Abstract
Targeted gene delivery vectors can enhance cellular specificity and transfection efficiency. We demonstrated previously that conjugation of Tet1, a peptide that binds to the GT1b ganglioside, to polyethylenimine results in preferential transfection of neural progenitor cells in vivo. In this work, we investigate the effect of Tet1 ligand density on gene delivery to neuron-like, differentiated PC-12 cells. A series of statistical, cationic peptide-based polymers containing various amounts (1—5 mol%) of Tet1 were synthesized via one-pot reversible addition-fragmentation chain transfer (RAFT) polymerization by copolymerization of Tet1 and oligo-l-lysine macromonomers with N-(2-hydroxypropyl)methacrylamide (HPMA). When complexed with plasmid DNA, the resulting panel of Tet1-functionalized polymers formed particles with similar particle size as particles formed with untargeted HPMA–oligolysine copolymers. The highest cellular uptake in neuron-like differentiated PC-12 cells was observed using polymers with intermediate Tet1 peptide incorporation. Compared to untargeted polymers, polymers with optimal incorporation of Tet1 increased gene delivery to neuron-like PC-12 cells by over an order of magnitude but had no effect compared to control polymers in transfecting NIH/3T3 control cells.
Keywords: Non-viral gene delivery, Peptide copolymers, Targeted delivery, Neuron delivery, HPMA polymer
1. Introduction
Gene delivery can potentially treat a range of neurological diseases inadequately addressed by current therapeutics. Delivery of genes expressing various neurotropic factors have been studied for neuroprotection and axonal regeneration following central nervous system (CNS) injury or for delayed progression of amyotrophic lateral sclerosis, Huntington's and Parkinson's [1–3]. Of the available delivery technologies, adeno-associated viral vectors have been most extensively explored for CNS gene delivery applications due to their high transduction efficiency and innate neural tropism [4], but immunogenicity, long-term safety, and cost of manufacturing remain significant concerns. Non-viral delivery systems, such as cationic polymers or liposomes, can potentially overcome such limiting barriers; however, relatively poor transfection efficiency and high cytotoxicity remain challenges [5].
The incorporation of targeting ligands into non-viral gene delivery vehicles has been shown to both increase gene delivery efficiency and specificity. Various ligands, such as folate [6], transferrin [7], and RGD sequences [8], have been used to mediate cellular binding and internalization. It has been postulated that to achieve specificity in vivo, the density of targeting ligands must be controlled [9]. Recent work reported that intermediate levels of ligand density in folate [10], transferrin [11], and antibody-targeted nanoparticles [12] conferred the highest level of tissue specificity in vivo.
Current approaches towards multivalent decoration of polymeric nanoparticles typically involve grafting of ligands onto the polymer carrier. However, control and reproducibility of synthesis remain a challenge for these approaches, leading to incomplete coverage or varied surface functionalization of nanoparticles [13]. We have developed an approach to controllably incorporate ligands into a polymeric construct through direct copolymerization of functionalized ligand monomers, allowing direct control over material properties. This method allows for control over ligand density, orientation of display, and architectural display in the final polymeric construct, overcoming some limitations of grafting approaches.
We recently reported the synthesis and optimization of well-defined, narrowly disperse oligo-l-lysine-HPMA cationic polymers utilizing reverse addition-fragmentation chain transfer (RAFT) polymerization to copolymerize HPMA with methacrylamido-functionalized peptide macromonomers [14]. Polymers displaying multiple peptide entities can be easily synthesized using this approach, and incorporation of water-soluble peptides can be controlled based on feed ratios [15,16]. This platform can therefore be used to probe the effect of targeting ligand density on polymer gene transfer efficiency. Tet1, a peptide identified by in vitro phage display, binds to GT1b gangliosides, sphingophospholipids highly expressed on neuronal cell types [17]. Tet1 has been used in applications such as peripheral neuron labeling and in the delivery of PLGA nanoparticles and polymersomes to neuronal targets [18–20]. Our group has previously shown neuronal transfection in vitro using Tet1-modified poly(ethylenimine) (PEI) polyplexes and specific gene delivery to neural stem and progenitor cells in vivo upon intraventricular administration using PEGylated versions of these materials [21,22].
We report here the synthesis and evaluation of a series of peptide-based polymers containing varying amounts of Tet1 for targeted gene delivery to neuron-like cells. We utilize RAFT polymerization for one-pot synthesis of three component peptide– polymers using Tet1 as a targeting sequence, oligolysine for DNA binding and condensation, and HPMA for colloidal stability. Polyplexes were formed by self-assembly of polycations with plasmid DNA, and characterized by YOYO-1 DNA packaging assay and particle sizing. Cellular uptake as a function of Tet1 modification as well as gene transfection efficiency was studied in cultured neuronlike cells.
2. Materials and methods
2.1. Materials
N-(2-Hydroxypropyl)methacrylamide (HPMA) was purchased from Poly-sciences (Warrington, PA). The initiator VA-044 was purchased from Wako Chemicals (Richmond, VA). Fmoc-protected amino acids and HBTU were purchased from AAPPTec (Louisville, KY), N-succinimidyl methacrylate from TCI America (Portland, OR), and Rink Amide Resin from EMD Biosciences (Darmstadt, Germany). All other materials were reagent grade or better and were purchased from Sigma—Aldrich (St. Louis, MO) unless otherwise stated. Endotoxin-free plasmid pCMV-Luc2 was prepared using the Qiagen Plasmid Giga kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations.
2.2. Synthesis of peptide monomers
Two peptide sequences were synthesized on solid support: K3-Tet1 (KKKHLNILSTLWKYR) and the oligolysine K10 (KKKKKKKKKK). Peptides were synthesized via solid phase peptide synthesis following standard Fmoc chemistry using an automated PS3 Peptide Synthesizer (Protein Technologies, Phoenix, AZ). 6-Aminohexanoic acid (Ahx) was added to the N-terminus of the peptide sequences. Prior to peptide cleavage from resin, the N-termini of the peptides were deprotected and coupled with N-succinimidyl methacrylate. These methacrylamido-functionalized peptides were cleaved from resin by treatment with trifluoroacetic acid (TFA)/triiso-propylsilane (TIPS)/1,3-dimethoxybenzene/ddH2O (90:2.5:5:2.5, v/v/v/v) for 3 h under gentle mixing. Cleaved peptide monomers were precipitated in ice-cold ether, dissolved in methanol and re-precipitated twice in ice-cold ether. Peptide monomers were analyzed by RP-HPLC and MALDI-TOF MS and purified as necessary.
2.3. Polymer synthesis
A series of copolymers were synthesized with varying amounts of Tet1 peptide in the feed (0%, 1%, 3%, 5%) while holding K10 peptide constant at 20%. Monomers were dissolved in acetate buffer (1 m, pH 5.1) with 10% ethanol (v/v) such that the final monomer concentration of the solution was 0.5 m. The RAFT chain transfer agent (CTA) used was ethyl cyanovaleric trithiocarbonate (ECT, molecular weight 263.4 g/mol) and the initiator (I) used was VA-044. The molar ratios of total monomer:CTA:I at the start of the polymerization were 190:1:0.1. The reaction solutions were transferred to round bottom flasks, capped with a rubber septa, purged with argon for 10 min, and then submerged in a 44 °C oil bath to initiate polymerization. The polymerization was allowed to proceed for 48 h. The flasks were removed from the oil bath and polymers dialyzed against distilled H2O to remove unreacted monomers and buffer salts. The dialyzed products were lyophilized dry.
2.4. Size exclusion chromatography
Molecular weight analysis was carried out by size exclusion chromatography. The copolymers were dissolved at 2 mg/mL in running buffer (1:1 MeOH:300 mm acetate buffer, pH 4.4) for analysis by size exclusion chromatography – multiangle laser light scattering (SEC—MALLS). Analysis was carried out on an OHpak SB-804 HQ column (Shodex, Kawasaki, Japan) in line with a miniDAWN TREOS multiangle laser light scattering detector (Wyatt, Santa Barbara, CA) and an OptiLab rEX refractive index detector (Wyatt). Absolute molecular weight averages (Mn, Mw) were calculated using ASTRA software (Wyatt).
2.5. Amino acid analysis
The actual incorporated amount of peptide and HPMA in the final copolymers was determined through modified amino acid analysis as previously reported [23]. Briefly, hydrolyzed polymers were derivatized with o-phthalaldehyde/β-mercap-topropionic acid and run on a Zorbax Eclipse X-18 (Agilent Technologies, Santa Clara, CA) HPLC column with pre-column derivatization to label hydrolyzed amino acids and 1-amino-2-propanol (hydrolysis product of HPMA). Calibration curves were generated using serial dilutions of (l)-lysine, (l)-histidine, and reagent grade 1-amino-2-propanol.
2.6. Polyplex formation
pCMV-Luc2 plasmid was diluted in ddH2O to a concentration of 0.1 mg/mL and mixed with an equal volume of polymer (in ddH2O) at the desired amino to phosphate (N:P) ratio. After mixing, polyplexes were allowed to form for 10 min at room temperature.
2.7. Polyplex sizing by dynamic light scattering (DLS)
Polyplexes (0.5 μg DNA, 10 μL) were formed with polymers pHT1K10, pHT3K10, pHT5K10, and pHK10 at N:P ratios of 2,3, and 4 and were mixed with either 90 μL of ddH2O or PBS such that the final salt concentration was 150 mm. Particle size was determined by dynamic light scattering (ZetaPLUS, Brookhaven Instruments Corp, Holtsville, NY).
2.8. DNA condensation using YOYO-1 fluorescence quenching assay
pCMV-Luc2 plasmid was mixed with the bis-intercalating dye YOYO-1 iodide (Invitrogen, Carlsbad, CA) at a dye/base pair ratio of 1:50 and incubated at room temperature for 1 h. Polyplexes were formed at N/P ratios of 0, 1, 2, 4, 6, and 10 by complexing YOYO-1-labeled DNA with pHT1K10, pHT3K10, pHT5K10, or pHK10. Ten microliters (containing 0.5 μg DNA) of polyplex was added to each well of a 96-well plate, followed by 90 μL of ddH2O. The fluorescence from each well was measured on a Tecan Safire2 plate reader with excitation at 491 nm and emission at 509 nm. The fluorescence signal for each N/P ratio was normalized to the N/P 0 (uncomplexed DNA) signal.
2.9. Cell culture
NIH/3T3 cells and PC-12 cells were grown according to the ATCC recommendations. For in vitro studies, PC-12 cells were seeded onto collagen-coated plates. PC-12 cells were differentiated in F12K media supplemented with 1% HS, 1% ABAM, and 100 ng/mL nerve growth factor. Media was changed every 2 days.
2.10. Polyplex uptake
pCMV-Luc2 plasmid was mixed with the bis-intercalating dye YOYO-1 iodide at a dye/base pair ratio of 1:50 and incubated at room temperature for 1 h. Polyplexes were formed at N/P 3 by complexing YOYO-1 labeled DNA with pHK10, pHT1K10, pHT3K10, and pHT5K10 polymers. Labeled polyplexes were added to cells for 2 h at 37 °C. Cells were washed 3 times with PBS, detached by treatment with collagenase, and then washed twice more before labeling with propidium iodide (PI) stain. Cells were analyzed for fluorescence intensity by flow cytometry using the MACSQuant Analyzer (Miltenyi Biotec, Cologne, Germany) and gated for PI- (live) cells.
2.11. In vitro transfection efficiency
PC-12 and NIH/3T3 cells were transfected as previously described [14]. Briefly, polyplexes (1 μg DNA) were formed at 2.5, 3, and 4 N:P, diluted to 200 μL in Opti-MEM (Invitrogen), and added to cells for 4 h. After an additional 44 h, luciferase expression was quantified using a luciferase assay kit (Promega, Fitchburg, WI) according to the manufacturer's instructions, except with an additional freeze–thaw cycle at −20 °C to ensure complete cell lysis. Luminescence intensity was measured on a Tecan Safire2 plate reader (Männerdorf, Switzerland) with 1 s integration; total protein content was measured using a BCA Protein Assay Kit (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions so luciferase activity could be normalized to the total protein content. Each sample was tested in triplicate.
2.12. Hemolysis assay
A hemolysis assay evaluating the membrane-lytic activity of the materials was performed as previously described [24]. Briefly, plasma was removed from freshly-drawn human blood via centrifugation. The erythrocyte layer was washed 3× with 150 mm NaCl and resuspended in PBS. Polymers and polyplexes at various N/P with 1% Triton X-100 as a control were added to the erythrocyte suspensions in 96-well plates and incubated at 37 °C for 1 h. The plate was then centrifuged to pellet intact cells and released hemoglobin from lysed cells was measured spectrophotometrically at 541 nm absorbance. Percent hemolysis was calculated relative to Triton X-100.
3. Results and discussion
3.1. Polymer and polyplex characterization
A series of peptide–HPMA copolymers were synthesized via RAFT polymerization of peptide monomers with HPMA to investigate the effect of Tet1 peptide density on gene transfection using oligolysine-co-HPMA copolymers (Scheme 1). Monomer feed ratio and degree of polymerization were based on previous optimization studies suggesting a 4:1 HPMA to MaAhxK10 ratio at a degree of polymerization (DP) 150—190 yielded efficient vectors [14].
Scheme 1.
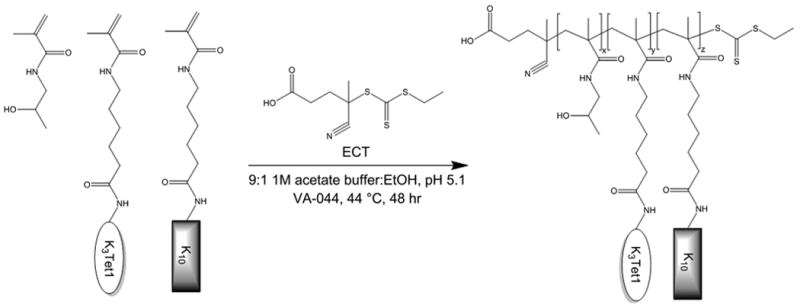
Synthetic scheme of Tet1-co-oligolysine-co-HPMA copolymers.
The molecular weight and composition of synthesized copolymers are summarized in Table 1. Copolymers were synthesized with narrow polydispersities (<1.2) and molecular weights in the 55−80 kDa range. Copolymers containing ∼ 1, 3 and 5 mol% Tet1 and ∼ 13%—23% K10, as determined by amino acid analysis, were prepared. pHK10 was used as an untargeted control for all studies.
Table 1.
Properties of HPMA–oligolysine copolymers.
| HPMA–oligolysine copolymer | % Tet1 mole feed | Determined Mn (kDa)a | Mw/Mna | Mole % Tet1b | Mole % K10b |
|---|---|---|---|---|---|
| pHT1K10 | 1% | 54.53 | 1.07 | 0.72 | 13.3 |
| pHT3K10 | 3% | 79.51 | 1.04 | 2.86 | 14.3 |
| pHT5K10 | 5% | 65.92 | 1.11 | 4.87 | 23.2 |
| pHK10 | – | 65.5 | 1.14 | – | 20.5 |
Determined by SEC–MALLS.
Determined by amino acid analysis.
3.2. Polyplex characterization
Polymers were used to formulate polyplexes (cationic polymer/plasmid DNA complexes) in either ddH2O or PBS buffer containing physiological salt concentrations. The average hydrodynamic diameter of polyplexes was measured using dynamic light scattering (Fig. 1). Polymers showed comparable polyplex size (100− 150 nm) in ddH2O regardless of Tet1 incorporation, suggesting that Tet1 incorporation does not affect particle formation. In physiological salt, 3% and 5% Tet1 polymers showed improved salt stability relative to 1% Tet1 and untargeted polymers. Inclusion of hydrophobic domains has previously been shown to improve colloidal stability [25]. Overall, relatively good salt stability is observed, with particles increasing to 200−300 nm in size in the presence of physiological salt. Particles show slight increase in size over time in PBS (Supplementary Fig. S1) but are stable against flocculation in PBS at polyplex concentrations both 20-fold higher and 50-fold lower than transfection formulation concentrations. This correlates well with previous work optimizing oligolysine-co-HPMA copolymers where polymers in the 60−80 kDa mass range showed good colloidal stability [14].
Fig. 1.
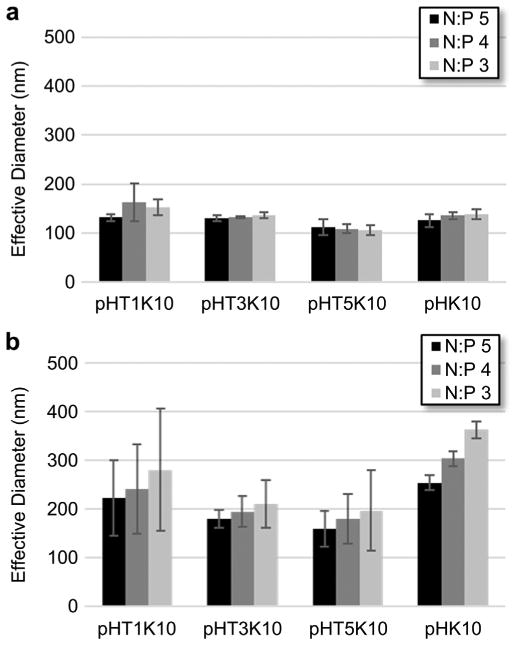
Particle size of polyplexes at various N/P ratios in (a) ddH2O and (b) 150 mM PBS.
To determine the effects of Tet1 incorporation on plasmid complexation and condensation, DNA condensation was monitored using YOYO-1 fluorescence quenching assay. In this assay, plasmid DNA is labeled with a DNA-intercalating dye fluorophore YOYO-1. DNA condensation results in self-quenching of the YOYO-1 fluorescence due to electronic interactions between nearby dyes [26]. The YOYO-1 fluorescence, normalized to uncondensed plasmid, is shown in Fig. 2 as a function of polymer DNA N/P ratio. Complexation of plasmid DNA with the various peptide–HPMA copolymers resulted in increasing YOYO-1 quenching with higher N/P ratios up to ∼ N/P = 6. Comparable trends in YOYO-1 fluorescence quenching are observed between the polymers, suggesting Tet1 functionali-zation does not affect plasmid complexation.
Fig. 2.
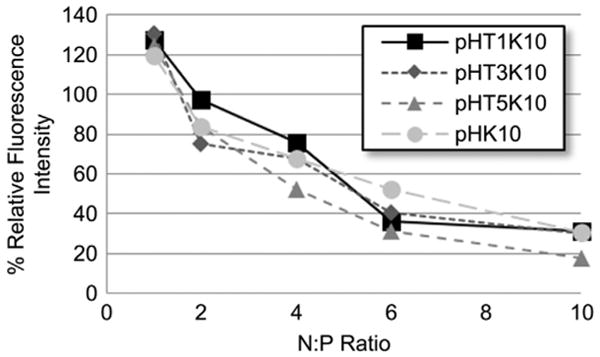
YOYO-1 fluorescence quenching assay as a measure of DNA complexation and condensation in ddH2O.
3.3. In vitro cellular uptake
Treatment of PC-12 pheochromocytoma cells with nerve growth factor (NGF) results in neuronal differentiation; cells acquire a neuron-like phenotype with extended neurites and reduced cell proliferation [27,28]. PC-12 cells differentiated for 6 days with NGF were therefore treated with fluorophore-labeled polyplexes (via YOYO-1 labeling) for 2 h at 37 °C to determine relative cellular uptake of Tet1-targeted polyplexes. Cells were washed, stained with propidium iodide for dead cells, and then analyzed by flow cytometry to determine the relative polyplex uptake of live cells based on mean fluorescence intensity.
There is a 1.41-fold increase in mean fluorescence intensity at 2 h with pHT3K10 and 1.92-fold increase in intensity with pHT5K10 in 6-day differentiated PC-12 cells. pHT1K10 shows comparable cellular uptake to pHK10 (0.91-fold increase), suggesting that at very low modifications (∼1−2 peptides per polymer chain), we observe no increase in cellular uptake. However, at higher levels (4−10 peptides per polymer), we observe increased cellular uptake as a function of increasing Tet1 modification. Similar transferrin-and bFGF-targeted poly-l-lysine nanoparticles have also shown around 2−4 fold increased cellular association while displaying over an order of magnitude increased transgene expression in vitro [7].
Multivalent display of targeting ligands by polymers and nanoparticles has been shown to give significantly enhanced binding due to avidity [29]. Dendrimeric display of folate, for example, has been able to show decreased effective Kd by over 4 orders of magnitude [30]. However, recent work suggests optimal cellular specificity based on differential receptor expression benefits most from using low-affinity ligands [31]. While multivalent display of antibodies leads to effective ∼pm Kd, cellular specificity is lost to high avidity due to increased binding to both target and non-target cells [12]. Instead, multivalent display of weak targeting ligands allows for super-selective targeting based on particle ligand density and cellular receptor density [31]. The observed increased cellular uptake with pHT3K10 and pHT5K10 relative to pHT1K10 suggests insufficient ligand density in pHT1K10 for effective binding to target PC-12 cells.
3.4. In vitro gene delivery
Gene transfection efficiency of the various Tet1-modified oligolysine–HPMA copolymers was then evaluated in neuron-like PC-12 cells and in control NIH/3T3 murine fibroblast cells. Polyplexes formulated at various charge ratios (N/P 2.5–4) were used to transfect 6-day differentiated PC-12 cells. Several charge ratios (N/P 2.5–4) were evaluated. The transfection efficiency of pHT1K10 is comparable to pHK10 for all N:P ratios tested, showing that low levels of Tet1 incorporation have no effects on transfection efficiency or cytotoxicity (Fig. 3a). pHT3K10 and pHT5K10 showed significantly increased transfection efficiency at all three N/P ratios tested, with 11.5- and 10.7-fold higher luciferase expression at N/P 3 for pHT3K10 and pHT5K10 compared to pHK10, respectively, with no observed increase in toxicity (Fig. 3c). At N/P 4, there was a 54.1-and 89.3-fold increase in transfection efficiency in pHT3K10 and pHT5K10 but that corresponded to a 30% and 60% reduction in cell viability, respectively, compared to a 15% drop in viability of pHK10.
Fig. 3.
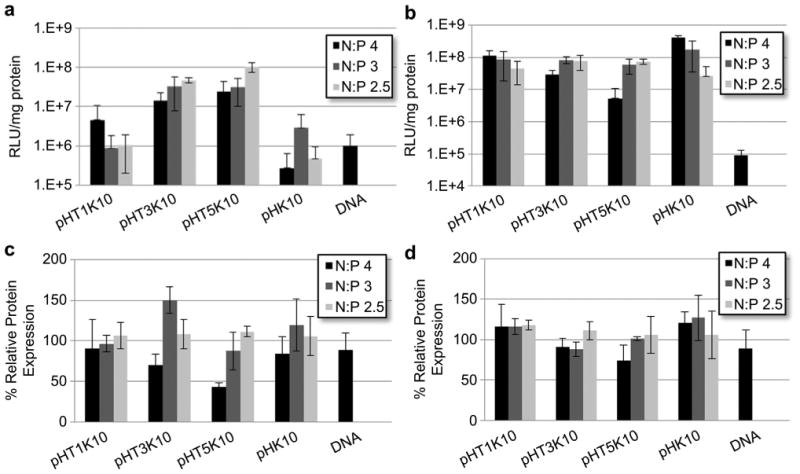
Normalized luminescence per mg protein as a measure of transfection efficiency for (a) 6-day differentiated PC-12 cells and (b) NIH/3T3 cells. Protein content normalized to untreated cells for (c) 6-day differentiated PC-12 cells and (d) NIH/3T3 cells as a measure of cellular viability.
The Tet1-modified oligolysine–HPMA copolymers were next tested in control NIH/3T3 cells, murine fibroblasts that express gangliosides at low levels. At N/P 3, the transfection efficiency of all materials was comparable, with no noticeable toxicity observed for any of the materials (Fig. 3b, d). pHT1K10 and pHK10 copolymers transfected NIH/3T3 cells with increasing efficiency as a function of increasing N/P with no observable toxicity at the highest N/P tested. In contrast, polymers with higher Tet1 peptide functionalization, pHT3K10 and pHT5K10, show decreased transfection efficiency at N/P 4, likely a result of decreased cellular viability at high N/P. This increased toxicity as a function of Tet1 peptide incorporation is consistent with what was observed for differentiated PC-12 cells.
Higher cytotoxicity of 3% and 5% Tet1 polymers compared to untargeted and 1% Tet1 polymer was observed in both control NIH/3T3 and target differentiated PC-12 cells, suggesting non-specific toxicity related to Tet1 peptide incorporation. Higher incorporation of Tet1 (pHT5K10 > pHT3K10) correlated with increased toxicity in a concentration-dependent manner. The Tet1 peptide sequence is relatively hydrophobic and suffers from poor solubility in water. Incubation with free Tet1 peptide alone shows no toxicity at concentrations >100 μM (data not shown), suggesting that toxicity is not due toTet1 peptide alone but in combination with the lysine-based polymers. Inclusion of hydrophobic domains has been shown to increase toxicity in several cationic vectors, postulated through increased membrane lyticity [32]. Polycationic polymers with increasing hydrophobicity demonstrated increasing hemolytic and antimicrobial activities [33]. Polylysines have been implicated in mitochondrial membrane disruption in vitro and have been implicated in induction of apoptosis via release of cytochrome c and induction of the caspase cascade [34].
Cationic polymers typically suffer from high cytotoxicity [35], but our group has recently reported several strategies incorporating enzymatically-labile or reducible motifs to improve biocompatibility [36,37]. Additionally, oligo-l-lysine copolymers undergo proteolytic degradation, with up to a 60% reduction in molecular weight thereby allowing for renal clearance of systemically-administered materials [36]. These strategies can be combined with optimized Tet1-targeted materials to produce efficient, biocompatible materials.
3.5. Hemolysis assay
To determine if Tet1 peptide was increasing transfection efficiency through increased endosomal escape, the membrane lyticity of the polymer materials was determined via hemolysis assay. Free polymer and polyplexes were incubated with freshly isolated human erythrocytes and tested for plasma membrane disruption by detecting hemoglobin release (Fig. 4). Less than 2% hemolysis was observed at all polymer concentrations used during transfection studies. Likewise, erythrocytes treated with polyplexes showed membrane lysis was around 1%–2%, suggesting that insignificant membrane disruption is occurring even at 5% Tet1 incorporation. Therefore, observed increased transfection efficiency in 3% and 5% Tet1 copolymers is likely through increased cellular uptake and not through enhanced endosomal escape.
Fig. 4.
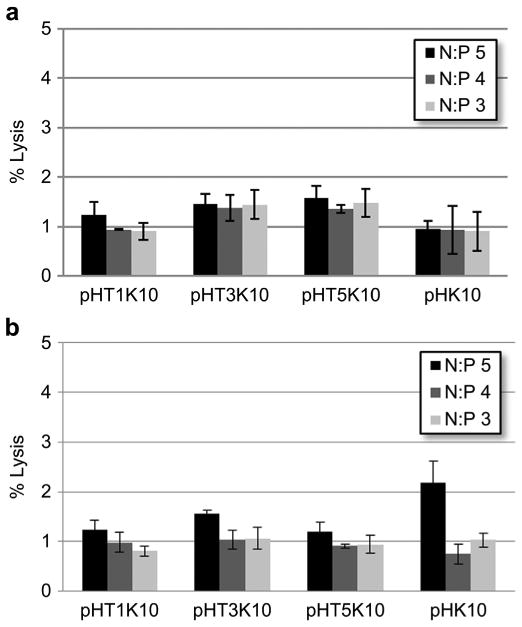
Percent hemolysis of (a) free polymer at concentrations equivalent to N/P ratios and (b) polyplexes relative to Triton X-100 control.
4. Conclusions
In this work, we have demonstrated the synthesis, characterization, and evaluation of Tet1-targeted HPMA–oligolysine co-polymers for targeted gene delivery to neuronal cells. Incorporation of Tet1 peptide did not affect polyplex size or DNA condensation. At moderate levels of peptide incorporation, increased cellular uptake and transfection efficiency was observed in neuron-like differentiated PC-12 cells but not in NIH/3T3 fibroblast cells. However, increased toxicity not related to plasma membrane disruption was observed with pHT3K10 and pHT5K10 polymers, suggesting a balance between enhanced transfection efficiency and increased cytotoxicity. At N/P 3, over an order of magnitude increase in luciferase expression was achieved without significant increased toxicity. Overall, incorporation of Tet1 targeting ligand enhanced gene delivery efficiency to neuron-like cells.
Supplementary Material
Acknowledgments
This work was supported by NIH/NINDS 1R01 NS064404. David Chu was supported by NIH T32 CA138312. We thank Profs. Anthony Convertine and Patrick Stayton for generous donation of the ECT chain transfer agent.
Footnotes
Appendix A. Supplementary data: Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.biomaterials.2013.08.045.
References
- 1.Hermens WT, Verhaagen J. Viral vectors, tools for gene transfer in the nervous system. Prog Neurobiol. 1998;55:399–432. doi: 10.1016/s0301-0082(98)00007-0. [DOI] [PubMed] [Google Scholar]
- 2.Lo Bianco C, Schneider BL, Bauer M, Sajadi A, Brice A, Iwatsubo T, et al. Lentiviral vector delivery of parkin prevents dopaminergic degeneration in an α-synuclein rat model of Parkinson's disease. Proc Natl Acad Sci U S A. 2004;101:17510–5. doi: 10.1073/pnas.0405313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Åkerud P, Canals JM, Snyder EY, Arenas E. Neuroprotection through delivery of glial cell line-derived neurotrophic factor by neural stem cells in a mouse model of Parkinson's disease. J Neurosci. 2001;21:8108–18. doi: 10.1523/JNEUROSCI.21-20-08108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, et al. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10:302–17. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 5.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–93. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 6.Morris VB, Sharma CP. Folate mediated l-arginine modified oligo (alkylaminosiloxane) graft poly (ethyleneimine) for tumor targeted gene delivery. Biomaterials. 2011;32:3030–41. doi: 10.1016/j.biomaterials.2010.11.048. [DOI] [PubMed] [Google Scholar]
- 7.Fisher KD, Ulbrich K, Subr V, Ward CM, Mautner V, Blakey D, et al. A versatile system for receptor-mediated gene delivery permits increased entry of DNA into target cells, enhanced delivery to the nucleus and elevated rates of transgene expression. Gene Ther. 2000;7:1337–43. doi: 10.1038/sj.gt.3301230. [DOI] [PubMed] [Google Scholar]
- 8.Park J, Singha K, Son S, Kim J, Namgung R, Yun CO, et al. A review of RGD-functionalized nonviral gene delivery vectors for cancer therapy. Cancer Gene Ther. 2012;19:741–8. doi: 10.1038/cgt.2012.64. [DOI] [PubMed] [Google Scholar]
- 9.Haun JB, Hammer DA. Quantifying nanoparticle adhesion mediated by specific molecular interactions. Langmuir. 2008;24:8821–32. doi: 10.1021/la8005844. [DOI] [PubMed] [Google Scholar]
- 10.Yamada A, Taniguchi Y, Kawano K, Honda T, Hattori Y, Maitani Y. Design of folate-linked liposomal doxorubicin to its antitumor effect in mice. Clin Cancer Res. 2008;14:8161–8. doi: 10.1158/1078-0432.CCR-08-0159. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Tian S, Petros RA, Napier ME, DeSimone JM. The complex role of multivalency in nanoparticles targeting the transferrin receptor for cancer therapies. J Am Chem Soc. 2010;132:11306–13. doi: 10.1021/ja1043177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zern BJ, Chacko AM, Liu J, Greineder CF, Blankemeyer ER, Radhakrishnan R, et al. Reduction of nanoparticle avidity enhances the selectivity of vascular targeting and PET detection of pulmonary inflammation. ACS Nano. 2013;7:2461–9. doi: 10.1021/nn305773f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakem IF, Leech AM, Johnson JD, Donahue SJ, Walker JP, Bockstaller MR. Understanding ligand distributions in modified particle and particlelike systems. J Am Chem Soc. 2010;132:16593–8. doi: 10.1021/ja107139c. [DOI] [PubMed] [Google Scholar]
- 14.Johnson RN, Chu DS, Shi J, Schellinger JG, Carlson PM, Pun SH. HPMA-oligo-lysine copolymers for gene delivery: optimization of peptide length and polymer molecular weight. J Control Release. 2011;155:303–11. doi: 10.1016/j.jconrel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson RN, Burke RS, Convertine AJ, Hoffman AS, Stayton PS, Pun SH. Synthesis of statistical copolymers containing multiple functional peptides for nucleic acid delivery. Biomacromolecules. 2010;11:3007–13. doi: 10.1021/bm100806h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, Schellinger JG, Johnson RN, Choi JL, Chou B, Anghel EL, et al. Influence of histidine incorporation on buffer capacity and gene transfection efficiency of HPMA-co-oligolysine brush polymers. Biomacromolecules. 2013;14:1961–70. doi: 10.1021/bm400342f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu JK, Teng Q, Garrity-Moses M, Federici T, Tanase D, Imperiale MJ, et al. A novel peptide defined through phage display for therapeutic protein and vector neuronal targeting. Neurobiol Dis. 2005;19:407–18. doi: 10.1016/j.nbd.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Zhang W, Johnston AH, Newman TA, Pyykko I, Zou J. Targeted delivery of Tet1 peptide functionalized polymersomes to the rat cochlear nerve. Int J Nanomed. 2012;7:1015–22. doi: 10.2147/IJN.S28185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathew A, Fukuda T, Nagaoka Y, Hasumura T, Morimoto H, Yoshida Y, et al. Curcumin loaded-PLGA nanoparticles conjugated with Tet-1 peptide for potential use in Alzheimer's disease. PLoS ONE. 2012;7:e32616. doi: 10.1371/journal.pone.0032616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Federici T, Liu JK, Teng Q, Yang J, Boulis NM. A means for targeting therapeutics to peripheral nervous system neurons with axonal damage. Neurosurgery. 2007;60:911–8. doi: 10.1227/01.NEU.0000255444.44365.B9. [DOI] [PubMed] [Google Scholar]
- 21.Park IK, Lasiene J, Chou SH, Horner PJ, Pun SH. Neuron-specific delivery of nucleic acids mediated by Tet1-modified poly(ethylenimine) J Gene Med. 2007;9:691–702. doi: 10.1002/jgm.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon EJ, Lasiene J, Jacobson BE, Park IK, Horner PJ, Pun SH. Targeted nonviral delivery vehicles to neural progenitor cells in the mouse subventricular zone. Biomaterials. 2010;31:2417–24. doi: 10.1016/j.biomaterials.2009.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bidlingmeyer BA, Cohen SA, Tarvin TL. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984;336:93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- 24.Schellinger JG, Pahang JA, Johnson RN, Chu DS, Sellers DL, Maris DO, et al. Melittin-grafted HPMA-oligolysine based copolymers for gene delivery. Biomaterials. 2013;34:2318–26. doi: 10.1016/j.biomaterials.2012.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filippov SK, Konak C, Kopeckova P, Starovoytova L, Spirkova M, Stepanek P. Effect of hydrophobic interactions on properties and stability of DNA–polyelectrolyte complexes. Langmuir. 2010;26:4999–5006. doi: 10.1021/la9036716. [DOI] [PubMed] [Google Scholar]
- 26.Krishnamoorthy G, Duportail G, Mély Y. Structure and dynamics of condensed DNA probed by 1,1′-(4,4,8,8-tetramethyl-4,8-diazaundecamethylene)bis[4-[[3- methylbenz-1,3-oxazol-2-yl]methylidine]-1,4-dihydroquinolinium] tetraiodide fluorescence. Biochemistry. 2002;41:15277–87. doi: 10.1021/bi020440y. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg ME, Greene LA, Ziff EB. Nerve growth factor and epidermal growth factor induce rapid transient changes in proto-oncogene transcription in PC12 cells. J Biol Chem. 1985;260:14101–10. [PubMed] [Google Scholar]
- 28.Greene L. Nerve growth factor prevents the death and stimulates the neuronal differentiation of clonal PC12 pheochromocytoma cells in serum-free medium. J Cell Biol. 1978;78:747–55. doi: 10.1083/jcb.78.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7:771–82. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 30.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Jr, Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem Biol. 2007;14:107–15. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Veracoechea FJ, Frenkel D. Designing super selectivity in multivalent nano-particle binding. Proc Natl Acad Sci U S A. 2011;108:10963–8. doi: 10.1073/pnas.1105351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Incani V, Lavasanifar A, Uludag H. Lipid and hydrophobic modification of cationic carriers on route to superior gene vectors. Soft Matter. 2010;6:2124–38. [Google Scholar]
- 33.Kuroda K, Caputo GA, DeGrado WF. The role of hydrophobicity in the antimicrobial and hemolytic activities of polymethacrylate derivatives. Chemistry. 2009;15:1123–33. doi: 10.1002/chem.200801523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A, Moghimi SM. Low and high molecular weight poly(l-lysine)s/poly(l-lysine)–DNA complexes initiate mitochondrial-mediated apoptosis differently. FEBS Lett. 2005;579:6191–8. doi: 10.1016/j.febslet.2005.09.092. [DOI] [PubMed] [Google Scholar]
- 35.Chu DS, Schellinger JG, Shi J, Convertine AJ, Stayton PS, Pun SH. Application of living free radical polymerization for nucleic acid delivery. Acc Chem Res. 2012;45:1089–99. doi: 10.1021/ar200242z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu DS, Johnson RN, Pun SH. Cathepsin B-sensitive polymers for compartment-specific degradation and nucleic acid release. J Control Release. 2012;157:445–54. doi: 10.1016/j.jconrel.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J, Johnson RN, Schellinger JG, Carlson PM, Pun SH. Reducible HPMA-co-oligolysine copolymers for nucleic acid delivery. Int J Pharm. 2012;427:113–22. doi: 10.1016/j.ijpharm.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


