross-ring cleavage products (0,2X2 and 0,2A2) and T23:0 ions, reported previously,27 appeared in the lower m/z region. HCD of deprotonated GM3(18:1/23:0) (Figure 3B) yielded a similar array of fragment ions, mostly Y and Z ions. The HCD spectrum, unlike the CID spectrum, contained a single dominant B1 ion (representing a glycosidic bond cleavage between the GM3 sialic acid and hexose residues). T23:0 was the only fragment ion diagnostic for the GM3 ceramide structure observed in both the CID and HCD spectra. In contrast, UVPD yielded the most diverse array of fragment ion types with the glycosidic cleavages producing complementary B/Y and C/Z ions (Figure 3C with m/z values reported in Supplemental Table 1). These ceramide and glycan terminal fragments provided the most confident structural assignment of the oligosaccharide portion. UVPD also afforded several new and unique ion types via cross-ring cleavages. These UVPD-specific A/X ions were most useful for elucidation of branching patterns of oligosaccharide chains, and mirrored the same types of fragment ions observed for oligosaccharides upon 193 nm UVPD.59 Furthermore, C-C and C-N cleavages upon UVPD led to G23:0, O18:1 and E fragment ions that revealed the structure of the hydrophobic ceramide of GM3(18:1/23:0). Several different G and O fragment ions were observed in Figure 3C, indicating a mixture of isobaric GM3 gangliosides. In particular, the O17:1/G24:0 and O19:1/G22:0 ions correspond to GM3(17:1/24:0) and GM3(19:1/22:0) isobars. CID and HCD only led to a single low abundance diagnostic ion (T23:0) which confirmed a single ganglioside species. Based on the UVPD mass spectrum, there is evidence for three isobaric gangliosides (GM3(17:1/24:0), GM3(18:1/23:0) and GM3(19:1/22:0), all with a nominal mass of 1250 Dalton) and yielding complementary G/O ion fragment ion pairs. CID-MS3 was undertaken on the Y0 fragment ion (from Figure 3A) and resulted in the dominant neutral loss of 30 Da (Y0-CH2O) as well as several other ceramide cleavages (producing P, T22:0 T23:0, T24:0, S, R, V, and U ions) (see Supplemental Figure 2). These MS3 results for the Y0 fragment ion confirmed that the G and O ions observed in the UVPD spectrum (Figure 3C) provided direct evidence of the isobaric mixture. The G and O ions have low abundances upon UVPD, but the signal-to-noise ratios are sufficiently high to allow them to be used as diagnostic ions for revealing the presence of isobaric contributions.
Figure 3.
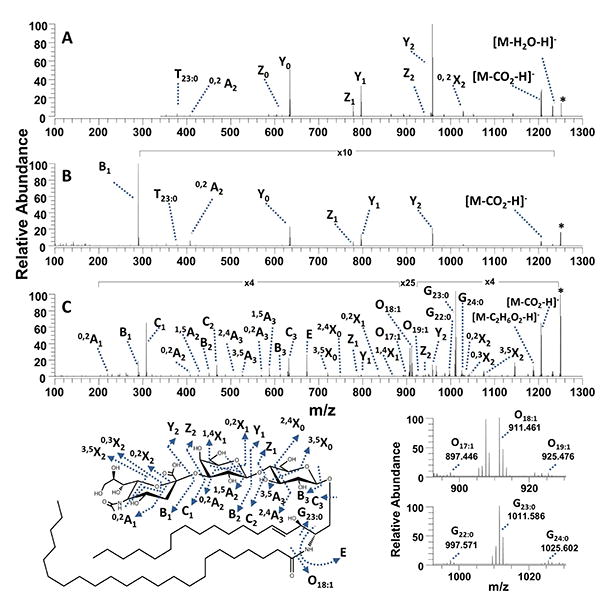
(A) CID, (B) HCD and (C) UVPD mass spectra of the singly deprotonated bovine milk ganglioside GM3(18:1/23:0). The precursor ion is labelled with an asterisk. A fragmentation map is shown at the bottom left. Expanded regions of the UVPD spectrum are shown on the bottom right to highlight the key O and G ions. The m/z values of the fragment ions in C (UVPD) are summarized in Supplemental Table 1.
To systematically evaluate the analytical utility of UVPD relative to CID and HCD for differentiating isobaric gangliosides, several GM3 milk gangliosides were analyzed and the diagnostic fragments compared for each activation method. The fatty acid hydrophobic chain lengths vary in gangliosides, and typically the sphingoid base length is assumed to be 18 carbons long with a single saturation site (18:1) because it is the most common base length observed in mammals.65 However there are reports of variation in ganglioside sphingoid base hydrocarbon chain lengths depending on the type of biological sample, and in particular milk sphingoid bases and gangliosides have been reported with odd carbon chain lengths and structures.65–67 Several GM3 gangliosides with nominal masses of 1179 Da, 1207 Da, 1235 Da, and 1263 Da, corresponding to the gangliosides GM3(18:1/18:0), GM3(18:1/20:0), GM3(18:1/22:0), and GM3(18:1/24:0) (assuming the gangliosides have a sphingoid base composition of 18:1), respectively, were analyzed. The CID and HCD mass spectra of these species are shown in Supplemental Figures 3 and 4, respectively. CID produced mainly ceramide terminal Y and a few Z ions. The HCD spectra showed a single dominant B1 ion with low abundance Y and Z ions. (If this prominent B1 ion had also been formed upon CID, its low m/z value would prohibit its detection due to the low mass cutoff associated with CID in ion traps.) The only changes observed for each GM3 species in the HCD and CID spectra were shifts of 28 Da in the Y and Z ions, indicating additions of C2H4 groups in the alkyl chains. Without more elaborate MSn experiments, the HCD and CID fragmentation patterns were not conducive to elucidation of the sphingoid base, and the variations in the MS/MS patterns might be errantly ascribed to changes within the fatty acid moieties. The UVPD spectra of the same GM3 gangliosides (Supplemental Figure 5) showed the widest range of fragment ions and thus were most distinctive for characterization of the milk gangliosides. For the UVPD spectra, glycan terminal A, B, C and D ions and many complementary X, Y, and Z fragment ions were observed, and the types of G/O ion pairs were very diagnostic for each GM3 species. In particular the presence of the G20:0 and O16:1 ions arose from ganglioside GM3(16:1/20:0) that is isobaric to the expected GM3(18:1/18:0) precursor. For each ganglioside, the M and H ions revealed the presence of isobaric structures. In fact, both GM3(16:1/22:0) and GM3(18:1/20:0) were identified as components of the mixture in Supplemental Figure 5b (m/z 1207); GM3(18:1/22:0), GM3(16:1/24:0) and GM3(17:1/23:0) were identified as components of the mixture in Supplemental Figure 5c (m/z 1235); and likewise GM3(18:1/24:0) and GM3(19:1/23:0) in Supplemental Figure 5d (m/z 1263). The types and abundance of each G varied for each selected GM3. There was some variation in the abundances of the T ions in the HCD and CID spectra that appeared to correlate with the ceramide structure; however the abundances of the T ions were very low and there was no consistent indication of ceramide heterogeneity via production of different T ions. In contrast, UVPD produced dominant G and O fragment ions which revealed both the sphingoid base and the fatty acid hydrophobic chains of each of the isobaric gangliosides. In general, UVPD provided the most diverse and robust catalog of fragment ions for characterizing both the acidic oligosaccharide and ceramide moieties as well as producing a wider range of A, B, C, X, Y, and Z ions for elucidation of the glycan core.
The charge state of the ganglioside also had a significant impact on the observed fragmentation pathways. GM3 gangliosides only contain a single highly acidic site and thus naturally favored the formation of singly deprotonated species (1-). However, addition of 0.05% NH4OH to the 1 μM 50:50 methanol/water solutions allowed generation of 2- ions upon ESI, albeit of low abundance. The CID and HCD mass spectra of the resulting doubly deprotonated GM3(18:1/23:0) species (Supplemental Figure 6A and Supplemental Figure 6B) were similar to each other but strikingly different from those obtained for the singly deprotonated species. CID and HCD of the doubly deprotonated GM3(18:1/23:0) yielded abundant A, B and C ions in each spectrum and a few low abundance Z and Y ions, contrasting with the dominant Y and Z ions produced for the singly deprotonated precursors. There were also noticeable differences in the in the abundance and number of the T fragment ions upon CID or HCD. CID and HCD of doubly deprotonated GM3(18:1/23:0) yielded a mixture of T23:0, T22:0, and T24:0 fragment ions, whereas the latter two product ions were not observed upon activation of the singly charged precursor (Figure 3A and Figure 3B). Interestingly, the ceramide-terminal Y ions which were prominent for the singly charged precursor had low abundances or were absent from the CID and HCD spectra of the doubly deprotonated precursors. UVPD, unlike CID and HCD, did not exhibit any significant charge state dependence on the types of ganglioside fragment ions. The UVPD spectrum of the doubly deprotonated GM3(18:1/23:0) (Supplemental Figure 6C with m/z values summarized in Supplemental Table 1) displayed a diverse mixture of A, B, C, X, Y, Z, G, E and O fragment ions and had significant overlap with the UVPD spectrum of the singly charged GM3(18:1/23:0) species (Figure 3C). Additionally, the photodissociation efficiency (i.e. conversion of precursor to product ions and calculated by dividing the sum of the abundances of all product ions by the sum of the abundances of all product ions plus the surviving precursor) was greater for the doubly deprotonated precursor (84% conversion) relative to the singly deprotonated species (56% conversion). One reason for the apparent increase in photodissociation efficiency is the relatively large abundance of the electron photodetachment product ([M – 2H]-) observed upon UVPD of the doubly charged precursor. This analogous photodetachment product would be a neutral species for the single charged precursor and thus not detectable. It is also conceivable that the charge reduced products may dissociate to other informative fragments, and these secondary products would be neutral and not detectable for the singly charged precursor but could be the origin of some of the singly charged fragment ions for the doubly deprotonated precursor. The theoretical monoisotopic exact masses of the fragment ions for GM3 are summarized in Supplemental Table 1 along with the experimental masses of the ions in Figure 3C and Supplemental Figure 6C obtained upon UVPD. The experimental masses are generally consistent with even electron fragment ions for both singly and doubly deprotonated precursors. However, some of the G and A (cross-ring cleavage) fragment ions are shifted by 1 to 2 Da, suggesting that they are ones that could evolve from secondary dissociation after the electron photodetachment process.
MS/MS and LC-MS of Brain Gangliosides
The composition of gangliosides found in brain tissue varies considerably with respect to the number of appended sugar residues. Moreover, brain gangliosides contain mixtures of ceramide species comprised of a sphingoid base of 18 or 20 carbons long and fatty acid chains which vary in length. To evaluate the utility of UVPD for characterization of brain gangliosides, a mixture was first screened by direct infusion ESI-MS. A doubly deprotonated ganglioside of m/z 917.479 (corresponding to a mixture of the branched GD1a(18:1/18:0) and GD1b(18:1/18:0)) was analyzed by HCD, CID and UVPD. Based on the MS/MS spectra, there were fragment ions which could be assigned to either GD1a or GD1B. The spectral analysis was streamlined by basing the interpretation on the GD1a structure (fragment ions labelled in black font) and annotating fragment ions specific to GD1b in red font. The CID and HCD mass spectra of GD1(18:0/18:0) are presented in Figure 4A and Figure 4B, respectively. Each MS/MS spectra exhibited two major fragments, B1α/1β and Y4α/4β, attributed to cleavages of the sialic acid glycosidic bonds. The other diagnostic fragment ions were low abundance, and the only fragment ions that distinguished GD1a from GD1b were the two low abundance B2β (colored red for GD1b) and B3β (colored black for GD1b) fragment ions. The HCD mass spectrum displayed a very minor fatty acid cleavage product, T18:0 that aided in the elucidation of ceramide structure. The high resolution/high mass accuracy of the Orbitrap mass spectrometer was essential for resolving the C1β and T18:0 ions (Supplemental Figure 7). The UVPD spectrum of GD1, presented in Figure 4C, exhibited a broader range of diagnostic fragment ions that proved key for characterization of the branched ganglioside. The oligosaccharide moiety was readily mapped via complementary A/X, B/Y and C/Z ion pairs, and UVPD was also effective for differentiating the GD1a specific fragment ions (labelled red) from GD1b fragment ions via the complete series of A, B and C fragment ions. The UVPD spectrum was particularly distinctive as it exhibited several internal fragment ions (B3-Z3β, C4-Z4, B4-Z4β, and C4-Z4) and dual branch fragment ions (Y2α-Y4β, Y3α-Y4β and Y4α-Y4β). Abundant fragments (G, O and E ions) arising from cleavages of the sphingoid base C2-C3 bond and the fatty acid-sphingoid C-N bond were essential for characterization of the ceramide moiety. Unlike the milk gangliosides, the brain gangliosides only showed evidence of GD1(18:1/18:0) and no other ceramide structural variants.
Figure 4.
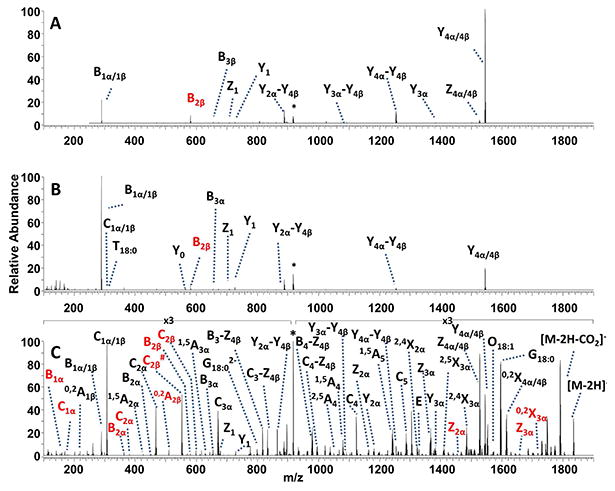
(A) CID, (B) HCD and (C) UVPD mass spectra of the doubly deprotonated porcine brain ganglioside GD1(18:1/18:0). The precursor ion is labelled with an asterisk. Fragment ions corresponding to GD1a(18:1/18:0) are labelled in black. Fragment ions corresponding to GD1b(18:1/18:0) are labelled in red.
GT1b(18:1/18:0), a branched ganglioside with three sialic acids, was analyzed using CID, HCD and UVPD. This ganglioside generated ions in two charges states (2-, 3-) upon ESI. HCD and CID (Supplemental Figure 7A and 7B) of triply deprotonated GT1b primarily resulted in cleavages at sialic acid-containing residues (B1β/1α, Y4α/4β) and few diagnostic ions to characterize the structure of GT1b. UVPD (Supplemental Figure 8C) yielded a wide range of fragments, including cleavages at the ceramide moiety, dual cleavages at both glycan branches, and a few internal ions. The UVPD mass spectrum also exhibited some multiply charged fragment ions (1,5A52-, C52-, D2-, Z4α/4β2-, M18:02- and 0,2X4β). Neutral loss of CO2 from the C2β product and the electron photodetachment product, [M-3H]2-, were also observed as non-informative pathways, but overall the UVPD mass spectrum of GD3b was more diagnostic than the HCD or CID spectra. There was no evidence in the UVPD spectrum to suggest an isomeric mixture of GT1a and GT1b.
A HPLC-UVPD-MS method was developed to examine the array of gangliosides in a brain tissue extract. Details about the LCMS conditions are described in the experimental section, the resulting total ion chromatogram is shown in Figure 5, and the identified gangliosides are listed in Table 1. The UVPD-MS methodology allowed identification of 27 gangliosides among five classes, as summarized in Supplemental Table 2. The most abundant gangliosides were GD1(18:1/18:0), GD1(20:1/18:0), GM1(18:1/18:0) and GM1(20:1/18:0). Highly branched GT1 and GQ1 species and several modified species including acetylated and fucosylated gangliosides were identified. A number of GD1 and GM1 gangliosides co-eluted, but the rich UVPD spectra provided sufficient unique diagnostic fragment ions to assign the structures. Several sulfotides, a type of sulfoglycolipid, were also found in the mixture (Supplemental Table 3). The UVPD mass spectra and structures of the sulfotides are shown in Supplemental Figures 9 and 10. The UVPD patterns of the sulfoglycolipids contained several distinctive fragments arising from cleavages adjacent to the carbonyl or hydroxyl groups along with UVPD-specific G, E and O fragment ions. UVPD of (3'-sulfo)Galβ-Cer(d18:1/24:0(2OH)) resulted in a unique cleavage adjacent to the hydroxyl-modified fatty acid chain (labelled J). Similarly cleavages adjacent to the C=C bond of the fatty acid chain of (3'-sulfo)Galβ-Cer(d18:1/24:1(15Z)) (yielding the ions labeled F(15Z) and m/z 750.490 and m/z 774.491) aided in the elucidation of the double bond position within the fatty acid chain. Each UVPD mass spectrum consisted of a single scan (comprised of five microscans) which is identical to the scan parameters used for CID and HCD data collection and allows approximately 40 MS/MS events per chromatographic peak. LC-UVPD-MS is thus a versatile alternative to LC-CID-MS or LC-HCD-MS and does not necessitate the use of MSn protocols that may require significant signal averaging.
Figure 5.
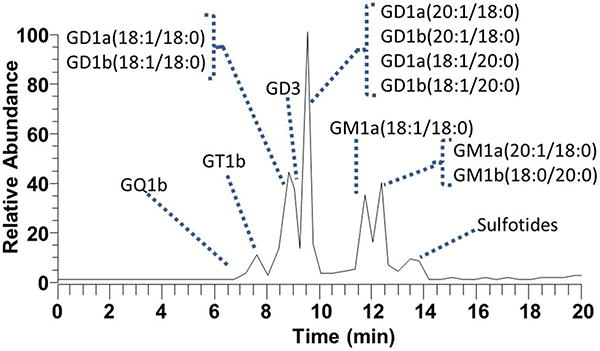
LC-MS total ion chromatogram of porcine brain ganglioside extract.
Conclusions
Ultraviolet photodissociation (UVPD) produced a wider and more diverse array of fragment ions than CID or HCD for the structural characterization of gangliosides and glycosphingolipids. CID and HCD resulted predominantly in glycan fragments which limited the ability to characterize the ceramide moiety. Several unique UVPD-specific cleavages at ceramide C-C and C-N bonds as well as additional cross ring cleavages of the glycan moieties contributed to the richer fragmentation patterns observed upon UV photoactivation, ultimately aiding the differentiation of isobaric ceramide and glycan moieties in both milk and brain gangliosides and alleviating the reliance on more elaborate MSn methods. Implementation of UVPD in an LCMS workflow allowed the high throughput separation and analysis of brain gangliosides.
Supplementary Material
Acknowledgments
Funding from the NIH (R01GM103655) and the Welch Foundation (F1155) is acknowledged. We thank Thermo Fisher Scientific with helping on the modifications to the Orbitrap Elite mass spectrometer to allow UVPD.
References
- 1.Merrill AH, Jr, Wang MD, Park M, Sullards MC. Trends Biochem Sci. 2007;32:457–468. doi: 10.1016/j.tibs.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Merrill AH, Stokes TH, Momin A, Park H, Portz BJ, Kelly S, Wang E, Sullards MC, Wang MD. J Lipid Res. 2009;50:S97–S102. doi: 10.1194/jlr.R800073-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu RK, Tsai YT, Ariga T. Neurochem Res. 2012;37:1230–1244. doi: 10.1007/s11064-012-0744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Posse de Chaves E, Sipione S. FEBS Lett. 2010;584:1748–1759. doi: 10.1016/j.febslet.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Van Echten-Deckert G, Walter J. Prog Lipid Res. 2012;51:378–393. doi: 10.1016/j.plipres.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Ariga T, McDonald MP, Yu RK. J Lipid Res. 2008;49:1157–1175. doi: 10.1194/jlr.R800007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Kiechle FL. Ann Clin Lab Sci. 2004;34:3–13. [PubMed] [Google Scholar]
- 8.Bobowski M, Cazet A, Steenackers A, Delannoy P. Carbohydrate Chemistry: Volume 37. Vol. 37. The Royal Society of Chemistry; 2012. pp. 1–20. [Google Scholar]
- 9.Wenk MR. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 10.Blanksby SJ, Mitchell TW. Annu Rev Anal Chem. 2010;3:433–465. doi: 10.1146/annurev.anchem.111808.073705. [DOI] [PubMed] [Google Scholar]
- 11.Wenk MR. Cell. 2010;143:888–895. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 12.Murphy RC, Gaskell SJ. J Biol Chem. 2011;286:25427–25433. doi: 10.1074/jbc.R111.233478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fhaner CJ, Liu S, Ji H, Simpson RJ, Reid GE. Anal Chem. 2012;84:8917–8926. doi: 10.1021/ac302154g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Fhaner CJ, Ferguson-Miller SM, Reid GE. Int J Mass Spectrom. 2012;316–318:100–107. doi: 10.1016/j.ijms.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X, Yang K, Gross RW. Mass Spectrom Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han X, Gross RW. Expert Rev Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 17.Lee H, Lerno LA, Jr, Choe Y, Chu CS, Gillies LA, Grimm R, Lebrilla CB, German JB. Anal Chem. 2012;84:5905–5912. doi: 10.1021/ac300254d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flangea C, Serb A, Sisu E, Zamfir AD. Biochim Biophys Acta BBA - Mol Cell Biol Lipids. 2011;1811:513–535. doi: 10.1016/j.bbalip.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Schiopu C, Vukelić Ž, Capitan F, Kalanj-Bognar S, Sisu E, Zamfir AD. Electrophoresis. 2012;33:1778–1786. doi: 10.1002/elps.201200044. [DOI] [PubMed] [Google Scholar]
- 20.Richards AL, Lietz CB, Wager-Miller J, Mackie K, Trimpin S. J Lipid Res. 2012;53:1390–1398. doi: 10.1194/jlr.D019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oradu SA, Cooks RG. Anal Chem. 2012;84:10576–10585. doi: 10.1021/ac301709r. [DOI] [PubMed] [Google Scholar]
- 22.Paglia G, Ifa DR, Wu C, Corso G, Cooks RG. Anal Chem. 2010;82:1744–1750. doi: 10.1021/ac902325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JI, Tao WA, Cooks RG. Anal Chem. 2011;83:4738–4744. doi: 10.1021/ac1030946. [DOI] [PubMed] [Google Scholar]
- 24.Angel PM, Spraggins JM, Baldwin HS, Caprioli R. Anal Chem. 2012;84:1557–1564. doi: 10.1021/ac202383m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis SR, Hughes JR, Mitchell TW, in het Panhuis M, Blanksby SJ. The Analyst. 2012;137:1100. doi: 10.1039/c1an15864c. [DOI] [PubMed] [Google Scholar]
- 26.Vaikkinen A, Shrestha B, Nazarian J, Kostiainen R, Vertes A, Kauppila TJ. Anal Chem. 2012;85:177–184. doi: 10.1021/ac302432h. [DOI] [PubMed] [Google Scholar]
- 27.Domon B, Costello CE. Biochemistry (Mosc) 1988;27:1534–1543. doi: 10.1021/bi00405a021. [DOI] [PubMed] [Google Scholar]
- 28.Meisen I, Peter-Katalinić J, Müthing J. Anal Chem. 2003;75:5719–5725. doi: 10.1021/ac0347617. [DOI] [PubMed] [Google Scholar]
- 29.Vukelić Ž, Zamfir A, Bindila L, Froesch M, Peter-Katalinić J, Usuki S, Yu R. J Am Soc Mass Spectrom. 2005;16:571–580. doi: 10.1016/j.jasms.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Sørensen LK. Rapid Commun Mass Spectrom. 2006;20:3625–3633. doi: 10.1002/rcm.2775. [DOI] [PubMed] [Google Scholar]
- 31.Hsu FF, Turk J. J Am Soc Mass Spectrom. 2010;21:657–669. doi: 10.1016/j.jasms.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu FF, Turk J. J Am Soc Mass Spectrom. 2008;19:1681–1691. doi: 10.1016/j.jasms.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serb A, Schiopu C, Flangea C, Sisu E, Zamfir AD. J Mass Spectrom. 2009;44:1434–1442. doi: 10.1002/jms.1625. [DOI] [PubMed] [Google Scholar]
- 34.McFarland MA, Marshall AG, Hendrickson CL, Nilsson CL, Fredman P, Månsson JE. J Am Soc Mass Spectrom. 2005;16:752–762. doi: 10.1016/j.jasms.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Liang X, Liu J, LeBlanc Y, Covey T, Ptak AC, Brenna JT, McLuckey SA. J Am Soc Mass Spectrom. 2007;18:1783–1788. doi: 10.1016/j.jasms.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James PF, Perugini MA, O'Hair RAJ. J Am Soc Mass Spectrom. 2008;19:978–986. doi: 10.1016/j.jasms.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Thomas MC, Mitchell TW, Harman DG, Deeley JM, Murphy RC, Blanksby SJ. Anal Chem. 2007;79:5013–5022. doi: 10.1021/ac0702185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas MC, Mitchell TW, Blanksby SJ. J Am Chem Soc. 2005;128:58–59. doi: 10.1021/ja056797h. [DOI] [PubMed] [Google Scholar]
- 39.Sun C, Zhao YY, Curtis JM. Anal Chim Acta. 2013;762:68–75. doi: 10.1016/j.aca.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Thomas MC, Mitchell TW, Harman DG, Deeley JM, Nealon JR, Blanksby SJ. Anal Chem. 2007;80:303–311. doi: 10.1021/ac7017684. [DOI] [PubMed] [Google Scholar]
- 41.Ellis SR, Hughes JR, Mitchell TW, het Panhuis M, Blanksby SJ. The Analyst. 2012;137:1100. doi: 10.1039/c1an15864c. [DOI] [PubMed] [Google Scholar]
- 42.Brown SHJ, Mitchell TW, Blanksby SJ. Biochim Biophys Acta BBA - Mol Cell Biol Lipids. 2011;1811:807–817. doi: 10.1016/j.bbalip.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 43.Ly T, Julian RR. Angew Chem Int Ed. 2009;48:7130–7137. doi: 10.1002/anie.200900613. [DOI] [PubMed] [Google Scholar]
- 44.Diedrich JK, Julian RR. Anal Chem. 2011;83:6818–6826. doi: 10.1021/ac201647w. [DOI] [PubMed] [Google Scholar]
- 45.Thompson MS, Cui W, Reilly JP. J Am Soc Mass Spectrom. 2007;18:1439–1452. doi: 10.1016/j.jasms.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Reilly JP. Mass Spectrom Rev. 2009;28:425–447. doi: 10.1002/mas.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon SH, Moon JH, Kim MS. J Mass Spectrom. 2010;45:806–814. doi: 10.1002/jms.1773. [DOI] [PubMed] [Google Scholar]
- 48.Morgan JW, Russell DH. J Am Soc Mass Spectrom. 2006;17:721–729. doi: 10.1016/j.jasms.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Guan Z, Kelleher NL, O'Connor PB, Aaserud DJ, Little DP, McLafferty FW. Int J Mass Spectrom Ion Process. 1996;157–158:357–364. [Google Scholar]
- 50.Rosu F, Gabelica V, De Pauw E, Antoine R, Broyer M, Dugourd P. J Phys Chem. 2012;116:5383–5391. doi: 10.1021/jp302468x. [DOI] [PubMed] [Google Scholar]
- 51.Racaud A, Antoine R, Dugourd P, Lemoine J. J Am Soc Mass Spectrom. 2010;21:2077–2084. doi: 10.1016/j.jasms.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 52.Brodbelt JS. J Am Soc Mass Spectrom. 2011;22:197–206. doi: 10.1007/s13361-010-0023-6. [DOI] [PubMed] [Google Scholar]
- 53.Shaw JB, Madsen JA, Xu H, Brodbelt JS. J Am Soc Mass Spectrom. 2012;23:1707–1715. doi: 10.1007/s13361-012-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madsen JA, Boutz DR, Brodbelt JS. J Proteome Res. 2010;9:4205–4214. doi: 10.1021/pr100515x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devakumar A, O'Dell DK, Walker JM, Reilly JP. J Am Soc Mass Spectrom. 2008;19:14–26. doi: 10.1016/j.jasms.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Pham HT, Trevitt AJ, Mitchell TW, Blanksby SJ. Rapid Commun Mass Spectrom. 2013;27:805–815. doi: 10.1002/rcm.6503. [DOI] [PubMed] [Google Scholar]
- 57.Pham HT, Ly T, Trevitt AJ, Mitchell TW, Blanksby SJ. Anal Chem. 2012;84:7525–7532. doi: 10.1021/ac301652a. [DOI] [PubMed] [Google Scholar]
- 58.Madsen JA, Cullen TW, Trent MS, Brodbelt JS. Anal Chem. 2011;83:5107–5113. doi: 10.1021/ac103271w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ko BJ, Brodbelt JS. Anal Chem. 2011;83:8192–8200. doi: 10.1021/ac201751u. [DOI] [PubMed] [Google Scholar]
- 60.Hankins JV, Madsen JA, Giles DK, Childers BM, Klose KE, Brodbelt JS, Trent MS. Mol Microbiol. 2011;81:1313–1329. doi: 10.1111/j.1365-2958.2011.07765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hankins JV, Madsen JA, Giles DK, Brodbelt JS, Trent MS. Proc Natl Acad Sci. 2012;109:8722–8727. doi: 10.1073/pnas.1201313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. J Am Chem Soc. 2013;135:12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams J, Ann Q. Mass Spectrom Rev. 1993;12:51–85. [Google Scholar]
- 64.Eur J Biochem. 1998;257:293–298. doi: 10.1046/j.1432-1327.1998.2570293.x. [DOI] [PubMed] [Google Scholar]
- 65.Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes CA, Sullards MC, Liotta DC, Merrill AH. J Lipid Res. 2008;49:1621–1639. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martín MJ, Martín-Sosa S, Hueso P. Lipids. 2001;36:291–298. doi: 10.1007/s11745-001-0720-x. [DOI] [PubMed] [Google Scholar]
- 67.Merrill AH. Chem Rev. 2011;111:6387–6422. doi: 10.1021/cr2002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


