Abstract
Tomatoes are a principal dietary source of carotenoids and flavonoids, both of which are highly beneficial for human health1,2. Overexpression of genes encoding biosynthetic enzymes or transcription factors have resulted in tomatoes with improved carotenoid or flavonoid content, but never with both3–7. We attempted to increase tomato fruit nutritional value by suppressing an endogenous photomorphogenesis regulatory gene, DET1, using fruit-specific promoters combined with RNA interference (RNAi) technology. Molecular analysis indicated that DET1 transcripts were indeed specifically degraded in transgenic fruits. Both carotenoid and flavonoid contents were increased significantly, whereas other parameters of fruit quality were largely unchanged. These results demonstrate that manipulation of a plant regulatory gene can simultaneously influence the production of several phytonutrients generated from independent biosynthetic pathways, and provide a novel example of the use of organ-specific gene silencing to improve the nutritional value of plant-derived products.
Plant-based food offers a diverse mixture of nutrients that are essential for human nutrition and contribute to the promotion of good health. Epidemiological studies show that increased consumption of fruits and vegetables is correlated with a reduced risk of several diseases, including cancer and cardiovascular disease8. There is considerable interest in the development of food products rich in vitamins, flavonoids and carotenoids because it is generally thought that they will be more beneficial to human health than dietary supplements9. Although conventional breeding is one means of achieving this goal, the genetic diversity available within sexually compatible species of any given crop will limit the extent of improvement. Transgenic approaches can provide an alternative, although there is currently public concern about their use in contemporary agriculture, particularly when genes derived from organisms other than plants are used.
Tomato fruit and tomato-derived products are the principal dietary sources worldwide of lycopene and also contain large amounts of β-carotene. Increased lycopene gives the fruit a more appealing color and has proven nutritional value as an antioxidant1. Increased lycopene in the diet is associated with reduced rates of heart attack and is also a promising cancer chemopreventative, particularly for prostate cancer1,10,11. β-carotene is the most potent precursor of vitamin A—deficiency of which is the most common dietary problem affecting children worldwide. UNICEF has estimated that improved vitamin A nutrition could prevent up to 2 million deaths annually among children aged between one and four years12. Tomato fruit are also an important dietary source of other health-promoting phyto-chemicals besides carotenoids, such as flavonoids. Flavonoids are hydrophilic antioxidants13 complementing the hydrophobic nature of carotenoids. Diets rich in flavonoids have been associated with reduced risk of coronary heart disease, certain cancers and other age-related diseases2.
Several attempts have been made to increase the carotenoid content of tomato products using bacterial genes encoding biosynthetic enzymes3,4. These approaches have generally resulted in increases of only one or a few metabolites, and not in increased flux through the entire carotenoid pathway. Conversely, flavo noid levels have been elevated in tomato either by amplifying biosynthetic steps5,6 or by using known flavonoid transcription factor genes7. Although such approaches were successful in elevating flavonoids, carotenoid content remained unaffected in these transgenic lines.
One strategy to obtain more general increases in several metabolites could be to modulate regulatory genes whose products control flux through several biosynthetic pathways14. These genes would be of plant origin and so such strategies may also be more acceptable to the general public. The role of the gaseous hormone ethylene in the regulation of fruit ripening is well known15, and in addition it has recently emerged that genes encoding components of the light-signal-transduction machinery can influence tomato fruit quality16. Such genes may therefore represent promising genetic tools to improve nutritional value. DE-ETIOLATED1 (DET1) is a regulatory gene which represses several signaling pathways controlled by light17. Mutations in this gene are responsible for high pigment-2 (hp-2) phenotypes in tomato, characterized by exaggerated photoresponsiveness18,19. Light-grown hp-2 mutants display high levels of anthocyanins, are shorter and darker than wild-type plants and have more deeply pigmented fruits. The higher pigmentation of mature fruits from these mutants is due to elevated levels of both flavonoids and carotenoids18–21.
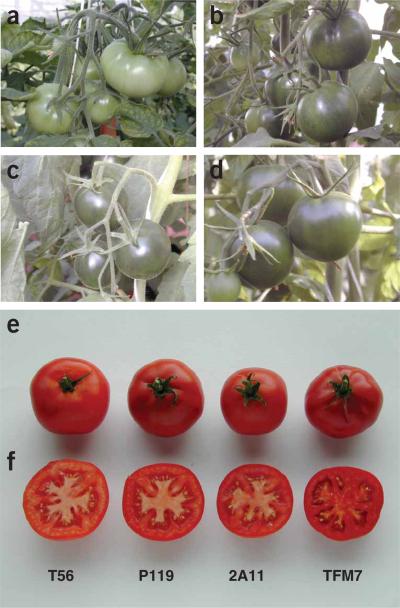
Constitutive silencing of the tomato DET1 (TDET1) gene in tomato was shown to cause elevations in the contents of β-carotene and lycopene in mature fruits, although severe developmental defects such as reduced plant stature, bushiness and dwarfing were also observed22. Such phenotypes are also observed in hp-2 mutants, which is why the mutation has not been successfully exploited by breeders. To harness the positive effects of TDET1 gene suppression in fruits without the collateral negative effects on plant growth, we attempted to inhibit TDET1 mRNA accumulation by RNA interference (RNAi) specifically in fruits using TDET1-derived inverted-repeat constructs driven by three different fruit-specific promoters, P119, 2A11 and TFM7 (refs. 23–25) (see Methods). Indeed, we found that transgenic plants containing each of these constructs appeared normal and healthy, but developed dark green immature fruits that subsequently became deep red at the mature stage (Fig. 1), reminiscent of hp-2 mutants18. With the P119 promoter driving the transgene, the darkest fruits were often observed on the lower trusses and fruits had a grainy appearance (Fig. 1b). With the 2A11 promoter the dark green fruits developed uniformly on all trusses all the way up the plant. Furthermore, phenotypes were not grainy but more uniformly dark green (Fig. 1c). With the TFM7 promoter, dark green fruits were also observed on trusses of all ages and were again not grainy (Fig. 1d). Immature fruits of plants containing the 2A11 construct were generally even darker than the fruits of transgenic plants containing the other two promoter constructs. Because loss of TDET1 function causes darker green foliage and bushy phenotypes22, it was easy to assess whether its expression had been modulated also in the vegetative parts of the plant. Aberrant phenotypes were not observed in any of the transgenic plants, neither in the primary transformants nor in subsequent generations (data not shown).
Figure 1.
Fruit-specific phenotypes of T2 generation transgenic plants containing a TDET1 inverted-repeat transgene driven by different promoters. (a) Immature fruits from wild-type (T56) plants. (b) Immature fruits from plant containing P119 promoter construct. (c) Immature fruits from plant containing 2A11 promoter construct. (d) Immature fruits from plant containing TFM7 promoter construct. (e) Fully red-ripe fruits from wild-type plants and transgenic plants containing the three promoter constructs. (f) Cross-sections of fruits shown in e.
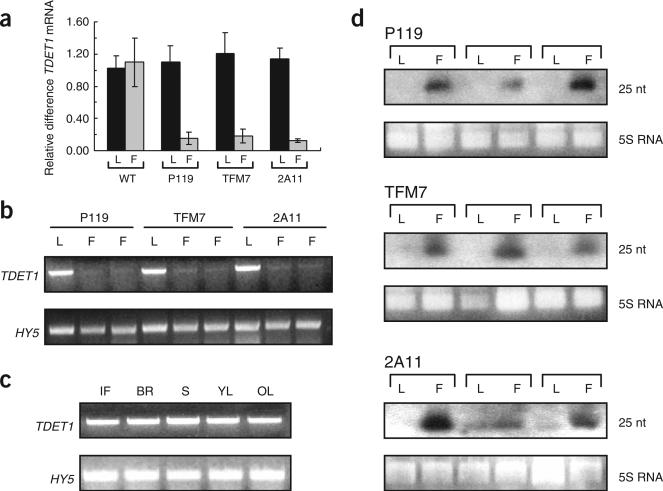
The fruit-specific phenotypes of transgenic plants containing all three promoter constructs suggested strongly that TDET1 gene expression had been suppressed specifically in the fruits. Due to the inverted-repeat design of our constructs, this suppression had most likely been achieved by RNAi26. A diagnostic feature of RNAi is the degradation of target mRNAs into low molecular weight products26,27. We therefore examined endogenous TDET1 mRNA abundance in the transgenic plants using a range of techniques. Real-time quantitative RT-PCR analysis of RNA extracted from the mature green fruits and leaves of all plants examined (n = for each construct) indicated that TDET1 mRNA in the fruits was much less abundant than in the leaves (Fig. 2a). In RNA extracted from leaves, the relative amounts of TDET1 mRNA were, however, much the same as in wild-type plants. To further analyze TDET1 mRNA levels we carried out semi-quantitative RT-PCR with RNA isolated from both fruits and leaves of the same plant. These analyses revealed that TDET1 mRNA abundance in the fruits was drastically reduced compared to the leaves of the same plants and demonstrated that this was the case for all three of the fruit-specific promoter constructs (Fig. 2b). This was not observed in nontransformed plants, where TDET1 mRNA abundance in different parts of the plant were similar, being present at low levels at all stages of plant development (Fig. 2c).
Figure 2.
Analysis of TDET1 RNA in transgenic plants containing fruit-specific promoter constructs. (a) Real time RT-PCR analysis of leaves (L) and mature green fruits (F) from T56 (wild type) and T3 generation transgenic plants. TDET1 mRNA abundance is shown relative to mRNA levels in leaves of wild-type plants. Each bar represents three repetitions from each RNA sample (derived from pools of three fruits or leaves per plant, and from two individual plants for each genotype). Error bars representing standard deviations are shown in each case. (b) Semi-quantitative RT-PCR analysis of TDET1 expression in leaves (L) and mature green fruits (F) from T3 generation plants transformed with fruit-specific promoter constructs. Full length TDET1 was synthesized from RNA extracted from leaves and fruits from the same plants. The HY5 gene was used as a control for cDNA synthesis. (c) Semi-quantitative RT-PCR analysis of TDET1 mRNA levels in different tissues from wild-type (T56) plants. IF, immature green fruit; BR, breaker stage fruit; S, stem; YL, young leaf; OL, old leaf. The HY5 gene was used as control for cDNA synthesis. (d) siRNA analysis of mature green fruits (F) and leaves (L) from transgenic plants containing fruit-specific promoter constructs. TDET1-derived siRNAs migrated the same as a 25-nucleotide DNA oligonucleotide marker. 5S rRNA is shown as loading control. In each panel, the results from three independent T3 plants are shown.
To better estimate the extent of TDET1 gene silencing in fruits we performed northern blot analysis of low molecular weight RNA to detect small interfering (si)RNAs, which are a hallmark of RNAi26,27. TDET1-derived siRNAs were indeed detected in the mature green fruits but not in the leaves (Fig. 2d) or in material from untrans-formed plants (data not shown). This was the case in all lines expressing the transgene from each of the three different fruit-specific promoters. This result demonstrated the successful silencing of the TDET1 gene in the fruits and revealed that gene silencing did not occur or did not spread to other parts of the plant.
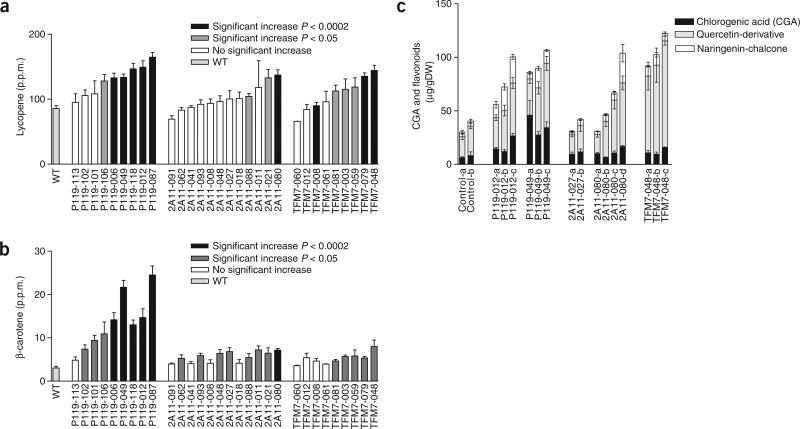
Chemical analysis of fruit samples selected from transgenic T2 generation plants containing all three promoter constructs revealed that both lycopene and β-carotene were present at higher levels than in wild-type fruits (Fig. 3a,b). All three promoter constructs generated up to twofold higher levels of lycopene although the P119 promoter was the most impressive. β-carotene levels in fruits from transgenic lines were also higher than in wild-type fruits, and in general the increases correlated with the increases in lycopene content in the different lines (Fig. 3a,b). Interestingly, the P119 promoter construct also provided the most dramatic increases in β-carotene (up to tenfold), which was readily apparent from visual inspection because fruits from these plants were more evidently orange (Fig. 1f).
Figure 3.
Quantification of carotenoid and flavonoid contents in red-ripe fruits from T56 (wild type, WT) and different lines of T2 generation plants containing fruit-specific promoter constructs. (a) Lycopene content. (b) β-carotene content. (c) Flavonoid content. In a and b the data represent mean values and s.e.m. and are derived from samples of three fruits taken from three to five sibling T2 plants per line (except 2A11-011 (n = 2), TFM7-012 (n = 2) and WT (n = 6)). In c the data represent mean values and s.e.m. and are derived from three to five fruits per plant. Statistical analysis of differences in transgenic lines with respect to WT was performed using Student's t-test.
Between two and four independent transgenic lines for each construct were then selected for further study in the T3 generation. T3 generation plants were phenotypically normal, indicating that siRNAs had not been transmitted through the seed to vegetative organs. Notwithstanding, these plants developed dark green fruits that subsequently became dark red, as in the T2 generation. Analyses of carotenoid content in mature fruits from these plants confirmed the observations from the T2 generation that both lycopene and β-carotene could be increased using all three promoter constructs, and that the P119 promoter outperformed the other two, particularly with regard to increased β-carotene content (Table 1). To examine whether the increased carotenoid content resulted in negative collateral effects on fruit quality, we examined two important parameters, fruit weight and brix (soluble solids). The results in Table 1 show that fruit weight was largely unaffected, except to a small extent in TFM7 promoter lines, and that brix values were also unchanged, except for small reductions in fruits from the P119 promoter lines.
Table 1.
Carotenoid content, fruit weight and brix of red-ripe fruits from T3 generation transgenic plants
| Line ID | Lycopene (p.p.m.) | n | β-carotene (p.p.m.) | n | Fruit weight (g) | n | Brix | n |
|---|---|---|---|---|---|---|---|---|
| WT (56) | 55.9 ± 2.5 | 30 | 2.4 ± 0.15 | 30 | 126.3 ± 3.1 | 59 | 4.8 ± 0.1 | 59 |
| P119-006-04 | 71.8 ± 9.5 | 5 | 5.7 ± 0.3b | 5 | 126.4 ± 8.0 | 5 | 4.4 ± 0.2a | 5 |
| P119-006-02 | 79.2 ± 7.2a | 3 | 12.4 ± 0.9b | 3 | 169.0 ± 20.5a | 2 | 3.4 ± 0.4b | 3 |
| P119-006-05 | 84.8 ± 11.4a | 4 | 12.0 ± 1.3b | 4 | 128.2 ± 9.2 | 4 | 4.1 ± 0.2a | 5 |
| P119-006-03 | 89.2 ± 5.8a | 5 | 13.3 ± 0.8b | 5 | 106.1 ± 5.5 | 5 | 4.0 ± 0.2b | 5 |
| P119-106-05 | 85.8 ± 6.4b | 13 | 12.4 ± 0.9b | 13 | 122.8 ± 7.4 | 14 | 4.1 ± 0.2b | 5 |
| P119-049-01 | 94.4 ± 10.5b | 9 | 16.6 ± 0.5b | 9 | 130.8 ± 7.0 | 9 | 3.8 ± 0.2b | 3 |
| P119-049-04 | 111.6 ± 9.1b | 12 | 20.5 ± 0.7b | 12 | 134.7 ± 7.9 | 12 | 3.9 ± 0.2b | 3 |
| P119-012-05 | 74.9 ± 4.9a | 5 | 11.1 ± 0.5b | 5 | 126.5 ± 9.4 | 5 | 3.8 ± 0.1b | 5 |
| P119-012-08 | 114.8 ± 4.7b | 3 | 13.2 ± 1.3b | 3 | 116.5 ± 9.9 | 3 | 4.0 ± 0.3a | 3 |
| P119-012-07 | 115.3 ± 14.0b | 5 | 9.9 ± 1.5b | 5 | 135.1 ± 11.7 | 5 | 3.9 ± 0.2b | 5 |
| P119-012-06 | 122.7 ± 13.6b | 4 | 10.1 ± 1.5b | 4 | 135.8 ± 13.4 | 4 | 4.1 ± 0.2a | 4 |
| 2A11-011-08 | 67.9 ± 6.1 | 11 | 4.4 ± 0.3b | 11 | 110.1 ± 8.5a | 11 | 4.7 ± 0.3 | 4 |
| 2A11-011-04 | 71.0 ± 5.5a | 13 | 4.9 ± 0.2b | 13 | 114.2 ± 6.4 | 13 | 4.7 ± 0.1 | 5 |
| 2A11-080-01 | 76.6 ± 3.3b | 16 | 4.5 ± 0.2b | 16 | 107.1 ± 3.2a | 16 | 4.6 ± 0.1 | 5 |
| 2A11-080-03 | 99.8 ± 5.3b | 18 | 5.8 ± 0.4b | 18 | 113.0 ± 5.5 | 18 | 5.1 ± 0.2a | 5 |
| 2A11-027-05 | 92.6 ± 6.7b | 11 | 6.1 ± 0.4b | 11 | 106.6 ± 5.8 | 11 | 4.6 ± 0.2 | 5 |
| 2A11-027-01 | 96.4 ± 5.2b | 15 | 7.5 ± 0.4b | 15 | 81.0 ± 4.8b | 11 | 4.2 ± 0.1b | 3 |
| 2A11-027-06 | 103.5 ± 7.4b | 12 | 8.9 ± 0.5b | 12 | 108.8 ± 4.8a | 12 | 4.6 ± 0.1a | 5 |
| TFM7-012-03 | 101.0 ± 5.2b | 19 | 6.3 ± 0.4b | 19 | 101.9 ± 5.3b | 20 | 4.5 ± 0.1a | 5 |
| TFM7-048-04 | 120.0 ± 8.0b | 13 | 9.0 ± 0.8b | 13 | 92.6 ± 5.4b | 14 | 4.5 ± 0.1a | 4 |
Table shows mean values ± s.e.m., together with the number of plants examined (n). Statistical analysis of differences in transgenic lines with respect to wild type was done using Student's t-test and significance of differences are indicated
P < 0.05
P < 0.0002.
To our knowledge, transgenic approaches have not increased both carotenoids and flavonoids simultaneously, except in a recent, rather limited study of one transgenic tomato plant (ectopically over-expressing the CRYPTOCHROME2 gene), that also showed several developmental defects28. Because hp mutant fruits have previously been shown to contain increased flavonoids as well as carotenoids20,21, we determined the content of three major fruit flavonoids in several transgenic plants containing the different constructs. Flavonoid levels higher than those of wild-type plants were found in almost all cases (Fig. 3c), with increases ranging from 1.9- to 3.5-fold. Plants containing the TFM7 promoter construct showed the most dramatic increases (2.6- to 3.5-fold), whereas the 2A11 promoter construct produced the lowest increases (typically 1.9-fold). Although in most cases both phenylpropanoids and flavonoids increased, qualitative differences were also observed. For example, P119 promoter lines had greater increases in chlorogenic acid and naringenin-chalcone, whereas the TFM7 promoter lines showed a greater elevation in quercetin derivatives.
These results therefore demonstrate that each of the three fruit-specific promoter constructs tested were able to increase both carotenoid and flavonoid concentrations to levels substantially higher than are observed in fruits from wild-type plants and similar or greater than those found in the different alleles of the hp-2 mutants18,21. When bacterial genes encoding carotenoid biosynthetic enzymes were overexpressed in tomato, more moderate increases in carotenoid levels were observed3,4. Furthermore, increases in one carotenoid were found to occur at the expense of others, which was proposed to be a consequence of rate-limiting steps within the carotenoid biosynthetic pathway, or perhaps of saturation of the carotenoid storage capacities within tomato fruits4. Our results demonstrate that this is not the case and reveal that manipulation of regulatory genes controlling carotenoid biosynthesis can generate impressive increases in flux through the whole pathway. It would therefore appear likely that even greater increases in carotenoid content can be achievable through the judicious manipulation of key regulatory genes rather than through manipulation of genes encoding biosynthetic enzymes. This finding also has implications for increasing the pro-vitamin A content of Golden Rice29.
In addition to elevated carotenoids, the flavonoid content of the transgenic plants was also increased and, in general, these increases paralleled the increases in carotenoid content in the transgenic lines examined. Furthermore, all pathway phenylpropanoids and flavonoids analyzed were elevated to varying degrees. Thus the whole biosynthetic pathway has been affected and no depletion in precursors or differential pathway partition is apparent.
The high efficiency of fruit-specific silencing of TDET1 in these experiments resulted at worst in only minor reductions in other quality traits such as fruit weight and brix. Overall yield, although not examined experimentally, also appeared to be unchanged (unpublished observations). The lack of negative collateral effects was likely a consequence of the combination of fruit-specific promoters with high-efficiency silencing constructs, that is, made up of an aberrant (truncated and mutant) TDET1 coding sequence in an inverted-repeat configuration. The highly fruit-localized nature of TDET1 silencing indicates that silencing signals did not propagate from the fruits to the rest of the plant. This is probably because fruits are sink rather than source organs and so do not possess efficient mechanisms for transport of substances out of them. Of importance for understanding the mechanisms of systemic transfer of gene silencing signals is that the silencing signal was not passed through the seed, because seeds derived from TDET1-silenced fruits gave rise to wild-type seedlings that developed like normal plants up until fruit development, when silencing was again activated.
In summary, this study shows that the judicious manipulation of a plant regulatory gene can result in dramatic simultaneous increases in fruit carotenoid and flavonoid content. To our knowledge, there have been no previous examples where metabolic engineering has resulted in the simultaneous elevation of flux through two independent health-related biosynthetic pathways with essentially no negative collateral effects on fruit yield or quality. Such transgenic strategies may prove to be more acceptable to the general public than currently used genetically modified crops because they can be based solely on the use of plant-derived sequences. Furthermore, the combination of RNAi technology with organ- or tissue-specific expression systems is likely to generate phenotypes that cannot be achieved by conventional breeding approaches, as in the present case. The fruit-suppressed TDET1 transgenic plants generated in this study can be useful for studying the fruit ripening process, and can provide valuable new tomato-based products with enhanced benefits for human health.
METHODS
Construction of plasmids
Three different constructs were generated, containing a TDET1-derived inverted-repeat downstream of one of three different fruit-specific promoters. Inverted-repeat constructs were used because it has been shown that they can greatly enhance the efficiency of gene silencing30. Furthermore, we hypothesized that promoters active during the early stages of fruit development might be necessary to effectively silence TDET1 expression because the late-expressing E8 promoter was found to be ineffective22 and because the hp-2 mutation has strong effects in immature fruits18. All three promoters are known to be expressed early during fruit development23–25. Promoter P119 is expressed from the earliest stages in green fruit through to red ripe stages, with increased levels during ripening25. Promoter 2A11 is expressed at high levels in ripening fruits, but also transiently in 2- to 3-cm green fruits24. The TFM7 promoter is expressed mainly during immature green fruit development and becomes inactive as green fruits reach full size24.
PCR-based cloning was used to generate suitable restriction sites to fuse each of the tomato fruit-specific promoters with the TDET1 transgene. The promoter of the TFM7 gene24 is 2.5 kb in length and was cloned in two steps. The upstream 1-kb fragment was amplified with flanking NotI and SpeI restriction sites, and the remaining 1.5-kb fragment was amplified with flanking SpeI and NcoI sites. Ligation together at the SpeI site created a NotI-NcoI cassette. Similarly, the 2.5-kb P119 promoter fragment25 was cloned in a single step between introduced flanking NotI and NcoI sites. The 4-kb 2A11 promoter fragment23 was cloned in three steps. An upstream 1-kb fragment was cloned between NotI and AvaII sites, an internal 1.2-kb fragment was amplified between AvaII and HindIII sites, and the remaining 1.8-kb fragment was inserted between HindIII and NcoI sites. The TDET1 inverted-repeat transgene was assembled in two steps and ligated downstream of the three promoters. In all cases the TDET1 transgene contained the hp-2 mutant allele18, which was previously shown to improve the efficiency of TDET1 gene silencing22. The first 180 bp of the TDET1 open reading frame (ORF) was removed and the remaining 1.5 kb was cloned between introduced NcoI and AvrII restriction sites. This was done by converting codon 61 (TTT; phenylalanine) to ATG (methionine) and introducing an NcoI site at the new ATG, retaining the frame of translation. An AvrII site was introduced 108 nucleotides after the translation stop. Finally, a 520-bp 5′ fragment of the ORF was cloned in an antisense orientation using the AvrII site and a SacI site, to create an inverted-repeat construct possessing the 3′ region of the ORF as a spacer between the inverted-repeat domains. The TDET1 inverted-repeat transgene was ligated as an NcoI-SacI cassette downstream of each promoter to generate the constructs P119::hp2-IR::nos, 2A11::hp2-IR::nos and TFM7::hp2-IR::nos. All chimeric gene constructs were confirmed by DNA sequencing. Expression cassettes were transferred to the binary vector pSVS297nos31, which contains the kanamycin-resistance marker gene NPTII driven by the CaMV 35S promoter, and were transformed into Agrobacterium tumefaciens by electroporation.
Plant transformation
To generate transgenic tomato plants, cotyledons from two-week-old seedlings were used for protocols as described32,33. As wild-type backgrounds we used Money Maker and T56 genotypes. Briefly, tomato seeds were sterilized using 2% commercial bleach and germinated on Murashige and Skoog (MS) medium. After 2 weeks of germination the cotyledons were cut and cocultivated for 2 d in the dark with the A. tumefaciens strain LBA4404 containing the different constructs. After 2 d of cocultivation the cotyledons were collected for selection on MS plates containing 50 mg/l kanamycin. When the plantlets regenerated they were transferred to MS rooting medium32,33. After ~2 months the plants were transferred to the greenhouse where they were grown to maturity for harvest of selfed seed (that is, from self-pollinated plants). Thirty-nine independent lines were generated for each construct. Progeny plants were analyzed by quantitative PCR of genomic DNA for the presence of the transgene, and homozygous lines were identified. Dark green immature fruit phenotypes were observed in 26 out of 39 independent lines containing the P119 promoter construct, 17 out of 39 independent lines containing the 2A11 promoter construct and 22 out of 39 of the transgenic lines containing the TFM7 promoter construct.
Molecular analyses
For the isolation of RNA, tomato leaves or pericarp from immature fruits (0.2 g) were ground to a fine powder in liquid nitrogen and subsequently extracted using hot phenol followed by two phenol:chloroform extractions. The RNA was precipitated using 4 M lithium chloride at –80 °C for 2 h. From total RNA, low molecular weight RNA was isolated using the protocols described by Hamilton and Baulcombe27 with small modifications. Low molecular weight RNA was separated from total RNA using 10% PEG 8000 and 0.5 M NaCl. This solution was spun at 10,000g for 10 min at 4 °C to remove high molecular weight RNA. The supernatant containing the low molecular weight RNA was then precipitated with ethanol at –20 °C for 2 h. We separated 50 μg of low molecular weight RNA on 15% polyacrylamide gels containing 7 M urea in 0.5× tris-borate/EDTA (TBE) buffer and electroblotted onto Hybond NX membranes (Amersham Pharmacia Biotech UK) using a Bio-Rad electroblotter (Bio-Rad). An in vitro–transcribed full-length antisense TDET1 riboprobe, prepared using the Riboprobe in vitro Transcription System (Promega Corporation), was used for hybridization. The probes were hydrolyzed with sodium carbonate buffer at 60 °C for 3 h to generate ~50-bp fragments. Hybridizations were carried out at 60 °C in Church and Gilbert buffer as described27 for 24 h. The blots were washed with 0.1% SDS and 20 mM NaPO4 twice for 10 min each at 60 °C.
Real-time quantitative RT–PCR was performed in triplicate using 200 ng of total RNA extracted from both leaves and fruits of the same plant. Targets were amplified in 25 μl reaction volumes using TaqMan EZ RT-PCR kit (Applied Biosystems) as described in the user manual. PCR, carried out in a 7000 Sequence Detection System (Applied Biosystems), consisted of an initial uracil-N-glycosylase (UNG) pretreatment (50 °C, 2 min), followed by a reverse transcription step (60 °C, 30 min), deactivation step (95 °C, 5 min) and then 40 amplification cycles (94 °C, 20 s; 60 °C, 1 min). For a given target, differences among tissue samples were calculated using the amplification rate and the cycle threshold (Ct) difference, and were expressed as relative mRNA levels.
For RT-PCR analysis, poly(A+) mRNA was isolated using Dynal mRNA extraction kits (Dynal). We used 100 ng of poly(A+) RNA to synthesize first strand cDNA using Superscript first-strand cDNA synthesis kits (Invitrogen). From this reaction, 2 μl was used to synthesize full length TDET1 cDNA using specific primers (forward, 5′-GTATGATTCACTAGTTTAATGCTGCTGAAAG-3′ and reverse, 5′-CCCATACTAGTCGTCTTGGCACTCTATCAAG-3′). The HY5 gene was PCR-amplified as a control for cDNA synthesis using specific primers (forward, 5′-ATGCAAGAGCAAGCGACGGTTCTAT-3′ and reverse, 5′-GTCCACGTGTCCTTCCCTCCTTCA-3′).
Biochemical analysis of fruits
Fully red-ripe greenhouse fruits (three fruits per plant) were harvested between 15 and 20 d post-breaker stage for carotenoid analysis (breaker is defined as the first external appearance of red color at the blossom end of the fruit). Carotenoids were extracted from tomato puree using 15 ml of acetone/methanol (2:1) and 4 ml hexane. After the addition of ~21 ml cold saline water, samples were shaken vigorously and then centrifuged into separate phases. An aliquot of the hexane phase was taken and analyzed on an HP 1050 system (Hewlett-Packard) configured with a diode array detector. The carotenoids were separated on a Whatman Partisil 5 ODS-3 column using a solvent mix of 81.7% acetonitrile, 9.6% methanol, 5.4% isopropyl alcohol and 3.3% MQ water. Lycopene was detected at 504 nm and β-carotene at 450 nm. Brix values were measured on a Bellingham & Stanley RFM-91 refractometer using filtered tomato puree.
For flavonoid and phenylpropanoid analyses, 3–5 ripe tomato fruits (9 d post-breaker) were harvested per plant. Fruit were quartered and one quarter per fruit (including peel, pericarp, jelly and seeds) was freeze-dried to complete dryness (14 d). A homogenous powder was made by homogenization in a freezer-mill (6750) apparatus (Glen Creston), 1 min at 70% full power. Flavonoids (including glycone derivatives) and phenylpropanoids were extracted from freeze-dried powder (50 mg) with methanol (1 ml) containing salicylic acid (20 μg) as the internal standard. The mixtures were incubated at 90 °C for 60 min. After cooling, the suspensions were centrifuged at 3,000g for 5 min. The resulting supernatants were removed and passed through a 0.4 μm filter. Separation and identification of flavonoids/phenylpropanoids was performed using a Dionex high-performance liquid chromatography system with online photodiode array detection (Dionex). A C18 reversed phase column (250 × 4.6 mm with 25 × 4.6 mm guard column from Hichrom) maintained at 25 °C was used to perform separations. The mobile phases consisted of 2% aqueous methanol containing 0.015% HCl (A) or 0.015% acetonitrile (B). Initial chromatographic conditions consisted of 95% A and 5% B for 10 min, followed by a linear gradient to 50% B over 30 min. Identification was achieved by cochromatography and comparison of spectral characteristics with authentic standards (purchased from Extrasynthase). Quantification was achieved by comparison with the international standard at 320 nm. All samples were analyzed in triplicate.
ACKNOWLEDGMENTS
This work was supported by Seminis Vegetable Seeds Inc., and by funding from the European Union (contract QLK5-CT-2000-00357), the Italian Ministry for Research and Education (FIRB contract RBNE01CFKB) to C.B. P.D.F. and P.M.B. acknowledge financial support from the UK Biotechnology and Biological Sciences Research Council (C19322).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Miller EC, et al. Lycopene, tomato products, and prostate cancer prevention. Have we established causality? Pure Appl. Chem. 2002;74:1435–1441. [Google Scholar]
- 2.Ross JA, Kasum CM. Dietary flavonoids: bioavailability, metabolic effects and safety. Annu. Rev. Nutr. 2002;22:19–34. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- 3.Romer S, et al. Elevation of the provitamin A content of transgenic tomato plants. Nat. Biotechnol. 2000;18:666–669. doi: 10.1038/76523. [DOI] [PubMed] [Google Scholar]
- 4.Fraser PD, et al. Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. USA. 2002;99:1092–1097. doi: 10.1073/pnas.241374598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muir SR, et al. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 2001;19:470–474. doi: 10.1038/88150. [DOI] [PubMed] [Google Scholar]
- 6.Niggeweg R, Michael AJ, Martin C. Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 2004;22:746–754. doi: 10.1038/nbt966. [DOI] [PubMed] [Google Scholar]
- 7.Bovy A, et al. High-flavonol tomatoes resulting from heterologous expression of the maize transcription factor genes LC and C1. Plant Cell. 2002;14:2509–2526. doi: 10.1105/tpc.004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Key TJ, Allen NE, Spencer EA, Travis RC. The effect of diet on risk of cancer. Lancet. 2002;360:861–868. doi: 10.1016/S0140-6736(02)09958-0. [DOI] [PubMed] [Google Scholar]
- 9.Cooper DA. Carotenoids in health and disease: recent scientific evaluations, research recommendations and the consumer. J. Nutr. 2004;134:221–224. doi: 10.1093/jn/134.1.221S. [DOI] [PubMed] [Google Scholar]
- 10.Kucuk O, et al. Lycopene in the treatment of prostate cancer. Pure Appl. Chem. 2002;74:1443–1450. [Google Scholar]
- 11.Heber D, Lu QY. Overview of mechanisms of action of lycopene. Exp. Biol. Med. 2002;227:920–923. doi: 10.1177/153537020222701013. [DOI] [PubMed] [Google Scholar]
- 12.Humphrey JH, West KP, Jr., Sommer A. Vitamin A deficiency and attributable mortality among under 5 year olds. Bull. World Health Organ. 1992;70:225–232. [PMC free article] [PubMed] [Google Scholar]
- 13.Rice-Evans CA, Millier NJ, Paganga G. Antioxidant properties of phenolic compounds. Trends Plant Sci. 1997;2:2152–2159. [Google Scholar]
- 14.DellaPenna D. Plant metabolic engineering. Plant Physiol. 2001;125:160–163. doi: 10.1104/pp.125.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovannoni J. Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:725–749. doi: 10.1146/annurev.arplant.52.1.725. [DOI] [PubMed] [Google Scholar]
- 16.Adams-Phillips L, Barry C, Giovannoni J. Signal transduction systems regulating fruit ripening. Trends Plant Sci. 2004;9:331–338. doi: 10.1016/j.tplants.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Schafer E, Bowler C. Phytochrome-mediated photoperception and signal transduction in higher plants. EMBO Rep. 2002;3:1042–1048. doi: 10.1093/embo-reports/kvf222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C. Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell. 1999;11:145–157. doi: 10.1105/tpc.11.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin I, Frankel P, Gilboa N, Tanny S, Lalazar A. The tomato dark green mutation is a novel allele of the tomato homolog of the DEETIOLATED1 gene. Theor. Appl. Genet. 2003;106:454–460. doi: 10.1007/s00122-002-1080-4. [DOI] [PubMed] [Google Scholar]
- 20.Yen HC, et al. The tomato high-pigment (hp) locus maps to chromosome 2 and influences plastome copy number and fruit quality. Theor. Appl. Genet. 1997;95:1069–1079. [Google Scholar]
- 21.Bino RJ, et al. The light-hyperresponsive high pigment-2dg mutation of tomato: alterations in the fruit metabolome. New Phytol. 2005;166:427–438. doi: 10.1111/j.1469-8137.2005.01362.x. [DOI] [PubMed] [Google Scholar]
- 22.Davuluri GR, et al. Manipulation of DET1 expression in tomato results in photo-morphogenic phenotypes caused by post-transcriptional gene silencing. Plant J. 2004;40:344–354. doi: 10.1111/j.1365-313X.2004.02218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pear JR, Ridge N, Rasmussen R, Rose RE, Houck CM. Isolation and characterization of a fruit-specific cDNA and the corresponding genomic clone from tomato. Plant Mol. Biol. 1989;13:639–651. doi: 10.1007/BF00016019. [DOI] [PubMed] [Google Scholar]
- 24.Santino CG, Stanford GL, Conner TW. Developmental and transgenic analysis of two tomato fruit enhanced genes. Plant Mol. Biol. 1997;33:405–416. doi: 10.1023/a:1005738910743. [DOI] [PubMed] [Google Scholar]
- 25.Dunsmuir P, Stott J. P119 promoter and their uses. 1997 US patent 5,633,440.
- 26.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton AJ, Baulcombe DC. A novel species of small antisense RNA in post-transcriptional gene silencing. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 28.Giliberto L, et al. Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 2005;137:199–208. doi: 10.1104/pp.104.051987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paine JA, et al. Improving the nutritional value of Golden Rice through increased provitamin A content. Nat. Biotechnol. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- 30.Wesley SV, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 31.Burgess DG, et al. A novel, two-component system for cell lethality and its use in engineering nuclear male-sterility in plants. Plant J. 2002;31:113–125. doi: 10.1046/j.1365-313x.2002.01330.x. [DOI] [PubMed] [Google Scholar]
- 32.McCormick S. Transformation of tomato with Agrobacterium tumefaciens. Plant Tissue Culture Manual B. 1991;6:1–9. [Google Scholar]
- 33.Yoder JI, Palys J, Alpert K, Lassner M. Ac transposition in transgenic tomato plants. Mol. Gen. Genet. 1988;213:291–296. [Google Scholar]