Abstract
Perturbations in epigenetic mechanisms have emerged as cardinal features in the molecular pathology of major classes of brain disorders. We therefore highlight evidence which suggests that specific epigenetic signatures measurable in central—and possibly even in peripheral tissues—have significant value as translatable biomarkers for screening, early diagnosis, and prognostication; developing molecularly targeted medicines; and monitoring disease progression and treatment responses. We also draw attention to existing and novel therapeutic approaches directed at epigenetic factors and mechanisms, including strategies for modulating enzymes that write and erase DNA methylation and histone/chromatin marks; protein-protein interactions responsible for reading epigenetic marks; and non-coding RNA pathways.
Keywords: bromodomain, epigenomic, exosome, glioma, histone deacetylase, long non-coding RNA, microRNA
EPIGENETICS AND EPIGENETIC MEDICINE
While first-generation epigenetic medicines for cancer are already FDA approved and increasingly becoming available in the marketplace, the field of epigenetics is evolving rapidly and promises to deliver advanced technologies with more widespread applications, including novel diagnostic and therapeutic modalities for brain disorders. Basic neuroscience research is revealing that epigenetic processes play leading roles in generating the extraordinary structural and functional complexity of the nervous system. Epigenetic factors and mechanisms orchestrate brain development, adult neurogenesis, synaptic plasticity, stress responses, and aging and the transgenerational inheritance of cognitive and behavioral phenotypes [1]. Translational research efforts are similarly demonstrating how epigenetic mechanisms—and their deregulation—contribute to nervous system disease pathogenesis. Furthermore, targeting epigenetic processes in disease models shows the potential to dramatically reduce pathology and relieve symptoms, including mitigating neurodegeneration, promoting neural regeneration, and restoring cognitive functions [2-5]. Characterizing epigenetic and genome-wide epigenomic profiles and modulating epigenetic factors, therefore, represent novel and powerful paradigms for identifying and monitoring how nervous system diseases unfold and for halting, preventing or even reversing them. In this integrated overview, we highlight opportunities and challenges for developing these clinical applications.
EXAMINATION OF ABERRANT EPIGENETIC PROFILES IN NERVOUS SYSTEM DISEASES
The principal epigenetic factors mediate DNA methylation (Glossary) and hydroxymethylation, histone protein and chromatin modifications, and non-coding RNA (ncRNA) deployment (Table 1; Supplementary Table 1) [1]. These dynamic and environmentally responsive epigenetic processes collectively regulate the tissue- and cell-specific execution of genomic programs, including gene transcription; post-transcriptional RNA processing, transport and translation; X-chromosome inactivation; genomic imprinting; gene dosage; and maintenance of genomic integrity.
Table 1.
Key epigenetic factors.
| Key epigenetic factors |
|---|
| DNA methyltransferase (DNMT) enzymes |
| Methyl-CpG-binding domain (MBD) proteins |
| DNA excision repair enzymes |
| Gadd45 enzymes |
| Ten-eleven translocation enzymes |
| Histone modifying enzymes |
| Polycomb group (PcG) and Trithorax group (Tr×G) proteins |
| RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) |
| MicroRNAs (miRNA) |
| Long ncRNAs (lncRNA) |
Accumulating evidence demonstrates that genetic variation and functional abnormalities in epigenetic enzymes and related factors modify nervous system disease risk, onset, and progression. Epigenetic mechanisms regulate (and are regulated by) disease-associated genes and pathways. Accordingly, epigenetic profiles are ubiquitously abnormal in central and peripheral nervous system disease patient-derived tissues. Here, we draw attention to salient examples linking these signatures with clinicopathological features and relevant outcomes.
DNA methylation
DNA methylation profiles have been interrogated most extensively in cancer, and in brain tumors more so than in other nervous system diseases. These approaches unequivocally provide diagnostic and prognostic information and can guide therapeutic decision-making.
One key study utilizing Cancer Genome Atlas data revealed a DNA methylation pattern in adult glioblastoma multiforme (GBM) specimens referred to as the glioma CpG island methylator phenotype (G-CIMP; Glossary) [6]. This signature defines a molecular subgroup of GBM associated with secondary or recurrent tumors, isocitrate dehydrogenase 1 (IDH1) mutations, a distinct profile of copy number variation, and a specific proneural gene expression pattern. Clinically, GBM patients with G-CIMP-positive tumors are younger at diagnosis and have a more favorable prognosis (longer median survival) than those with G-CIMP-negative tumors. In addition, it was recently demonstrated that mutant IDH1 produces an oncometabolite, which deregulates DNA methylation and generates the G-CIMP, uncovering a causal link between genetic and epigenetic abnormalities in GBM [7]. Another study integrated global DNA methylation profiles with genomic and transcriptomic data from both pediatric and adult GBM specimens and identified six GBM subgroups associated with distinct clinical features, including patient age, tumor location, and overall survival [8]. A related study showed that 5-hydroxymethylcytosine levels in gliomas are inversely correlated with tumor grade and survival [9]. Together, these interesting observations imply that DNA (hydroxy)methylation profiles provide diagnostic and prognostic data.
In terms of management, a seminal study published in 2005 reported that O6-methylguanine-DNA methyltransferase (MGMT) gene promoter methylation status in GBM specimens determines whether adding the alkylating agent, temozolomide (TMZ), is beneficial for patients undergoing radiotherapy [10]. Large randomized trials have since confirmed that MGMT methylation is associated with treatment responsiveness and longer survival [11, 12]. This advantage is conferred because methylation is associated with lower levels of MGMT, a DNA repair enzyme that mitigates the effects of TMZ. Intriguingly, MGMT methylation status in tumor-derived tissue and in cell free circulating DNA in serum is highly concordant [13]. This finding suggests that serum MGMT methylation status can predict the efficacy of TMZ when tumor tissue is unavailable and, further, that there is active central-peripheral signaling in GBM coordinating DNA methylation states in the tumor and in serum. DNA methylation also provides clinically relevant data for solid tumors that metastasize to brain. Studies of lung, breast and renal cell carcinomas as well as melanomas suggest that the methylation status of MGMT and other genes influences metastatic potential, response to TMZ, and relapse rate [14-17].
Emerging data is revealing that, in addition to brain tumors, every major class of nervous system disease harbors either locus-specific or genome-wide alterations in DNA methylation [1]. In some cases, these abnormalities are present in disease-specific neural cell types and tissues and are clearly linked to pathogenic processes, particularly in certain neurodevelopmental and neurodegenerative disorders.
For other findings, such as DNA methylation abnormalities found in peripheral tissues, the mechanistic significance is unclear. At select loci, it appears that DNA methylation profiles in readily accessible peripheral tissues reflect those in brain directly. Alternatively, they can potentially have utility as indirect or surrogate markers. For example, monoamine oxidase A (MAOA) is an enzyme critical for neurotransmitter metabolism that exhibits variability in expression in the brain. One study found that MAOA gene promoter methylation in white blood cells predicts MAOA levels present in brain [18]. Another study identified a robust association between aging-related DNA methylation in multiple tissues, including blood and brain [19]. In addition, a large case-control study demonstrated that DNA methylation profiles at the frataxin gene locus in peripheral blood mononuclear cells and buccal cells from Friedreich's ataxia (FRDA) patients correlate with age of onset of symptoms and clinical disease severity [20]. Also, population-based studies have reported that methylation levels of repetitive elements (i.e., Alu and LINE-1)—de facto markers of global DNA methylation—in blood correlate with the risk for Alzheimer's disease (AD) [21] and for stroke and related diseases [22].
Histone and chromatin modifications
Impaired chromatin regulation is directly responsible for the pathogenesis of a spectrum of brain diseases. Indeed, there is a rapidly increasing inventory of mutations in genes encoding histones, histone modifying enzymes (Glossary), and other chromatin related factors known to cause forms of syndromic and non-syndromic intellectual and developmental disability, primary brain tumors, and other disorders [1, 8, 23-27]. However, the potential array of disruptions in histone modification and chromatin states, which result from these mutations and from additional defects in chromatin regulators found in other nervous system diseases, is much more diverse and multifaceted and, thus, less well characterized than DNA methylation abnormalities. It is nevertheless clear that these aberrations are also present in both central and peripheral tissues and associated with specific clinical phenotypes. For example, one study interrogated global levels of histone (H) 3 lysine (K) 18 (H3K18) and H3K9 acetylation, H3K4 dimethylation, and H4K20 trimethylation along with other characteristics in glioma specimens and identified subgroups in which specific histone modification patterns predict progression-free and overall survival [28]. In addition, disruptions in histone modification and chromatin states are prominent in neuropsychiatric disorders, such as depression, psychosis, and addiction [29, 30]. Although these have been investigated primarily in animal models and in post-mortem neuropathological specimens, preliminary data suggests that profiling peripheral tissues is informative. One study reported that levels of H3K9 dimethylation are elevated in lymphocytes of schizophrenic patients and correlate with the age of onset [31]. Also, the response to therapy can be mediated by specific histone modifications. A recent study showed that atypical antipsychotic agents decrease histone acetylation at the metabotropic glutamate 2 receptor gene promoter in mouse and human frontal cortex [32]. Similarly, in Huntington's disease (HD), genome-wide histone acetylation and methylation, histone variant expression, and the function of the master epigenetic regulatory factor, RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF; Glossary), are all aberrant in animal models of HD. These are consistent with abnormalities present in HD patient-derived tissues, including striatal and cortical brain regions and blood and potentially have value for diagnosis, prognostication and monitoring therapy [33, 34].
Non-coding RNAs
Many investigators have characterized microRNAs (miRNAs; Glossary) in brain diseases, and a small, but increasing, number has begun to interrogate the roles played by other classes of ncRNAs [35, 36]. Studying model systems demonstrates that ncRNA networks are highly integrated with disease-linked genes and pathways and that ncRNAs can directly modulate disease pathogenesis. For example, in Drosophila, miRNA-mediated RNA induced silencing complex (miRNA-RISC) pathways are perturbed by Parkinson's disease (PD)-causing mutations in leucine-rich repeat kinase 2; and resulting abnormalities in let-7 and mir-184 activity lead to degeneration of dopaminergic neurons [37]. mir-34 is expressed in adult Drosophila brain and significantly upregulated with aging; and loss of mir-34 leads to accelerated brain aging and neurodegeneration [4]. Levels of the skeletal muscle-specific miRNA, miR-206, are increased in amyotrophic lateral sclerosis (ALS) mouse models [38]. This miRNA mediates muscle-nerve crosstalk and slows the progression of ALS by enhancing regenerative responses at neuromuscular synapses.
Analyzing ncRNA profiles in tissues from patients with nearly every class of nervous system disease reveals alterations that are consistent with those in model systems, linked to pathogenesis, parallel between central and peripheral tissues, and indicative of clinicopathological phenotypes. For example, miRNAs, piwi-RNAs and long ncRNAs (lncRNAs; Glossary) are deregulated in the brain in animal models of stroke [39-42]. miRNA levels in the ischemic brain overlap with those in blood and have an evolving temporal profile. miRNA expression patterns in the blood of stroke patients correspond with this animal data, are suggestive of stroke mechanisms (i.e., large vessel, lacunar, or cardioembolic), and might provide a quantitative index for stroke severity and recovery potential. Expression patterns for other classes of ncRNAs have not been studied in stroke patients, with the exception of a single lncRNA, CDKN2B antisense RNA 1 (CDKN2B-AS1/ANRIL). Levels of CDKN2B-AS1/ANRIL in human carotid atherosclerotic plaques and peripheral blood T lymphocytes are linked to rates of ischemic and hemorrhagic stroke. Intriguingly, polymorphisms in the genomic locus for this lncRNA (9p21) modify the risk of developing a number of diseases, including atherosclerosis, coronary artery disease, and intracranial aneurysms [43-45]. These observations suggest that ncRNAs might represent the long sought after peripheral biomarkers for stroke risk stratification, rapid diagnosis, mechanistic classification, and prognostication.
Specific miRNA signatures predict time to relapse and overall survival in ependymomas [46]; leptomeningeal spread and responsiveness to chemotherapies in medulloblastomas [47]; event-free and overall survival and responsiveness to chemotherapies in neuroblastomas [48]; and recurrence rate in meningiomas [49]. Particular miRNA and lncRNA profiles in gliomas are associated with tumor grade, Karnofsky performance score, MGMT expression, radio- and chemo-sensitivities, comorbidities, and overall survival [50-55]. In addition to primary neuropathological specimens, cerebrospinal fluid (CSF) miRNA levels permit detection of GBM, differentiation of GBM from metastatic brain tumors, and monitoring of disease activity [56]. Particular miRNA signatures are also present in the blood of GBM patients, vary with treatment, and might be useful for monitoring recurrence [57]. In multiple sclerosis, miRNA profiles in white matter lesions discriminate between active and inactive lesions [58]. miRNA levels in blood differentiate between patients with a relapsing-remitting course and one that is progressive. miRNA expression also identifies patients treated with disease-modifying agents (e.g., glatiramer acetate and natalizumab) and those that are untreated. Similarly, miRNA expression in blood distinguishes between patients with PD and controls and medicated patients from non-medicated ones [59]. Carriers of huntingtin gene mutations exhibit high levels of miR-34b in plasma prior to the onset of symptoms [60]. Furthermore, patients with AD display increased expression of let-7 in CSF [61] . Extracellular let-7 provokes neurodegeneration by activating neuronal RNA-sensing Toll-like receptor 7 signaling and promotes the spread of pathology. Other miRNAs (i.e., miR-146a and miR-155) upregulated in AD patient CSF specifically have deleterious proinflammatory effects [62]. miRNAs, such as these, play a role in neuroimmune crosstalk locally and may do so systemically because they can be trafficked in blood and other fluids [63]. miRNAs and other ncRNAs likely mediate additional forms of central-peripheral communication, potentially accounting for their pervasive deregulation in peripheral tissues in brain diseases.
While ncRNA profiles can be sampled in readily accessible biological fluids, it is vital to understand how and why these factors might specifically reflect central pathological states. The recently recognized role of membrane-bound microvesicles, called exosomes (Glossary; Box 1), in mediating intercellular communication provides some insight [64]. Not only are ncRNAs present intracellularly; but, as highlighted above, they are also found extracellularly in biological fluids including CSF, blood (serum and plasma), lymphatics, urine, and saliva in health and disease [65-70]. These extracellular ncRNAs are fairly resistant to nucleases because they form stable complexes with proteins (i.e., Argonaute 2) or lipids (i.e., high-density lipoproteins), or alternatively they can be packaged into exosomes.
Integrated epigenetic and epigenomic profiling
There are still many unanswered questions regarding the causal relationship between epigenetic deregulation and nervous system disease pathogenesis and the biological significance of central and peripheral epigenetic profiles (Box 2). Nevertheless the range of observations cited above supports the advancement of epigenetic biomarker discovery, qualification, and validation for improving and personalizing nervous system disease screening, early diagnosis, and prognostication; developing molecularly targeted treatments; and monitoring progression and therapeutic responses.
There exist significant challenges, however [71]. Even the most well established epigenetic biomarker, MGMT methylation in gliomas, is not widely utilized. Persistent technical questions are one of the major obstacles that have prevented this and other epigenetic tests from being adopted clinically. Even leading next-generation sequencing platforms are relatively insensitive for measuring changes in epigenetic profiles in nervous system disorders, which are often subtle. For example, lncRNAs are low abundance transcripts with differential expression in disease in the two-fold range. However, rapid technological and methodological innovations are poised to overcome these issues by offering advantages in terms of sample preparation and quantity, speed, resolution, throughput, and cost-effectiveness. Not only do these emerging approaches permit the analysis of a single “candidate” epigenetic modification, but they also allow the characterization of highly integrated genome-wide epigenomic profiles utilizing single cells and molecules, by employing sophisticated technology platforms. Specific examples of such powerful tools and techniques that have recently been reported include the following: (i) a DNA array (fabricated via advanced soft-lithography) for high-resolution methylation profiling of single DNA molecules [72]; (ii) a quantum dot-enabled electrophoretic mobility shift assay for high-resolution quantitative epigenetic analysis [73]; (iii) RNA aptamers—RNAs that bind with very high affinity and selectivity to a particular target—engineered to recognize molecules harboring certain epigenetic modifications [74]; (iv) a nanofluidic device for real-time multiplexed detection and automated sorting of individual molecules based on their epigenetic states [75]; and (v) a chemical sensor array that can robustly discriminate between complex profiles of histone modifications (e.g., unmethylated, mono-, di-, and tri-methylated lysines) at a single histone and across different sites [76]. In addition, novel approaches for targeting ncRNAs and microvesicles include (i) improved methods for miRNA isolation from blood [77]; (ii) single-molecule nanopore platforms designed for quantitative measurement of ncRNA levels [78]; (iii) oligonucleotide molecular beacons and magnetic nanoparticles enabling the detection of ncRNAs, including those in live animals, by magnetic resonance imaging [79]; and (iv) microfluidic devices enabling the rapid isolation, sorting and detection of microvesicles from small volume samples [80].
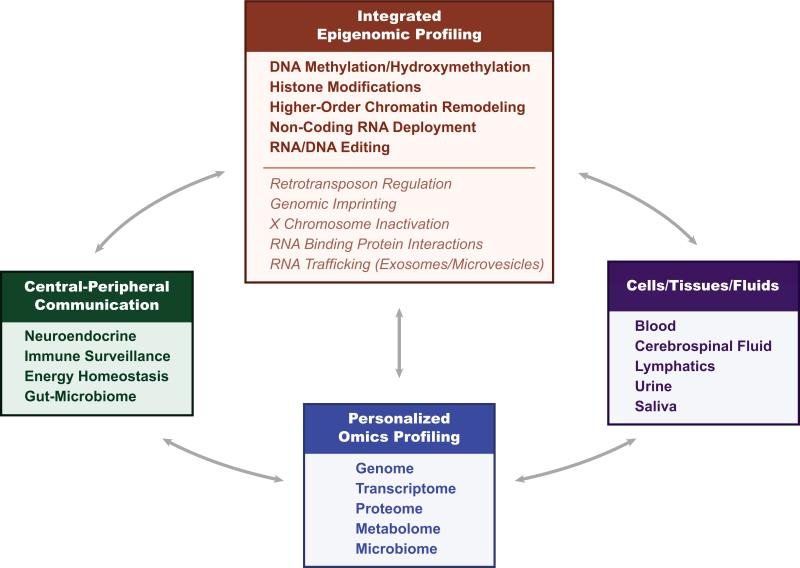
This integrated epigenomic profiling can be coupled with other “omics” profiling approaches for the genome, transcriptome, RNA editome, proteome, metabolome, and microbiome yielding a “systems” level view of pathology (along the spectrum from single cell to whole organism) and providing both mechanistic insights as well as combinatorial signatures with potential clinical applications (Figure 1) [81].
Figure 1. Integrated epigenomic and personalized “omics” profiling.
Utilizing a variety of accessible body fluids and associated cell and tissue sources, combinatorial epigenomic and personalized “omics” profiling are already providing physiological ‘readouts’ of dynamic multidimensional central-peripheral communications and an integrated systems biology analysis of nervous system functions in health and in various neurological disease states with an emerging spectrum of innovative diagnostic and therapeutic applications.
EVOLVING EPIGENETIC THERAPEUTIC STRATEGIES
Preclinical studies and clinical trials strongly suggest that agents targeting epigenetic factors and mechanisms, such as DNA methyltransferase (DNMT) and histone deacetylase (HDAC) enzyme inhibitors, have therapeutic efficacy in a very broad range of diseases, including brain disorders. However, the clinical utility of most first generation epigenetic agents is limited by their lack of specificity for individual enzymes, toxicities, poor bioavailability, and other factors. As such, there is considerable enthusiasm for identifying and designing additional compounds that modulate targets across the spectrum of epigenetic writer, eraser and reader classes and ncRNA pathways. These emerging agents have potential utility not only as biological probes for preclinical studies but also as lead compounds for optimization and clinical development. Here, we highlight examples of these evolving strategies and their relevance for brain disorders.
DNA methylation
Compounds available for modulating DNA methylation pathways include those that increase the supply of methyl donors (e.g., folic acid, betaine, vitamin B12, and creatine), inhibit DNMT enzymes (e.g., 5-azacytidine, 5-aza-2-deoxycytidine, and zebularine), and inhibit DNA demethylation enzymes (e.g., gemcitabine). Interestingly, supplementing with methyl donors in Rett and Angelman syndrome—disease mechanism-based treatment approaches—shows trends toward clinical benefits, meriting further study [82, 83]. Existing DNMT inhibitors have been evaluated in preclinical paradigms and modulate learning and memory, reward and addiction, ischemia, neurodegeneration, and epileptogenesis [84]. Thus, more efficient and specific DNMT inhibitors now being identified can potentially mitigate the pathogenesis and symptomatology in this wide range of disorders. Gemcitabine, an antimetabolite cancer therapeutic with neurotoxic effects, was recently found to inhibit Gadd45a-mediated DNA demethylation [85]. It is the first agent noted to have this ability and offers insights for developing drugs that target DNA demethylation enzymes. In addition, some commonly used drugs, such as hydralazine, procainamide and valproic acid (VPA), also influence DNA methylation profiles, and these drugs are prime candidates for epigenome-based repurposing/repositioning [86-89]. For example, preliminary studies demonstrate that adjunctive treatment with VPA enhances the efficacy of TMZ in GBM, by influencing MGMT promoter methylation status [90]. Notably, VPA might exert these beneficial effects via other mechanisms, which include the ability to inhibit HDAC enzymes. Conversely, toxicities and adverse effects associated with these and other commonly used drugs might result from an unintended impact on epigenetic pathways.
Histone and chromatin modifications
Compounds that modulate histone acetylation and methylation and other chromatin targets are increasingly becoming available. HDAC inhibitors are the most numerous amongst these. Their structural and mechanistic properties have been studied extensively, as have their context-specific biological functions, which include robust salutary effects in many different brain disease models. Notably, certain HDAC enzymes have roles in non-epigenetic pathways in the axon and mitochondria, and the mechanisms of action of HDAC inhibitors may include inhibiting these functions. Several HDAC inhibitors have advanced into clinical trials for nervous system disease indications, and more are likely to follow given the ongoing optimization of HDAC inhibitors, improving their selectivity for individual isoforms and pharmacokinetic and pharmacodynamic profiles. Some examples in clinical trials include (i) EVP-0334, a class I HDAC inhibitor being developed for AD (and other neurodegenerative disorders); (ii) SEN0014196 (selisistat), an HDAC inhibitor that targets SIRT1, being evaluated in HD; (iii) phenylbutyrate, a class I/II HDAC inhibitor, being studied in HD and ALS; (iv) suberoylanilide hydroxamic acid (vorinostat) and the related compound panobinostat, class I/II HDAC inhibitors, being analyzed individually and as adjunctive agents for treating gliomas [91, 92]. Of interest, emerging methodologies for analyzing histone acetylation levels in blood and other tissues are tools for monitoring and titrating therapy with these agents [93].
Efforts employing high-content and high-throughput screening of chemical libraries, structure based drug design, and other methods are focused on identifying and designing small molecules that target additional families of epigenetic factors with “writer/eraser” functions (e.g., protein methyltransferases/demethylases) as well as those with “reader” domains (e.g., bromodomains) [94]. For example, one study reported the discovery of a small molecule, GSK126, which is a highly selective and potent inhibitor of the enhancer of zeste homolog 2 (EZH2) histone methyltransferase [95]. EZH2 is implicated in the development of many cancers, including lymphoma and GBM; and preliminary data suggests that GSK126 is effective in treating these disorders. A complementary study reported finding a specific small molecule inhibitor of the histone demethylases, jumonji domain containing 3 and lysine-specific demethylase 6A, which is effective in modulating acute inflammatory responses, such as those that occur after spinal cord injury and contribute to secondary damage [96, 97]. In addition, because epigenetic enzymes often contain “reader” domains or they are components of complexes with other factors that fulfill these roles, efforts have focused on disrupting these protein-protein interactions. JQ1 is a first-in-class small molecule that selectively inhibits the interaction between bromodomain proteins and acetylated lysine residues [98] and shows promise for treating genetically diverse subgroups of GBM [99]. Emerging technologies for modulating protein-protein interactions, such as stapled peptides and other proteomimetic approaches, also hold significant promise in this regard [100].
Master epigenetic regulator: REST
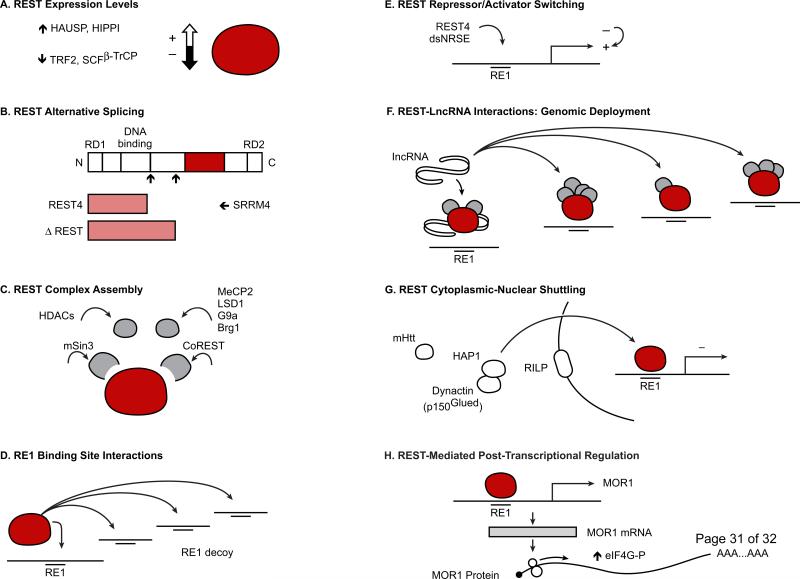
Forward-looking strategies can also be envisioned that target epigenetic factors with key roles in brain diseases, such as REST, in more complex and nuanced ways utilizing a variety of existing and novel approaches including small molecules, RNA interference (RNAi), decoy oligonucleotides, and synthetic peptide nucleic acid oligomers [101-103]. These include modulating REST expression, REST alternative splicing, macromolecular complex assembly, REST interactions with RE1 binding sites, regulators of REST activity (i.e., double stranded-RE1/-NRSE ncRNAs and the REST4 isoform) that mediate switching between REST transcriptional activator/repressor functions, interactions between REST and lncRNAs that mediate REST complex genomic site-specific deployment, REST cytoplasmic-nuclear shuttling, and REST post-transcriptional regulatory activity (Figure 2).
Figure 2. Novel epigenetic therapeutic targeting strategies.
Using a variety of existing and emerging approaches, it is possible to alter the functional roles of REST, a key transcriptional and epigenetic mediator of nervous system function in health and various disease states, by differentially modulating: (A) REST expression levels; (B) REST alternative splicing profiles; (C) REST macromolecular complex assembly; (D) REST/RE1 binding site interactions; (E) REST transcriptional activator/repressor activity and switching; (F) REST interactions with long non-coding RNAs that mediate REST genome-wide deployment: (G) REST cytoplasmic-nuclear shuttling; (H) REST post-transcriptional regulatory processes. Brg1=Brahma-related gene 1. CoREST=corepressor for element-1-silencing transcription factor. dsNRSE=double-stranded non-coding RNAs encoding the RE1 sequence. elF4G-P=elongation factor-4G, phosphorylated. G9a=histone H3-lysine 9 methyltransferase. HAP1=Huntingtin-associated protein 1. HAUSP=herpesvirus associated ubiquitin specific protease. HDACs=histone deacetylases. HIPPI=Huntingtin interacting protein 1 (HIP1) protein interactor. mHtt=mutant Huntingtin. lncRNA=long non-coding RNA. LSD1=lysine-specific demethylase-1. MeCP2=methyl-CpG binding protein 2. MOR1=mu opioid receptor-1. RD=repressor domain. RE1=repressor element-1. REST4=REST/NRSF truncated activator isoform 4. RILP=REST/NRSE-interacting LIM domain protein. SCF(βTrCP)=Skp1–Cullin1–F-box β-transducin repeat-containing protein. mSin3=mammalian component of histone deacetylase complex. SRRM4=splicing factor-encoding gene Ser/Arg repetitive matrix 4. TRF2=telomeric repeat-binding protein 2.
Non-coding RNAs
Considerable effort is now concentrated on developing ncRNA-based therapeutics and capitalizes on previously existing expertise in antisense oligonucleotides, RNAi, and related platforms. These approaches are most advanced for inhibiting pathogenic miRNA expression and function and employ antisense oligonucleotides, termed antimirs, engineered with various chemical modifications to increase their specificity, stability, nuclease resistance, and delivery across the blood brain barrier (e.g., locked nucleic acids [LNA; Glossary] and antagomirs). These molecules ameliorate nervous system pathology in preclinical models. For example, antagomir-mediated inhibition of miR-206, which is increased in human AD brains and targets brain derived neurotrophic factor (BDNF), enhances BDNF levels, hippocampal synaptic density and neurogenesis, and memory in an AD mouse model [104]. Also, a recent study presented data suggesting that utilizing an antimiR directed at miR-886-3p is a possible treatment for FRDA [105]. Alternative strategies to inhibit miRNAs include introducing oligonucleotides that interfere with miRNA-target mRNA interactions (i.e., miRNA “masks” and “sponges”). Modulating multiple miRNAs simultaneously with a single therapeutic oligonucleotide has also been proposed. Related strategies for lncRNAs are also being developed [106]. Specifically, antagoNATs are oligonucleotides designed to target natural antisense transcripts (NATs)—lncRNAs in antisense genomic configurations relative to protein-coding genes—including those that regulate genes with key roles in the nervous system, such as BDNF, glial-derived neurotrophic factor, and ephrin receptor B2 [107]. Strategies to inhibit lncRNA functions with oligonucleotides (or small molecules) that interfere with sequence-specific and structural interactions between lncRNAs with other molecules (i.e., DNA, RNA, and proteins [e.g., chromatin remodeling complexes]) are being considered. Although ncRNA-based therapeutics for brain disorders are in early stages of development, proof-of-principle is illustrated by the advancement of miravirsen, a LNA antisense oligonucleotide targeting miR-122, into phase 2 clinical trials for hepatitis C (NCT01727934).
Methods for introducing ncRNAs with salubrious effects are also being investigated. These include mimics or replacements for endogenous ncRNAs. The p137 RNA derived from cytomegalovirus is one particularly interesting example [108]. This ncRNA protects dopaminergic neurons from cell death provoked by mitochondrial dysfunction in PD models. Intriguingly, fusing this ncRNA with rabies virus glycoprotein peptide, a powerful technique for drug delivery to the brain, permits peripheral intravenous administration of this agent overcoming one of the greatest challenges for any of these potential therapies [109]. Another approach for restoring beneficial ncRNAs, such as those with tumor suppressor properties, involves using other epigenetic agents (e.g., DNMT and HDAC inhibitors) that upregulate ncRNA expression. Alternative strategies for modulating ncRNA functions include the identification of small molecule agents that influence ncRNA biogenesis and effector pathways (e.g., inhibitors of miRNA precursor processing and RISC complex loading) [110-112].
CONCLUDING REMARKS
We are at the vanguard of the era of epigenetic and epigenomic medicine, which is poised to revolutionize the diagnosis and treatment of nervous system diseases. This paradigm shift is being propelled by basic and translational discovery efforts that have revealed how epigenetic changes mediate central pathology or indirectly reflect it in the periphery. Epigenetic diagnostic and therapeutic innovations are, in turn, being driven by ongoing technological progress (i) allowing higher-resolution interrogation of epigenetic profiles and molecular imaging and (ii) promoting the discovery, design and optimization of novel compounds that can modulate epigenetic pathways and their delivery into the nervous system. We foresee these epigenetic clinical applications evolving in concert with complementary diagnostic and therapeutic platforms (e.g., microfluidics, RNA aptamers, nanotechnologies, oligonucleotide-based strategies, immunotherapies, and cellular reprogramming and regenerative medicine) that are revolutionary in their own right.
Supplementary Material
Box 1. Exosomes.
Exosomes are membrane-bound microvesicles derived from intracellular multivesicular bodies and the endosomal pathway that participate in local and systemic cell-cell communication. Exosomes are secreted by donor cells (e.g., neural, immune, and other cell types) into bodily fluids and release their contents into selectively targeted recipient cells [113, 114]. Exosomes can transport cargo including non-coding RNAs (ncRNAs) and other molecules that are functional in recipient cells. For example, microRNAs transferred via exosomes repress their target genes in recipient cells [115]. In terms of pathological roles, exosomes are released by primary brain tumors, such as gliomas, internalized by normal cells, and implicated in modulating the tumor microenvironment to promote tumor invasion and angiogenesis and preventing immune responses [113, 114]. Other classes of ncRNAs may similarly be found in exosomes and influence target cells. Indeed, exosomes harboring tumor-specific factors—mutant proteins, mutated and amplified oncogene sequences, and retrotransposable elements—are present in the serum, implicating them in mediating the cancer state systemically. Ongoing studies of exosome formation, contents and delivery further suggest that the spreading of pathology in a spectrum of other nervous system diseases is associated with the release of these microvesicles.
Therapeutic strategies aimed at exploiting exosomes are also being developed. Exosomes can be engineered to deliver cargo, including ncRNAs, into the brain through the periphery. Alternatively, endogenous exosomes can be harnessed to treat disease by activating endogenous immune responses. For example, one intriguing study reported the utilization of glioma-derived exosomes to generate CD8+ T-cells with glioma-specific cytotoxic activity in vitro and suggested this stratagem for immunizing against gliomas [116].
Box 2. Outstanding Questions.
Why do patients with central pathologies that are often brain region- and cell type-selective exhibit abnormal epigenetic profiles in peripheral tissues? Are these peripheral epigenetic signatures directly connected to disease mechanisms, indirect but potentially valuable markers thereof, or simply downstream phenomena that are neither sensitive nor specific?
In addition to exosome-mediated communication, what (if any) other mechanisms might link epigenetic states in the brain to those in the periphery? Can these also be exploited for diagnostic or therapeutic applications?
How can emerging technologies enabling high-resolution real-time in vivo interrogation of epigenetic and epigenomic states be used to (i) differentiate between pathogenic processes and those that are protective responses and (ii) develop corresponding clinically relevant and cost-effective assays?
Can epigenetic therapeutics be designed, developed, and delivered into the brain and specific neural cell types not only in animal models but also in patients? What are the explicit barriers that must be overcome?
Is the development of epigenetic drugs and associated companion diagnostics a biologically and commercially viable therapeutic strategy? What brain diseases and epigenetic targets might be most amenable to providing proof-of-concept for this approach?
Can epigenetic agents be combined with each other or with other classes of drugs to more effectively address the multiple layers of molecular pathology present in complex brain disorders?
Highlights.
Epigenetic signatures found in central and peripheral nervous system disease patient-derived tissues correlate with pathological features and clinical outcomes.
Emerging technological advances are allowing integrated epigenomic profiling.
Compounds for modulating targets across the entire spectrum of epigenetic writer, eraser and reader classes and non-coding RNA pathways are being developed.
ACKNOWLEDGEMENTS
We regret that space constraints have prevented the citation of many relevant and important references. M.F.M. is supported by grants from the National Institutes of Health (NS071571, HD071593, MH66290), as well as by the F.M. Kirby, Alpern Family, Mildred and Bernard H. Kayden and Roslyn and Leslie Goldstein Foundations.
Glossary
- Exosomes
membrane-bound microvesicles secreted by various cell types into different bodily fluids that contain signaling molecules, including non-coding RNAs, involved in intercellular communication.
- DNA methylation
the formation of 5-methylcytosine catalyzed by DNA methyltransferase enzymes that transfer methyl groups from S-adenosylmethionine to cytosine residues, which often occurs in gene regulatory regions and is associated with transcriptional repression.
- Glioma CpG island methylator phenotype (G-CIMP)
characteristic profile of DNA methylation alterations found in gene promoter regions in glioma specimens that define a subset of tumors with distinct genetic, pathological and clinical features.
- Histone modifying enzymes
complementary families of enzymes, including histone acetyltransferases/deacetylases and histone methyltransferases/demethylases, responsible for “writing” and “erasing” histone protein post-translational modifications.
- Locked nucleic acids (LNAs)
chemically modified nucleic acids that are locked or restricted in terms of their conformation, enhancing their stability, target specificity and pharmocokinetic profiles.
- Long ncRNAs (lncRNAs)
highly versatile non-coding RNA molecules greater than 200 nucleotides in length that have a broad range of regulatory, structural, and other emerging biological roles.
- MicroRNAs (miRNAs)
non-coding RNAs, 20-23 nucleotides in length, which participate in post-transcriptional regulation of target mRNAs through RNA interference pathways.
- RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF)
a master transcriptional and epigenetic regulatory factor within the nervous system, implicated in the pathogenesis of many brain diseases, that binds to the RE1 motif associated with many neural genes and non-coding RNAs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
We declare that we have no conflicts of interest.
REFERENCES
- 1.Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog. Neurobiol. 2008;86:305–341. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graff J, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliveira AM, et al. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 2012;15:1111–1113. doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- 4.Liu N, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482:519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivieccio MA, et al. HDAC6 is a target for protection and regeneration following injury in the nervous system. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19599–19604. doi: 10.1073/pnas.0907935106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sturm D, et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Orr BA, et al. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS ONE. 2012;7:e41036. doi: 10.1371/journal.pone.0041036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 11.Wick W, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13:707–715. doi: 10.1016/S1470-2045(12)70164-X. [DOI] [PubMed] [Google Scholar]
- 12.Malmstrom A, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13:916–926. doi: 10.1016/S1470-2045(12)70265-6. [DOI] [PubMed] [Google Scholar]
- 13.Balana C, et al. Tumour and serum MGMT promoter methylation and protein expression in glioblastoma patients. Clin. Transl. Oncol. 2011;13:677–685. doi: 10.1007/s12094-011-0714-x. [DOI] [PubMed] [Google Scholar]
- 14.Ingold B, et al. Homogeneous MGMT immunoreactivity correlates with an unmethylated MGMT promoter status in brain metastases of various solid tumors. PLoS ONE. 2009;4:e4775. doi: 10.1371/journal.pone.0004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto K, et al. Methylation status of O6-methylguanine-DNA-methyl transferase promoter region in non-small-cell lung cancer patients with brain metastasis. Clin. Transl. Oncol. 2012;14:31–35. doi: 10.1007/s12094-012-0758-6. [DOI] [PubMed] [Google Scholar]
- 16.Wu PF, et al. O(6)-Methylguanine-DNA methyltransferase expression and prognostic value in brain metastases of lung cancers. Lung Cancer. 2010;68:484–490. doi: 10.1016/j.lungcan.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Loss of expression of the differentially expressed in adenocarcinoma of the lung (DAL-1) protein is associated with metastasis of non-small cell lung carcinoma cells. Tumour Biol. 2012 doi: 10.1007/s13277-012-0452-x. [DOI] [PubMed] [Google Scholar]
- 18.Shumay E, et al. Evidence that the methylation state of the monoamine oxidase A (MAOA) gene predicts brain activity of MAO A enzyme in healthy men. Epigenetics. 2012;7:1151–1160. doi: 10.4161/epi.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath S, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13:R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans-Galea MV, et al. FXN methylation predicts expression and clinical outcome in Friedreich ataxia. Ann. Neurol. 2012;71:487–497. doi: 10.1002/ana.22671. [DOI] [PubMed] [Google Scholar]
- 21.Bollati V, et al. DNA methylation in repetitive elements and Alzheimer disease. Brain. Behav. Immun. 2011;25:1078–1083. doi: 10.1016/j.bbi.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baccarelli A, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21:819–828. doi: 10.1097/EDE.0b013e3181f20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012 doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Mut JV, et al. Aberrant epigenetic landscape in intellectual disability. Prog. Brain Res. 2012;197:53–71. doi: 10.1016/B978-0-444-54299-1.00004-2. [DOI] [PubMed] [Google Scholar]
- 25.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 27.Chan KM, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013 doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu BL, et al. Global histone modification patterns as prognostic markers to classify glioma patients. Cancer Epidemiol. Biomarkers Prev. 2010;19:2888–2896. doi: 10.1158/1055-9965.EPI-10-0454. [DOI] [PubMed] [Google Scholar]
- 29.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labrie V, et al. Epigenetics of major psychosis: progress, problems and perspectives. Trends Genet. 2012;28:427–435. doi: 10.1016/j.tig.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavin DP, et al. Dimethylated lysine 9 of histone 3 is elevated in schizophrenia and exhibits a divergent response to histone deacetylase inhibitors in lymphocyte cultures. J. Psychiatry Neurosci. 2009;34:232–237. [PMC free article] [PubMed] [Google Scholar]
- 32.Kurita M, et al. HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat. Neurosci. 2012;15:1245–1254. doi: 10.1038/nn.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 34.Marullo M, et al. Analysis of the repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy of non-neuronal genes in peripheral lymphocytes from patients with Huntington's disease. Brain Pathol. 2010;20:96–105. doi: 10.1111/j.1750-3639.2008.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 36.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gehrke S, et al. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams AH, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu DZ, et al. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J. Cereb. Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeyaseelan K, et al. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 41.Dharap A, et al. Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke. 2011;42:1105–1109. doi: 10.1161/STROKEAHA.110.598391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dharap A, et al. Effect of Focal Ischemia on Long Noncoding RNAs. Stroke. 2012;43:2800–2802. doi: 10.1161/STROKEAHA.112.669465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W, et al. Variants on Chromosome 9p21.3 Correlated With ANRIL Expression Contribute to Stroke Risk and Recurrence in a Large Prospective Stroke Population. Stroke. 2011 doi: 10.1161/STROKEAHA.111.625442. [DOI] [PubMed] [Google Scholar]
- 44.Cunnington MS, et al. Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foroud T, et al. Genome-Wide Association Study of Intracranial Aneurysms Confirms Role of Anril and SOX17 in Disease Risk. Stroke. 2012;43:2846–2852. doi: 10.1161/STROKEAHA.112.656397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costa FF, et al. Identification of microRNAs as potential prognostic markers in ependymoma. PLoS ONE. 2011;6:e25114. doi: 10.1371/journal.pone.0025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weeraratne SD, et al. miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 2011;13:165–175. doi: 10.1093/neuonc/noq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan J, et al. MicroRNA-204 increases sensitivity of neuroblastoma cells to cisplatin and is associated with a favourable clinical outcome. Br. J. Cancer. 2012;107:967–976. doi: 10.1038/bjc.2012.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhi F, et al. A microRNA expression signature predicts meningioma recurrence. Int. J. Cancer. 2012 doi: 10.1002/ijc.27658. [DOI] [PubMed] [Google Scholar]
- 50.You G, et al. Significance of miR-196b in Tumor-Related Epilepsy of Patients with Gliomas. PLoS ONE. 2012;7:e46218. doi: 10.1371/journal.pone.0046218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chistiakov DA, Chekhonin VP. Contribution of microRNAs to radio-and chemoresistance of brain tumors and their therapeutic potential. Eur. J. Pharmacol. 2012;684:8–18. doi: 10.1016/j.ejphar.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 52.Han L, et al. LncRNA pro fi le of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int. J. Oncol. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Chang C, et al. Correlation of microRNA-375 downregulation with unfavorable clinical outcome of patients with glioma. Neurosci. Lett. 2012 doi: 10.1016/j.neulet.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 55.Zhang W, et al. miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012;14:712–719. doi: 10.1093/neuonc/nos089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teplyuk NM, et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roth P, et al. A specific miRNA signature in the peripheral blood of glioblastoma patients. J. Neurochem. 2011;118:449–457. doi: 10.1111/j.1471-4159.2011.07307.x. [DOI] [PubMed] [Google Scholar]
- 58.Thamilarasan M, et al. MicroRNAs in multiple sclerosis and experimental autoimmune encephalomyelitis. Autoimmun. Rev. 2012;11:174–179. doi: 10.1016/j.autrev.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Margis R, Rieder CR. Identification of blood microRNAs associated to Parkinsons disease. J. Biotechnol. 2011 doi: 10.1016/j.jbiotec.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 60.Gaughwin PM, et al. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington's disease. Hum. Mol. Genet. 2011;20:2225–2237. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- 61.Lehmann SM, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012 doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 62.Lukiw WJ, et al. Spreading of Alzheimer's disease inflammatory signaling through soluble micro-RNA. Neuroreport. 2012;23:621–626. doi: 10.1097/WNR.0b013e32835542b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soreq H, Wolf Y. NeurimmiRs: microRNAs in the neuroimmune interface. Trends Mol. Med. 2011;17:548–555. doi: 10.1016/j.molmed.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 64.Gutierrez-Vazquez C, et al. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol. Rev. 2013;251:125–142. doi: 10.1111/imr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bryant RJ, et al. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer. 2012;106:768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallo A, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kucharzewska P, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rana S, et al. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15:281–295. doi: 10.1593/neo.122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duttagupta R, et al. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS ONE. 2011;6:e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat. Rev. Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 72.Cerf A, et al. Single DNA molecule patterning for high-throughput epigenetic mapping. Anal. Chem. 2011;83:8073–8077. doi: 10.1021/ac202506j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, et al. Mapping DNA quantity into electrophoretic mobility through quantum dot nanotethers for high-resolution genetic and epigenetic analysis. ACS Nano. 2012;6:858–864. doi: 10.1021/nn204377k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hyun S, et al. An RNA aptamer that selectively recognizes symmetric dimethylation of arginine 8 in the histone H3 N-terminal peptide. Nucleic acid therapeutics. 2011;21:157–163. doi: 10.1089/nat.2011.0300. [DOI] [PubMed] [Google Scholar]
- 75.Cipriany BR, et al. Real-time analysis and selection of methylated DNA by fluorescence-activated single molecule sorting in a nanofluidic channel. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8477–8482. doi: 10.1073/pnas.1117549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Minaker SA, et al. Antibody-free reading of the histone code using a simple chemical sensor array. J. Am. Chem. Soc. 2012;134:11674–11680. doi: 10.1021/ja303465x. [DOI] [PubMed] [Google Scholar]
- 77.Li Y, Kowdley KV. Method for microRNA isolation from clinical serum samples. Anal. Biochem. 2012;431:69–75. doi: 10.1016/j.ab.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gu LQ, et al. Detection of miRNAs with a nanopore single-molecule counter. Expert Rev. Mol. Diagn. 2012;12:573–584. doi: 10.1586/erm.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hwang do W, et al. Smart magnetic fluorescent nanoparticle imaging probes to monitor microRNAs. Small. 2010;6:81–88. doi: 10.1002/smll.200901262. [DOI] [PubMed] [Google Scholar]
- 80.Chen C, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab. Chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun J, et al. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Res. 2010;70:6128–6138. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freilinger M, et al. Effects of creatine supplementation in Rett syndrome: a randomized, placebo-controlled trial. J. Dev. Behav. Pediatr. 2011;32:454–460. doi: 10.1097/DBP.0b013e31822177a8. [DOI] [PubMed] [Google Scholar]
- 83.Peters SU, et al. Double-blind therapeutic trial in Angelman syndrome using betaine and folic acid. Am. J. Med. Genet. A. 2010;152A:1994–2001. doi: 10.1002/ajmg.a.33509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chestnut BA, et al. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 2011;31:16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schafer A, et al. Gemcitabine functions epigenetically by inhibiting repair mediated DNA demethylation. PLoS ONE. 2010;5:e14060. doi: 10.1371/journal.pone.0014060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Csoka AB, Szyf M. Epigenetic side-effects of common pharmaceuticals: A potential new field in medicine and pharmacology. Med. Hypotheses. 2009 doi: 10.1016/j.mehy.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 87.Coronel J, et al. A double-blind, placebo-controlled, randomized phase III trial of chemotherapy plus epigenetic therapy with hydralazine valproate for advanced cervical cancer. Preliminary results. Med. Oncol. 2011;28(Suppl 1):S540–546. doi: 10.1007/s12032-010-9700-3. [DOI] [PubMed] [Google Scholar]
- 88.Lee BH, et al. Procainamide is a specific inhibitor of DNA methyltransferase 1. J. Biol. Chem. 2005;280:40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dong E, et al. Valproate induces DNA demethylation in nuclear extracts from adult mouse brain. Epigenetics. 2010;5:730–735. doi: 10.4161/epi.5.8.13053. [DOI] [PubMed] [Google Scholar]
- 90.Berendsen S, et al. Valproic acid for the treatment of malignant gliomas: review of the preclinical rationale and published clinical results. Expert Opin. Investig. Drugs. 2012;21:1391–1415. doi: 10.1517/13543784.2012.694425. [DOI] [PubMed] [Google Scholar]
- 91.Lee EQ, et al. Phase I Study of Vorinostat in Combination with Temozolomide in Patients with High-Grade Gliomas: North American Brain Tumor Consortium Study 04-03. Clin. Cancer Res. 2012;18:6032–6039. doi: 10.1158/1078-0432.CCR-12-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drappatz J, et al. Phase I study of panobinostat in combination with bevacizumab for recurrent high-grade glioma. J. Neurooncol. 2012;107:133–138. doi: 10.1007/s11060-011-0717-z. [DOI] [PubMed] [Google Scholar]
- 93.Rigby L, et al. Methods for the analysis of histone H3 and H4 acetylation in blood. Epigenetics. 2012;7:875–882. doi: 10.4161/epi.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arrowsmith CH, et al. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 95.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012 doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 96.Kruidenier L, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee K, et al. Molecular mechanism of Jmjd3-mediated interleukin-6 gene regulation in endothelial cells underlying spinal cord injury. J. Neurochem. 2012 doi: 10.1111/j.1471-4159.2012.07786.x. [DOI] [PubMed] [Google Scholar]
- 98.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheng Z, et al. Inhibition of BET Bromodomain Targets Genetically Diverse Glioblastoma. Clin. Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jubb H, et al. Structural biology and drug discovery for protein-protein interactions. Trends Pharmacol. Sci. 2012;33:241–248. doi: 10.1016/j.tips.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 101.Rigamonti D, et al. Turning REST/NRSF dysfunction in Huntington's disease into a pharmaceutical target. Curr. Pharm. Des. 2009;15:3958–3967. doi: 10.2174/138161209789649303. [DOI] [PubMed] [Google Scholar]
- 102.Leone S, et al. SAR and QSAR study on 2-aminothiazole derivatives, modulators of transcriptional repression in Huntington's disease. Bioorg. Med. Chem. 2008;16:5695–5703. doi: 10.1016/j.bmc.2008.03.067. [DOI] [PubMed] [Google Scholar]
- 103.Rigamonti D, et al. Loss of huntingtin function complemented by small molecules acting as repressor element 1/neuron restrictive silencer element silencer modulators. J Biol Chem. 2007;282:24554–24562. doi: 10.1074/jbc.M609885200. [DOI] [PubMed] [Google Scholar]
- 104.Lee ST, et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 2012;72:269–277. doi: 10.1002/ana.23588. [DOI] [PubMed] [Google Scholar]
- 105.Mahishi LH, et al. miR-886-3p Levels Are Elevated in Friedreich Ataxia. J. Neurosci. 2012;32:9369–9373. doi: 10.1523/JNEUROSCI.0059-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qureshi IA, Mehler MF. Long Non-coding RNAs: Novel Targets for Nervous System Disease Diagnosis and Therapy. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics. 2013 doi: 10.1007/s13311-013-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Modarresi F, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kuan WL, et al. A novel neuroprotective therapy for Parkinson's disease using a viral noncoding RNA that protects mitochondrial complex I activity. J. Exp. Med. 2012;209:1–10. doi: 10.1084/jem.20111126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kumar P, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 110.Tan GS, et al. Small molecule inhibition of RISC loading. ACS Chem. Biol. 2012;7:403–410. doi: 10.1021/cb200253h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shan G, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 2008;26:933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Melo S, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Balaj L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 116.Bu N, et al. Exosome-loaded dendritic cells elicit tumor-specific CD8+ cytotoxic T cells in patients with glioma. J. Neurooncol. 2011;104:659–667. doi: 10.1007/s11060-011-0537-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.