Abstract
Cytokines stimulate biological responses by activating intracellular signaling pathways. We have been adapting flow cytometric techniques to measure the levels of expression and activation of signaling molecules within mixed populations containing NK cells and to characterize their differences within NK cell sub-populations. Approaches for evaluating the total levels of the signal transducers and activators of transcription STAT1 and STAT4, of STAT1 in cells expressing IFNγ, and of the type-1-interferon (type 1 IFN) activation by phosphorylation, i.e. induction of pSTAT1 and pSTAT4, have been developed. The results of experiments using these techniques have demonstrated that an unusual feature of NK cells is high basal expression of STAT4 but reduced STAT1 levels. The condition pre-disposes for pSTAT4 activation by type 1 IFNs. The work has also shown, however, that total STAT1 levels are induced during viral infections as a result of IFN exposure, and that this change acts to promote the activation of STAT1 but limit both the activation of STAT4 and IFNγ expression. The intracellular staining approaches used for these studies and described here have utility in characterizing other mechanisms regulating cytokine-mediated signaling, and defining additional pathways shaping cellular responses to cytokines.
Keywords: STAT1, STAT4, pSTATs, intracellular staining, type 1 IFN
1. Introduction
Cytokine Signaling
A range of cytokines use signal transducers and activators of transcription molecules (STATs) as intracellular intermediaries for eliciting cellular responses. A class of cytokines using STATs to signal is the type 1 interferon (IFN) family comprised of a single β and multiple αs. The factors are elicited in response to a variety of stimuli including viral infections (1, 2). As a result of binding to specific receptors, type 1 IFNs induce particular kinases to activate STATs by phosphorylation (3, 4). The best understood signaling pathway downstream of the type 1 IFN receptor uses STAT1 and STAT2, and this pathway is linked to the induction of antiviral defense mechanisms. Type 1 IFNs, however, have been reported to induce a wide range of biological functions and have important immunoregulatory effects (1, 2). Some of these are paradoxical, including enhancing and inhibiting IFNγ production as well as blocking and promoting cell proliferation (1, 5–8), but the mechanisms regulating selection of the sub-set functions are poorly understood. There are a total of seven STATs, 1 through 6 with two different 5s. Type 1 IFNs have been reported to conditionally activate all of these, including STAT4 (1, 9). Their negative effects on IFNγ expression and cell proliferation are dependent on STAT1 (5, 6, 10), whereas STAT4 contributes to IFNγ expression (7, 11, 12). Thus, conditions affecting the relative accessibility of STAT4 and STAT1 have the potential to contribute to the shaping of cellular responses to type 1 IFNs.
Approaches for Evaluating Access to Signaling Pathways
Until recently, evaluation of the availability and activation of intracellular signaling pathways has been limited to biochemical Western blot analyses using proteins extracted from cell populations. The technique does allow determination of total protein and phosphoprotein levels within samples, and has the advantage of revealing the molecular weights of proteins detected. Using this approach, we have analyzed samples from mouse splenic leukocytes to show that there is an inverse correlation between type 1 IFNs’ ability to activate STAT4 with total STAT1 levels (7). This method, however, does not allow characterization of differences in responses within mixed cell sub-populations, and the numbers of purified cells required can present a challenge for certain NK cell studies. Flow cytometric or fluorescent activated cell sorting (FACS) techniques can evaluate multiple-parameters within mixed cell subsets. The approach has become very powerful because of the development of: 1) specific monoclonal antibodies against a variety of cell determinants and cytokines; 2) an increasing range of fluorochromes with different excitation and emission spectra for coupling to the monoclonal antibodies; and 3) improved instrumentation (13). In addition to being used to evaluate the molecules expressed on the surfaces of cell subsets, flow cytometry is being used to characterize mixed cell subsets in regard to the range and levels of cytokines they can be induced to express in their cytoplasm. This is possible because of the development of different permeabilization protocols allowing intracellular access of antibodies detecting cytokines. Thus, there are indications that flow cytometric techniques may provide opportunities for studying intracellular expression of signaling molecules in different cells within mixed populations.
Technical Challenges to Flow Cytometic Approaches for Measuring STATs
Because of our interest in defining the intracellular signaling pathways activated by type 1 IFNs in NK cells, this group committed to developing protocols to evaluate the levels of STAT1, STAT4, pSTAT1, pSTAT4, IFNγ and/or combinations of these within NK cells (14). The approach required identification of multiple cell surface determinants with multiple intracellular molecules. Challenges included the availability of specific antibodies and/or specific antibodies for use with fluorochromes having emission spectra that could be separated. In addition, depending on their activation state, STATs can be found in both the cytoplasm and nucleus. Therefore, staining and permeabililization/fixation methods that could be used in combination needed to be identified to allow detection of the different STATs or pSTATs within cells without destroying the fluorescent function of the fluorochromes and/or the antigenic determinants being detected by the monoclonal antibodies. The methods developed are based on the following: commercially available protocols for staining incorporated nuclear analogues (BD Biosciences), earlier work from our group examining STAT1 levels in T cells (10), reports for detecting pSTATs (15–20), testing using wild type (WT), STAT1-deficient and STAT4-deficient cells (14), and comparisons of results to those obtained with Western blotting (14). Commercially available antibodies to detect pSTATs facilitated the work (BD Biosciences). Antibodies from a variety of sources were screened for intracellular staining of total STAT1 and STAT4 proteins. Monoclonal antibodies detecting full-length STAT1 and STAT4 were being commercially produced and identified (BD Biosciences), and were first used with specific secondary antibodies. Eventually custom reagents with directly-conjugated fluorochromes were made. Methanol permeabilization was optimal for intracellular detection of the STATs. It did, however, present problems for detection of cytoplasmic cytokines and the use of particular fluorochromes to identify cell surface markers.
Methods Developed
The protocols were developed with the goal of characterizing the signaling pathways and responses to type 1 IFN exposure in vivo. The focus was on measuring total STATs, pSTATs, and IFNγ levels immediately after isolation of the cells from uninfected mice and/or from mice at different times after viral infection. It became clear, however, that optimal protocols for detecting STATs were not compatible with detecting intracellular cytokines. Thus, a second method was developed for identifying STAT1 and IFNγ within the same cells. Finally, the detection of pSTAT activation in vivo was possible in only low frequencies of cells from immunocompetent mice. This is likely to be in part a result of the need to capture cells just after cytokine exposure because the biochemical evidence indicates that phosphorylated forms of the STATs are short-lived. In addition, however, the experiments carried out also demonstrated that a dynamic regulation of the STAT levels contributed to the selection of particular STATs for activation in vivo. This latter point was proven by examining cellular responsiveness for type 1 IFN activation of STAT1 or STAT4 under control ex vivo conditions after isolation, and by using cells from mice mutated in STAT1 or STAT2. The three methods are presented below: 1) detecting total STATs or total STAT1 with pSTAT4 within freshly isolated NK cells; 2) detecting STAT1 with IFNγ immediately after isolation; and 3) detecting ex vivo responsiveness to type 1 IFNs with the examination of single parameter pSTATs or total STAT1 with pSTAT4.
2. Materials
2.1 Mice
All protocols require the preparations of splenic leukocytes from wild type (WT) mice and mice genetically deficient for STAT1 or STAT2. They are available on the 129 background (21, 22) (Taconic Labs). For NK studies, it is best to use mice at 4-to-9 weeks of age. As indicated, mice can be treated to induce an immunological response. For the protocols detailed below, mice were either uninfected (D0) or infected intraperitoneally with lymphocytic choriomeningitis virus (LCMV) (10, 14).
2.2 General
6-well tissue culture plate (BD Biosciences).
96-well-V-bottom assay plate (Costar).
15 ml polypropylene conical tubes (BD Falcon).
24-well tissue culture plate (BD Biosciences).
FACS Tube: 1.2 ml polypropylene U-bottom tube (Costar).
Sterile frosted glass microscope slides (Fisher Scientific).
Nylon mesh (Sefar America).
Red Blood Cell Lysing Buffer (Sigma).
RPMI medium 1640 (GIBCO).
Assay Medium: RPMI-1640 containing 10% FBS, with 1× Penicillin-Streptomycin and 10 mM Hepes Buffer (GIBCO) at pH 7.4.
Brefeldin A (Sigma), dissolved in DMSO at 10 mg/ml, aliquoted and stored at −20°C.
Staining Buffer: PBS containing 2% fetal bovine serum (FBS, Hyclone).
Goat Block: PBS containing 20% FBS and 10% goat serum (Sigma).
2.4G2 antibody (anti-FcγRIII/II; BioXcell) at a working concentration of 1 mg/ml.
Cytofix/Cytoperm Buffer (BD Biosciences).
Perm Wash Buffer (BD Biosciences).
DNase I (Sigma), dissolved in PBS, aliquoted at 1 mg/ml concentration of stock solution and stored at −80°C. Dilute with PBS to 300 μg/ml for use.
Methanol (Fisher Scientific).
2.3 Stimulation
In Vivo: 2 X 104 plaque-forming units (PFUs) of LCMV.
Ex Vivo: Recombinant murine IFNβ (a gift from Biogen Idec, specific activity of 2×109 U/mg). Other type 1 IFNs, i.e IFNβ or IFNα, can be used (PBL InterferonSource).
2.4 Flow Cytometric Application and Fluorochromes
There are a number of instruments available for FACS, and these have increasing flexibility in extending parameters for measurement. The Methods described here, however, were developed using a FACSCalibur (BD Biosciences) with two lasers having outputs at 15 mW of 488- and 635 nm wavelengths. The results were analyzed using the CellQuest Pro Software (BD Biosciences). The experiments required staining with either three or four different antibodies identified by three or four fluorochromes that could be: 1) excited at these wavelengths, 2) result in emission wavelengths distinguishable using available filters, and 3) purchased from commercial sources either directly conjugated to, or available for use in secondary detection steps with, the antibodies needed. In the end, five different fluorochromes were used: fluorescein (FITC), Alexa Fluor 647 (Alexa 647), phycoerythrin (PE), allophycocyanin (APC), and peridinin chlorophyll protein (PerCP). The combinations of four that can be used in individual tests are: FITC, PerCP, PE, and Alexa 647; or FITC, PerCP, PE, and APC (see Note 1).
2.5 Antibodies
The reagents detecting mouse leukocyte cell surface markers were developed against mouse determinants. Because NK cells were being identified, two reagents are required for cell surface staining to identify the NK cells and exclude T cells. In the detailed methods, CD49b expression is used to identify NK cells and CD3ε expression is used to exclude T cells. As discussed in the Notes, however, other combinations of antibodies can be used, including expression of NK1.1 to identify NK cells from appropriate strains of mice and expression of TCRβ to exclude T cells. In contrast to the reagents identifying murine leukocyte subsets, many of the reagents being developed to detect total STATs or pSTATs were prepared against the human molecules, but cross-react with mouse because of the strong homologues between the species. Care should be taken to verify specific reactivity with mouse STATs or pSTATs when new reagents are use. The detailed Methods use a custom prepared anti-STAT1 conjugated to PE to detect total STAT1. An anti-STAT4 mouse monoclonal antibody of the IgG1 isotype is identified using a monoclonal anti-mouse IgG1. This secondary reagent specifically identifies the primary anti-STAT4 when: 1) the other reagents used are as listed in the Methods because no other is a mouse IgG1, and 2) the splenic leukocytes are prepared at times during infection that precede induction of B cell isotype switching. The Methods describe the use of antibodies recognizing STAT forms phosphorylated at particular tyrosine residues. STATs can be phosphorylated at different tyrosine or serine residues, and the targets of phosphorylation may vary depending on the conditions of stimulation. Thus, modifications of Methods to evaluate other sites of phosphorylation maybe of interest to investigators, but will require careful consideration of the biochemical literature. All of the antibodies purchased for flow cytometry and used in the Methods are available from BD Biosciences. Some of them can also be purchased from eBioscience. The amounts identified are based on the lots used for developing the protocols. Generally, 30 to 500 ngs of a particular antibody are used for a single test with 2 × 106 splenic leukocytes. All new lots should be evaluated by titration.
2.5.1 Detection of Total STAT1, Total STAT4, or Total STAT1 with pSTAT4
Commercial Specific Antibodies: FITC-anti-CD49b (clone DX5); Biotin-anti-CD49b (clone DX5); PerCP-anti-CD3ε (clone 145-2C11); Streptavidin-APC; Purified anti-STAT4 (clone 8); FITC-anti-mouse IgG1 (clone A85-1); Alexa 647-anti-STAT4 pY693 (clone 38/pSTAT4); all purchased from BD Biosciences.
Customized Antibody: PE-anti-STAT1 (C-terminal clone 42), prepared and conjugated with fluorescence dye by BD Biosciences, at a working stock of 2 μg/ml.
Isotype Controls: PE-Mouse IgG2b (clone 27-35) for PE-anti-STAT1; Purified mouse IgG1 (clone MOPC-21) for purified anti-STAT4; Alexa 647-Mouse IgG2b (clone 27-35) for Alexa 647-anti-STAT4 pY693; all purchased from BD Biosciences.
2.5.2 Detection of Total STAT1 with IFNγ
Commercial Specific Antibodies: FITC-anti-CD49b (clone DX5); PerCP-anti-CD3ε (clone 145-2C11); APC-anti-IFNγ (clone XMG1.2); all antibodies purchased from BD Biosciences.
Customized Antibody: PE-anti-STAT1 (C-terminal clone 42), prepared and conjugated with fluorescence dye by BD Biosciences, at a working stock of 2 μg/ml.
Isotype Controls: PE-Mouse IgG2b (clone 27-35) for PE-STAT1; APC-Rat IgG1 (clone R3-34) for APC-anti-IFNγ; from BD Biosciences.
2.5.3 Detection of Type 1 IFN Responsiveness for pSTAT1 or pSTAT4 Activation or Total STAT1 with pSTAT4 Activation
Commercial Specific Antibodies: FITC-anti-CD49b (clone DX5); PerCP-anti-CD3ε (clone 145-2C11); PE-anti-STAT1 pY701 (clone 4a); Alexa 647-anti-STAT4 pY693 (clone 38/p-STAT4); all purchased from BD Biosciences.
Customized Antibody: PE-anti-STAT1 (C-terminal clone 42), prepared and conjugated with fluorescence dye by BD Biosciences, at a working stock of 2 μg/ml.
Isotype Controls: PE-Mouse IgG2a (clone MOPC-173) for PE-anti-STAT1 pY701; Alexa 647-Mouse IgG2b (clone 27-35) for Alexa 647-anti-STAT4 pY693; all purchased from BD Biosciences.
3. Methods
3.1 Cell Preparation
Prepare splenic leukocytes (see Note 2) by a regular method (see Note 3). Spleen processing is carried out in a 6-well tissue culture plate, with RPMI medium 1640. Sterile frosted glass microscope slides and nylon mesh, 15 ml polypropylene conical tubes, and Red Blood Cell Lysing Buffer are used.
3.2 Detection of Total STAT1, Total STAT4 or Total STAT1 with pSTAT4
The methods provided below evaluate intracellular levels of total STATs or both STAT1 and pSTAT4 expression immediately after isolation of cells from mice (Fig. 1A). They all use methanol permeabilization. The methanol permeabilization is required for detection of STAT4 or pSTATs. The steps detailed will result in samples having been stained with: 1) FITC-anti-CD49b, PerCP-anti-CD3ε, and PE-anti-STAT1 antibodies; 2) biotin-anti-CD49b antibody detected with streptavidin-APC, PerCP-anti-CD3ε antibody, anti-STAT4 antibody detected with FITC-conjugated anti-mouse IgG1 antibody; and 3) FITC-anti-CD49b, PerCP-anti-CD3ε, PE-anti-STAT1 and Alexa 647-anti-STAT4 pY693 antibodies.
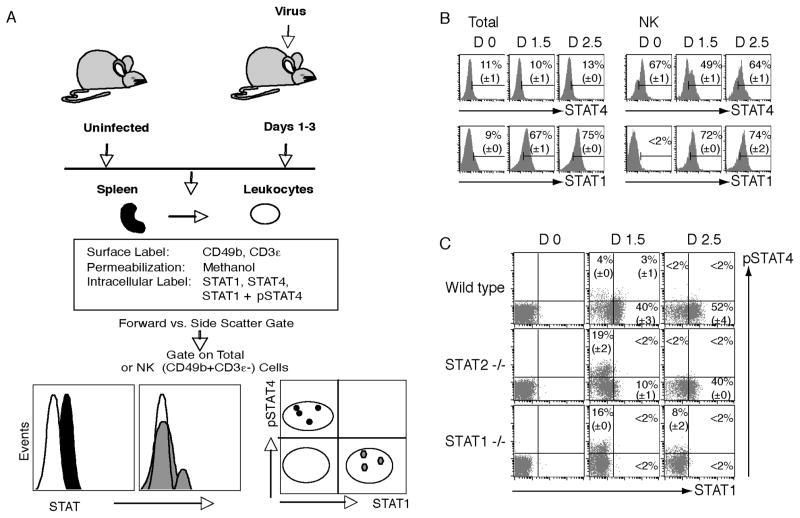
Figure 1. Methods for identifying intracellular total STATs or total STAT1 with pSTAT4.
(A) Leukocytes are prepared from spleens and stained to identify the CD49b+CD3ε− NK cells as defined in Methods 3.1. Intracellular levels of STAT1, STAT4 or pSTAT4 are evaluated using methanol permeabilization. Total cells are evaluated based on gates set by forward and side scatter. Flow cytometric gates set on CD49b+CD3ε− cell subset allows examination of the NK cell subset. Representation of results with STAT levels in histograms can reveal either general shifts in intensity or mixed populations of positive and negative cells. Dot plots of total STAT1 levels with pSTAT4 reveals diminishing STAT4 activation associated with increasing concentrations of STAT1. (B) Histograms show total levels of STAT1 and STAT4 evaluated separately in total and NK cells prepared from WT mice either uninfected (D0) or LCMV infected for 1.5 or 2.5 days (D1.5 or D2.5). (C) Dot plots show levels of STAT1 and pSTAT4 in NK cell subsets at D0, D1.5, and D2.5 after infection as evaluated in WT, STAT2-deficient, and STAT1-deficient mice. (Panels B and C were originally published in The Journal of Experimental Medicine, 2007, 204:2382–2396. © Miyagi et al., 2007. doi:10.1084/jem.20070401.)
Resuspend the cells to 2 × 107 cells/ml in cold Staining Buffer.
Use 96-well-V-bottomed plate and load 100 μl of cell suspension per well (or 2 X 106 cells per test).
Centrifuge at 700 g for 3 mins. with low brake. Remove buffer from plate by flicking plate.
To prevent non-specific binding of antibodies to Fc receptors during staining, add 200 μl of Goat Block with 0.25 μl of 2.4G2 antibody to each well and mix well (e.g. mix up and down 10 times) (see Note 4). Incubate for 15 mins. at 4°C. Centrifuge as described above and remove buffer from plate.
To label the cells with NK cell surface marker, add 50 μl of Staining Buffer containing 0.5 μl of FITC-anti-CD49b (for test leading to STAT1 detection) or biotin-conjugated anti-CD49b antibody (for test leading to STAT4 detection) (see Notes 1 & 5), to each well and mix well. Incubate for 15 mins. at 4°C.
To wash the cells, add 150 μl of Staining Buffer to each well, mix up and down 4 times. Then centrifuge and remove buffer.
To fix the cells, add 100 μl of Cytofix/Cytoperm to each well and mix well under the fume hood (see Note 6). Incubate for 20 mins. at 4°C.
Wash once with freshly prepared Perm Wash Buffer.
To permeabilize the cell, add 200 μl of pre-chilled (−20°C) pure methanol (see Note 7) to each well and mix well under the fume hood. Incubate for 15 mins. on ice under the fume hood (see Note 8).
Spin down the cells by centrifugation.
Flick plate and wash 2 times with Staining Buffer.
Add 50 μl of staining buffer containing 0.5 μl of PerCP-conjugated anti-CD3ε antibody (see Notes 1 & 9), with 15 μl of PE-conjugated anti-STAT1 antibody (for STAT1 detection), or 0.5 μl of streptavidin-APC and 2μl of anti-STAT4 antibody (for STAT4 detection), to each well. For control staining, add the same amount of corresponding isotype controls. In the case of simultaneous staining for STAT1 and pSTAT4, use 15μl of PE-conjugated anti-STAT1 antibody and Alexa 647-conjugated anti-STAT4 pY693 antibody or the same amount of corresponding isotype controls to tests that have been first labeled with FITC-anti-CD49b antibody.
Mix well and incubate for 20 mins. at room temperature. For STAT1 staining, proceed to the step 16.
Wash once with Staining Buffer.
For STAT4 staining, add 50 μl of Staining Buffer containing the secondary antibody, 0.5 μl of FITC-conjugated anti-mouse IgG1 antibody to each well and mix well. Incubate for 15 mins. at 4°C.
Wash once with Staining Buffer.
Resuspend in 250 μl of Staining Buffer and then transfer to FACS Tubes. The samples are ready to be acquired using FACSCalibur (BD Biosciences) with the CellQuest Pro software (BD Biosciences) (see Note 10). An example result in the single staining of total STAT1 or STAT4 is shown in Fig. 1B. Another example result in the staining of both total STAT1 and pSTAT4 is shown in Fig. 1C.
3.2 Detection of Total STAT1 with IFNγ
The methods presented here evaluate the levels of total STAT1 as compared to IFNγ expression in individual cells (Fig. 2A). Because detection of intracellular cytokines is not possible after treatment with methanol, the fixation and permeabilization method used is dependent on Cytofix/Cytoperm (BD Biosciences) (See Note 11). At this time, it appears that the only signaling molecule detectable by this approach is STAT1. The steps detailed will result in samples having been stained with FITC-anti-CD49b, PerCP-anti-CD3ε, PE-anti-STAT1, and APC-anti-IFNγ antibodies.
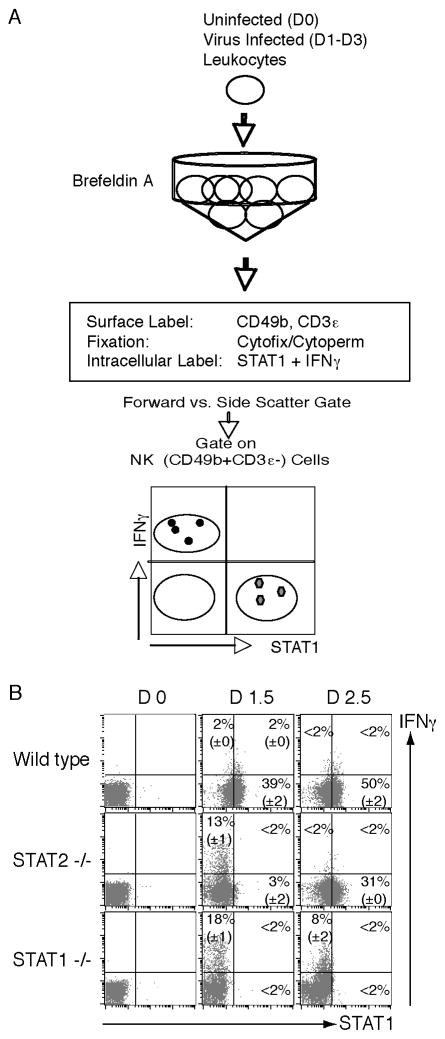
Figure 2. Methods for detecting total STAT1 and IFNγ within individual cells.
(A) Leukocytes are prepared from spleens, treated with brefeldin A, and stained to identify the CD49b+CD3ε− NK cells as defined in Methods 3.2. Intracellular levels of STAT1 along with IFNγ are evaluated using fixation/permeabilization with Cytofix/Cytoperm. Flow cytometric gates are set on CD49b+CD3ε− cell subset to identify the NK cell subset. Dot plots of total STAT1 levels with IFNγ reveals increases in STAT1 expression associated with decreasing cytokine expression. (B) Dot plots show NK cells levels of STAT1 and IFNγ expression at D0, D1.5, and D2.5 after infection as evaluated in WT, STAT2-deficient, and STAT1-deficient mice. (Panels B was originally published in The Journal of Experimental Medicine, 2007, 204:2382–2396. © Miyagi et al., 2007. doi:10.1084/jem.20070401.)
Prepare cells and resuspend at 1 × 107 cells/ml in Assay Medium containing brefeldin A (see Note 12) in a 15 ml conical tube.
Incubate for 4 hrs. at 37°C in a CO2 incubator, with mixing after 2hrs. by tapping the bottom of the tube.
To prepare cells for flow cytometry, use a 96-well-V-bottomed plate and load 200 μl of cell suspension per well (or 2 × 106 cells per test). Centrifuge and then flick buffer from plate.
Wash once with Staining Buffer.
To prevent non-specific binding of antibodies to Fc receptors, add 200 μl of Goat Block with 0.25 μl of 2.4G2 Ab to each well and mix well (see Note 4). Incubate for 15 mins. at 4°C. Centrifuge and then flick buffer from plate.
To label the cells with surface markers, add 50 μl of staining buffer containing 0.5 μl of FITC-conjugated anti-CD49b antibody and 0.5μl of PerCP-conjugated anti-CD3ε antibody to each well and mix well. Incubate for 15 mins. at 4°C.
Wash once with Staining Buffer.
To fix and permeabilize the cells, add 100 μl of Cytofix/Cytoperm to each well and mix well under a fume hood. Incubate for 20 mins. at 4°C.
Wash once with freshly prepared Perm Wash Buffer.
Add 100 μl of DNase solution to each well and mix well. Incubate (preferably with a lid such as aluminum foil) for 1 hr. at 37°C in an incubator.
Wash once with the Perm Wash Buffer.
Add 50 μl of the Perm Wash Buffer containing 0.5 μl of APC-conjugated anti-IFNγ antibody and 0.5 μl of PE-conjugated anti-STAT1 antibody, or corresponding isotype controls, to each well. Mix well and incubate for 20 mins. at room temperature.
Wash once with the Perm Wash Buffer.
Resuspend in 250 μl of staining buffer and then transfer to FACS Tubes. The samples are ready to be acquired using FACSCalibur with the CellQuest Pro software. An example result is shown in Fig. 2B.
3.3 Detection of Type 1 IFN Responsiveness for pSTAT1 or pSTAT4 Activation or Total STAT1 with pSTAT4 Activation
The methods presented below evaluate changes in responsiveness to type 1 IFN for activation of STAT1 or STAT4 resulting from conditioning during different ex vivo treatments (Fig. 3A). Thus, they require the ex vivo exposure of isolated populations to cytokines prior to staining. The steps detailed will result in samples having been stained with: 1) FITC-anti-CD49b, PerCP-anti-CD3ε, and PE-anti-STAT1 pY701 antibodies; 2) FITC-anti-CD49b, PerCP-anti-CD3ε, and Alexa 647-anti-STAT4 pY693 antibodies; and/or 3) FITC-anti-CD49b, PerCP-anti-CD3ε, PE-anti-STAT1, and Alexa 647-anti-STAT4 pY693 antibodies.
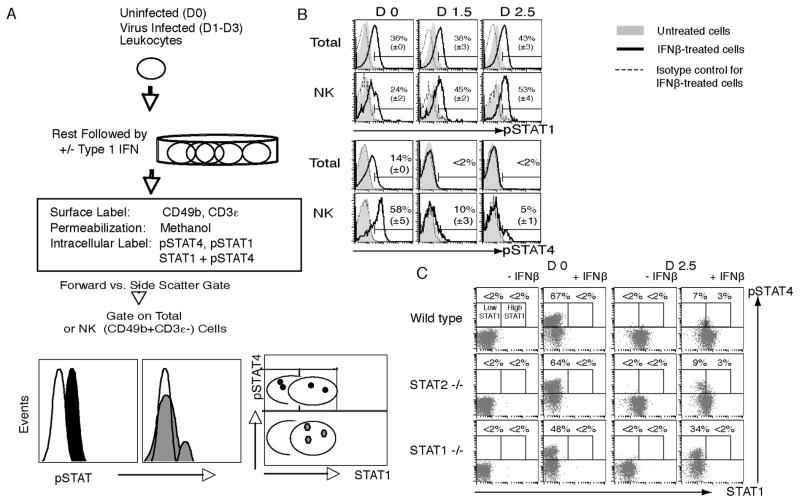
Figure 3. Methods for evaluating type 1 IFN responsiveness with pSTAT1 or pSTAT4 activation.
(A) Leukocytes are prepared from spleens, rested in culture to clear cytokine receptors, and treated with type 1 IFN to induce STAT activation. The cells are then stained to identify the CD49b+CD3ε− NK cells as per Methods 3.3. Intracellular levels of pSTAT1 or pSTAT4 are evaluated using methanol permeabilization. Total cells are evaluated based on gates set by forward and side scatter. Flow cytometric gates set on CD49b+CD3ε− cells allows examination of the NK cell subset. Dot plots of total STAT1 levels with pSTAT4 reveals differences in STAT4 activation associated with increasing concentrations of STAT1. (B) Ex vivo responsiveness of total and NK cells prepared from WT mice on D0, D1.5 and D2.5 of LCMV infection for STAT1 or STAT4 activation in response to type 1 IFN treatment. (C) Evaluation of ex vivo responsiveness of NK cells to type 1 IFN treatment in association with STAT1 levels on D0 and D2.5 of LCMV infection. Populations for analysis were prepared from WT, STAT2-deficient, and STAT1-deficient mice. (Panels B and C were originally published in The Journal of Experimental Medicine, 2007, 204:2382–2396. © Miyagi et al., 2007. doi:10.1084/jem.20070401.)
Resuspend the prepared cells to 2 × 107 cells/ml in Assay Medium.
Use 24-well-flat-bottomed plate and load 500 μl of cell suspension per well. To clear receptors of cytokines bound in vivo and allow cells to return to basal states, incubate for 4 hrs. at 37°C in an incubator before testing for responsiveness to type 1 IFNs for STAT1 or STAT4 activation.
Add 500 μl of Assay Medium with or without a type 1 IFN (e.g. recombinant murine IFNβ at a final concentration of 10,000 U/ml) for stimulated or unstimulated cells, respectively, to each well. Mix and incubate with a lid for 90 mins. at 37°C in a CO2 incubator.
To prepare cells for transfer and staining, mix the content in each well by pipeting up and down 10 times on ice.
For the analysis of flow cytometry, use 96-well-V-bottomed plate and load 200 μl of medium with stimulated or unstimulated cells to each well (or 2 X 106 cells per test). Centrifuge and then flick buffer from plate.
Wash once with cold Staining Buffer.
To prevent non-specific binding of antibodies for surface staining, add 200 μl of Goat Block with 0.25 μl of 2.4G2 Ab to each well and mix well (see Note 4). Incubate for 15 mins. at 4°C. Centrifuge and then flick buffer from plate.
To label the cells with NK cell surface marker, add 50 μl of Staining Buffer containing 0.5 μl of FITC-conjugated anti-CD49b antibody to each well and mix well. Incubate for 15 mins. at 4°C.
Wash once with Staining Buffer.
To fix the cells, add 100 μl of Cytofix/Cytoperm to each well and mix well under a fume hood. Incubate for 20 mins. at 4°C.
Wash once with freshly prepared Perm Wash Buffer.
To permeabilize the cells, add 200 μl of pre-chilled (−20°C) pure methanol to each well and mix well under a fume hood. Incubate for 15 mins. on ice.
Spin down the cells by centrifugation.
Flick and wash 2 times with Staining Buffer.
Add 50 μl of Staining Buffer containing 0.5 μl of PerCP-conjugated anti-CD3ε antibody with 15 μl of PE-conjugated anti-STAT1 pY701 antibody and/or Alexa 647-conjugated anti-STAT4 pY693, or the same amount of corresponding isotype controls, to each well. If the experiment is to detect total STAT1 and pSTAT4, add 50 μl of Staining Buffer containing 0.5 μl of PerCP-conjugated anti-CD3ε antibody with 15 μl of PE-conjugated anti-STAT1 antibody and Alexa 647-conjugated anti-STAT4 pY693, or the same amount of corresponding isotype controls, to each well. Mix well and incubate for 20 mins. at room temperature.
Wash once with Staining Buffer.
Resuspend in 250 μl of Staining Buffer and then transfer to FACS Tubes. The samples are ready to be acquired using FACSCalibur with the CellQuest Pro software. Examples of single parameter analyses of pSTAT1 and pSTAT4 are shown in Fig. 3B and of total STAT1 with pSTAT4 analysis are shown in Fig. 3C.
Acknowledgments
The authors thank D. Ashley Feldman for review of the manuscript. This work was supported by RO1 grants CA041268 and AI055677 from the National Institutes of Health, a Canadian Institutes of Health Research Fellowship, and funding from the Shinya Foundation.
Footnotes
The PE and PerCP fluorochromes are sensitive to methanol and cannot be used to label cell surface determinants prior to this treatment for cell permeabilization. This may require staining of cell determinants with PE- or PerCP-conjugated antibodies after methanol treatment, but it is not always possible to do so because antigenic determinants can also be sensitive to fixation and permeablization treatments. Thus, it is necessary to evaluate expression before and after treatments.
This protocol can be adapted for leukocytes from murine liver, and also for human mononuclear cells from peripheral blood with appropriate surface markers (data not shown).
The basic protocol for preparation of leukocytes from murine spleen is as follows. Sacrifice mice and remove spleen. Place spleens in a 6-well tissue culture plate containing 5 ml of RPMI medium on ice. Grind spleen between the rough surfaces of frosted glass slides. Filter cell suspension through a nylon mesh and transfer into a 15-ml centrifuge tube. Wash the well with additional 5 ml of cold RPMI medium. Centrifuge at 300 g for 10 mins. at 4°C, and discard supernatant. Disturb pellet by tapping the bottom of the tube, add 1 ml of Red Blood Cell lysing buffer and vortex briefly. Incubate for 1 min. at room temperature. Fill the tube to 10 ml with cold RPMI medium. Filter cell suspension again through nylon mesh and transfer into a 15-ml centrifuge tube and count cells. Centrifuge at 300 g for 10 mins. at 4°C, discard supernatant and resuspend cells at 2 × 107 cells/ml in cold RPMI medium.
The combination of goat serum and the 2.4G2 antibody, with directed against receptors for immunoglobulins (FcRs), in the Goat Block is required because it is difficult to inhibit non-specific binding of the antibodies for flow cytometry to the FcRs, particularly when the cells are isolated from infected mice.
The APC- as well as FITC- and biotin-conjugated but not PE-conjugated anti-CD49b antibodies (eBioscience), were resistant to methanol exposure. Therefore, APC-conjugated anti-CD49b antibody can be adapted in this protocol. Using cells from C57BL/6 background mice, the FITC-, APC- or biotin-conjugated but not PE-conjugated anti-NK1.1 antibody (PK136) were also resistant to methanol exposure. Moreover, a FITC-, APC- or biotin-conjugated but not PE-conjugated anti-TCRβ (clone H57-597) antibodies were found to be resistant to methanol exposure.
To fix cells, we take advantage of the Cytofix/Cytoperm from BD because it contains paraformaldehyde (PFA). We found that 4% PFA provided almost identical results with those by Cytofix/Cytoperm so that 4% PFA can be used to fix cells at this step instead of Cytofix/Cytoperm. As PFA is an evaporating toxin, this step must be performed under a fume hood.
The treatment with cold pure methanol for the permeabilization of cells was found to provide the best results for intracellular staining of total STATs and pSTATs. The treatment with Cytofix/Cytoperm alone, which is normally used for intracellular staining of cytokines, did not demonstrate a sufficient level of intracellular total STAT4 or pSTATs.
Pure methanol is also an evaporating toxin. Thus, this step must be performed under a fume hood.
In the Methods, anti-CD3ε staining is done after methanol treatment because the fluorochromes available for the combinations required in individual tests limits the commercially available regent to PerCP-anti-CD3ε, and because the CD3ε determinant is still detectable after methanol treatment.
At least 100,000 events should be collected within the leukocytes gated for analysis of intracellular molecules in NK cells because the cells are generally at lower than 5% of the total populations.
The treatment of methanol following Cytofix/Cytoperm treatment did not allow detection of intracellular IFNγ expression. It was possible, however, to analyze both STAT1 and IFNγ because in contrast to STAT4 or the pSTATs, STAT1 levels could be identified following fixation/permeabilization with Cytofix/Cytoperm and DNase treatment. This treatment was already reported to be useful to identify intracellular STAT1 level (10).
The treatment with brefeldin A is required for detecting the intracellular cytokine under these conditions. The best final concentration and time of incubation was found to be 5–10 μg/ml and 4-to-6-hrs. of incubation, respectively, in this system.
References
- 1.Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 2.Biron CA, Sen GC. Innate Immune Responses to Viral Infection. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Walter Kluwer/Lippincott: Williams & Wilkins; 2007. pp. 249–278. [Google Scholar]
- 3.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 4.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 5.Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen KB, Cousens LP, Doughty LA, Pien GC, Durbin JE, Biron CA. Interferon alpha/beta-mediated inhibition and promotion of interferon gamma: STAT1 resolves a paradox. Nat Immunol. 2000;1:70–76. doi: 10.1038/76940. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297:2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- 8.Tanabe Y, Nishibori T, Su L, Arduini RM, Baker DP, David M. Cutting edge: role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J Immunol. 2005;174:609–613. doi: 10.4049/jimmunol.174.2.609. [DOI] [PubMed] [Google Scholar]
- 9.Brierley MM, Fish EN. Review: IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J Interferon Cytokine Res. 2002;22:835–845. doi: 10.1089/107999002760274845. [DOI] [PubMed] [Google Scholar]
- 10.Gil MP, Salomon R, Louten J, Biron CA. Modulation of STAT1 protein levels: a mechanism shaping CD8 T-cell responses in vivo. Blood. 2006;107:987–993. doi: 10.1182/blood-2005-07-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 12.Lawless VA, Zhang S, Ozes ON, Bruns HA, Oldham I, Hoey T, Grusby MJ, Kaplan MH. Stat4 regulates multiple components of IFN- gamma-inducing signaling pathways. J Immunol. 2000;165:6803–6808. doi: 10.4049/jimmunol.165.12.6803. [DOI] [PubMed] [Google Scholar]
- 13.Herzenberg LA, De Rosa SC. Monoclonal antibodies and the FACS: complementary tools for immunobiology and medicine. Immunol Today. 2000;21:383–390. doi: 10.1016/s0167-5699(00)01678-9. [DOI] [PubMed] [Google Scholar]
- 14.Miyagi T, Gil MP, Wang X, Louten J, Chu WM, Biron CA. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J Exp Med. 2007;204:2383–2396. doi: 10.1084/jem.20070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleisher TA, Dorman SE, Anderson JA, Vail M, Brown MR, Holland SM. Detection of intracellular phosphorylated STAT-1 by flow cytometry. Clin Immunol. 1999;90:425–430. doi: 10.1006/clim.1998.4654. [DOI] [PubMed] [Google Scholar]
- 16.Uzel G, Frucht DM, Fleisher TA, Holland SM. Detection of intracellular phosphorylated STAT-4 by flow cytometry. Clin Immunol. 2001;100:270–276. doi: 10.1006/clim.2001.5078. [DOI] [PubMed] [Google Scholar]
- 17.Krutzik PO, Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A. 2003;55:61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- 18.Krutzik PO, Irish JM, Nolan GP, Perez OD. Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol. 2004;110:206–221. doi: 10.1016/j.clim.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Krutzik PO, Clutter MR, Nolan GP. Coordinate analysis of murine immune cell surface markers and intracellular phosphoproteins by flow cytometry. J Immunol. 2005;175:2357–2365. doi: 10.4049/jimmunol.175.4.2357. [DOI] [PubMed] [Google Scholar]
- 20.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 21.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 22.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]