Abstract
The present study examined the influences of the neovestitol-vestitol (NV) containing fraction isolated from Brazilian red propolis on biofilm development and expression of virulence factors by Streptococcus mutans using saliva-coated hydroxyapatite surfaces. In addition, NV was tested in a rodent model of dental caries to assess its potential effectiveness in vivo. Topical applications of NV (800μg/ml) significantly impaired the accumulation of S. mutans biofilms by largely disrupting the synthesis of glucosyltransferase-derived exopolysaccharides and the expression of genes associated with the adaptive stress response, such as copYAZ and sloA. Of even greater impact, NV was as effective as fluoride (positive control) in reducing the development of carious lesions in vivo. NV is a promising natural anti-biofilm agent that targets essential virulence traits in S. mutans, which are associated with cariogenic biofilm formation and the subsequent onset of dental caries disease.
Keywords: propolis, vestitol, neovestitol, Streptococcus mutans, biofilm, dental caries
Introduction
Virulent biofilms are responsible for a range of infections, including those occurring in the mouth. Dental caries is one of the most common and costly biofilm-dependent oral diseases, which afflicts children and adults worldwide (Dye, 2007; Baelum et al., 2007). This ubiquitous disease is a prime example of the consequences arising from interactions among microorganisms (and their products), host saliva, and dietary carbohydrates, which leads to the development of virulent biofilms on susceptible tooth surfaces (Marsh, 2003). The assembly of cariogenic biofilms is a dynamic process that is dependent on the production of a bacterial-derived exopolysaccharide (EPS)-rich matrix.
Streptococcus mutans and sucrose are key modulators of the development of cariogenic biofilms (Paes Leme et al., 2006). The bacterium is a chief producer of exopolysaccharides, and is highly acidogenic and tolerant of environmental stresses(Quivey et al., 2000; Lemos and Burne, 2008). Sucrose serves as a substrate for EPS synthesis and acid production (Bowen and Koo, 2011). Within the complex oral microbiome, S. mutans is not always the most abundant organism. However, it can rapidly orchestrate the formation of cariogenic biofilms when sucrose is available. S. mutans-derived glucosyltransferases (Gtfs) are present in the pellicle (GtfC) and on bacterial surfaces (GtfB), both of which produce large amounts of EPS in situ by utilizing sucrose as a substrate (Bowen and Koo, 2011). EPS formed on surfaces provides bacterial binding sites for the subsequent colonization and local accumulation of S. mutans and other organisms on the tooth surface (Schilling and Bowen, 1992). Over time, the accumulated EPS forms a highly cohesive and diffusion-limiting polymeric matrix that protects embedded bacteria (Xiao et al., 2012).
In parallel, various sugars, including sucrose, are fermented by S. mutans and other acidogenic organisms enmeshed in the EPS-rich milieu, creating acidic microenvironments (Bowen and Koo, 2011). The low pH environment further selects for the growth of acidogenic and acid tolerant microorganisms (Quivey et al., 2000; Marsh, 2003; Lemos and Burne, 2008), which leads to the formation of acidic regions throughout the biofilm and at the substratum (Xiao et al, 2012). The accumulation of acids eventually leads to the demineralization of the adjacent tooth enamel (expressed clinically as carious lesions) (Bowen and Koo, 2011).
Therapeutic agents that compromise the ability of virulent organisms to assemble and maintain biofilms on tooth surface could be an effective strategy to prevent dental caries and could likely be specifically accomplished by disrupting EPS synthesis, acid production, and/or stress survival mechanisms (Jeon et al., 2011). Fluoride continues to be the most effective anti-caries agent, but it does not offer complete protection against dental caries and may not effectively address the infectious character of the disease (Clarkson and McLoughlin, 2000). Thus, novel and effective therapies are warranted.
Natural products have been widely investigated as a potential source of new and active therapeutic agents (Newman and Cragg, 2012). Roughly 74% of all approved therapeutic agents developed between 1981 and 2010 have been of natural origin (Newman and Cragg, 2012); many more have been investigated in vivo and in clinical trials against a number of human maladies, including cancer and infectious diseases (Butler 2005). In contrast, comparatively fewer studies have been conducted to evaluate the potential use of natural products in antibiofilm and anticaries chemotherapy using a clinically relevant treatment regimen in vivo (Jeon et al. 2011).
Among the natural products, propolis (a resinous beehive product) appears to be a rich source of biologically active compounds that display potential antibiofilm and anticaries activities (Koo et al., 2002). Recently, Brazilian red propolis has been identified as a source of novel bioactive compounds, based on its distinctive chemical composition and potent antimicrobial and antioxidant activities (Alencar et al. 2007; Silva et al. 2008; Righi et al. 2011). Using bioassay guided fractionation coupled with spectroscopic analysis, a highly-purified neovestitol and vestitol-containing bioactive fraction (NV) was isolated from red propolis. Both neovestitol and vestitol are isoflavonoids and their biological properties against biofilms remain to be fully characterized (Bueno-Silva et al., 2013).
In this study, we have evaluated whether short-term topical applications of NV inhibit biofilm development by S. mutans in vitro and/or disrupt the development of dental caries in vivo. We found that NV treatments significantly reduced the accumulation of S. mutans biofilms on saliva-coated apatitic surfaces. The NV-treated biofilms contained less EPS (vs. vehicle-treated biofilms), possibly due to partial inhibition of Gtf activity. Brief exposures of NV also affected the expression of S. mutans genes associated with biofilm formation and stress tolerance. Furthermore, topical applications of NV were effective in reducing the incidence and severity of dental caries in a rodent model.
Material and methods
Chemical analysis of bioactive red propolis fraction
Red propolis samples were collected from Maceio, Alagoas State, Northeast of Brazil SL 09.40 and WL 35.41 at the end of summer, during the month of March. A previous study, using different chromatographic methods (including TLC), identified a sub-fraction of the chloroform fraction as the most bioactive using bioassay-guided fractionation (Bueno-Silva et al., 2013; Supplemental Figure 1). The sub-fraction was then selected for further chemical characterization with GC-MS as described in Bueno-Silva et al., 2013.
Biofilm preparation and treatment
Biofilms of S. mutans strain UA159 were formed on hydroxyapatite (HA) discs (1.25 cm diameter, surface area of 2.7±0.2 cm2, Clarkson Chromatography Products, Inc., South Williamsport, PA) vertically suspended in 24-well plates using a custom-made disc holder, as detailed elsewhere (Koo et al. 2005; 2010). Each HA disc was coated with clarified and filter-sterilized whole human saliva for 1h at 37°C. Saliva was collected from a single donor according to protocols approved by the Ethics Committee in Research of the Piracicaba Dental School, University of Campinas (Protocol #033/2008). The saliva-coated HA (sHA) discs were placed in ultra-filtered (10kDa cutoff, Millipore, Billerica, MA) yeast tryptone extract broth (UFYTE) with 1% sucrose containing 2 × 106 S. mutans CFU/ml, and incubated at 37°C and 5% CO2 (Koo et al., 2005; 2010). During the first 19 h, the organisms were grown undisturbed to allow initial biofilm formation. From this point on (19 h), the medium was changed every 24 hours and the biofilms were treated twice daily (21 h, 27 h, 45 h and 51 h) for a 60 s exposure until the end of experimental period (67 h) with the following solutions: NV (800 μg/mL) or vehicle-control (20% ethanol, v/v). During the treatments with the above solutions, biofilm discs were removed from the 24-well plate, dip-washed in saline solution and then transferred to a 24-well plate containing the treatment solutions. After 60 s exposure, the biofilm discs were dip-washed in saline solution and immediately returned to the 24-well plate containing the culture medium
The concentration of NV was selected based on preliminary dose-response biofilm studies and solubility in the vehicle system. The biofilms were retrieved at specific time points (43h and 67h) for confocal microscopy to examine the effects on the overall 3D architecture after exposure to the topical treatments. The gene expression profile was determined after the topical treatment at 44 h plus 1 h (Supplemental Figure 2), which represents the most active period of the biofilm development process using our model (Klein et al., 2010; Falsetta et al., 2012).
Laser scanning confocal fluorescence microscopy imaging of biofims
Extracellular polysaccharides (EPS) and bacterial cells were simultaneously labeled as described by Klein et al. (2009) and Xiao et al., (2012). EPS was labeled via incorporation of the Alexa Fluor 647 dextran conjugate (MW 10 kDa; absorbance/fluorescence emission maxima of 647/668 nm), while cell of S. mutans were stained with SYTO 9 (485/498 nm) (Molecular Probes Inc., Eugene, OR). Confocal imaging of intact biofilms was performed using an Olympus FV 1000 two photon laser scanning microscope (Olympus, Tokyo, Japan) equipped with a 10X (0.45 numerical aperture) water immersion objective lens. The excitation wavelength was 810 nm, and the emission wavelength filter for SYTO 9 was a 495/540 OlyMPFC1 filter, while the filter for Alexa Fluor 647 was an HQ655/40M-2P filter. Each biofilm was scanned at ten randomly selected positions, and a confocal image series was generated by optical sectioning at each of these positions (Xiao et al. 2012). The confocal image stacks were analyzed with COMSTAT, which generates several measurements for quantifying and characterizing the 3D structure of biofilms (Heydorn et al., 2000). In this study, the biomass of EPS and bacterial cells, as well as the average thickness, was calculated to determine the structural differences among treated biofilms. Amira 5.0.2 (Mercury Computer Systems Inc., Chelmsford, MS) was used to create 3D renderings of each structural component (EPS and bacteria) of the biofilms for visualization of the morphology and 3D architecture (Klein et al. 2009, Koo et al. 2010).
Isolation of biofilm RNA
RNA was extracted and purified using protocols optimized for biofilms formed in vitro (Cury and Koo, 2007). Briefly, the RNAs were purified and DNAse treated on the column using the Qiagen RNeasy Micro kit (Qiagen, Valencia, CA). The RNAs were then subjected to a second DNaseI treatment with Turbo DNase (Applied Biosystems/Ambion) and purified using the Qiagen RNeasy MinElute Cleanup kit (Qiagen). The RNAs were quantified using the NanoDrop ND1000 Spectrophotometer (Thermo Scientific\NanoDrop, Wilmington, DE). The RNA quality was evaluated using an Agilent 2100 Bionalazyer (Agilent Technologies Inc., Santa Clara, CA) and all RNAs used to prepare cDNAs for microarray analysis were determined to have an RNA integrity number (RIN) of 9.0 or above.
Microarray analysis
Whole genomic profiling was conducted using version 3 microarrays for S. mutans strain UA159 provided by the J. Craig Venter Institute (JCVI). A detailed description of these slides may be found at http://pfgrc.jcvi.org/index.php/microarray/array_description/streptococcus_mutans/version3.html. Reference RNAs were prepared as previously described (Klein et al. 2010), and cDNAs for both experimental and reference samples were synthesized following protocols provided by JCVI athttp://pfgrc.jcvi.org/index.php/microarray/protocols.html. Experimental cDNAs were labeled with indocarbocyanine (Cy3)-dUTP, while reference cDNAs were labeled with indodicarbocyanine (Cy5)-dUTP (Amersham Biosciences, Piscataway, NJ). Hybridizations were performed using the MAUI hybridization system (BioMicro Systems, Salt Lake City, UT), and slides were then washed and scanned using a GenePix scanner (Axon Instruments Inc., Union City, CA) following JCVI protocols. After scanning, single-channel images were simultaneously uploaded into JCVI Spotfinder321 (http://www.tm4.org/spotfinder.html), creating an overlay image with both Cy5 and Cy3 channels. A Spotfinder grid file was downloaded from http://pfgrc.jcvi.org/index.php/microarray/array_description/streptococcus_mutans/version3.html and adjusted to remove empty spots from the analysis. Spotfinder321 was used to identify spots and assign relative spot intensities in a .mev file for upload into JCVI MIDAS (http://www.tm4.org/midas.html). MIDAS software was used to perform a lowess normalization to flag and remove spots with low intensity values (less than 1000). Statistical analysis was performed using BRB-Array Tools, which is available as freeware from http://linus.nci.nih.gov/BRB-ArrayTools.html. An unpaired class comparison was performed with a P value cutoff of 0.05 for four replicates of NV-treated and four replicates of vehicle-treated samples
Analysis and classification of microarray data
MDV (available from LANL Oralgen http://www.oralgen.lanl.gov/) was used to assign gene names and functional classes to genes identified with BRB-Array Tools, as described previously (Klein et al. 2010). Genes were then sorted in Microsoft Excel to identify those with an absolute fold-change of 1.5 or greater. These genes were then organized into functional categories to identify the genes of interest (GOI) as described in Falsetta et al., (2012). These data were used to evaluate the impact of treatment by identifying the major themes affected.
Quantitative PCR (qRT-PCR) of selected genes for microarray data validation
The same RNAs also served as templates for cDNA synthesis using the Bio-Rad iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA) for RT-qPCR. cDNAs were amplified with specific primers using a MyiQ real-time PCR detection system with iQ SYBR Green Supermix (Bio-Rad). The following genes were amplified in qRT-PCR assays with primer sets that were used in previous studies: gtfBCD (Koo et al. 2006), copY and sloA (Falsetta et al., 2012). Primers for amyA (5′-CCAAGCTGACAAGGAAGC and 5′-TGGTGTGGCTGTCATCATA), copA (5′-CTAGAACAGGCTCAAGCAGATT and 5′-CCGCAATCGTAATCAGTCCTAA) and copZ (5′-CGCTGACAATGTCACCAA and 5′-AGAGACCACTTGCTTGGA) were designed using Beacon Designer 2.0 software (Premier Biosoft International, Palo Alto, CA). Standard curves were used to determine the relative number of cDNA molecules, which were normalized to the relative number of 16S rRNA cDNA molecules in each sample, as previously described (Koo et al. 2006). These values were used to determine the fold of change between each treated sample and the vehicle control.
Glucosyltranferase (Gtf) activity assays
GtfB, GtfC and GtfD were prepared purified to near homogeneity by hydroxyapatite column chromatography as detailed by Venkitaraman et al. (1995) and Wunder and Bowen, (1999). Glucosyltransferase activity was measured by the incorporation of [14C] glucose from labeled sucrose (NEN Research Products, Boston, MA) into glucans (Venkitaraman et al., 1995). The enzymes were evaluated in solution and adsorbed to saliva-coated hydroxyapatite (sHA) surface. Purified GtfB, GtfC and GtfD (1.0–1.5 U), in solution or immobilized on sHA beads, was mixed with NV (800 μg/mL) or vehicle control (20% ethanol, v/v) and incubated with a [14C] glucose-sucrose substrate (0.2 μCi/ml; 100 mmol of sucrose per liter, final concentration) as described by Venkitaraman et al. (1995). The samples were incubated at 37°C for 2h. After incubation, radiolabeled glucan was analyzed by scintillation counting.
Animal Study
The animal experiment protocol was reviewed and approved by the Ethical Committee on Animal Research at the University of Campinas, SP, Brazil – UNICAMP (Protocol # 1485-1) and was performed according to methods described previously (Bowen et al. 1988; Koo et al. 1999). A total of 39 SPF female pups free of S. mutans and SDA virus from 11 litters of SPF Wistar rats were provided by CEMIB (UNICAMP). At age 21 days, the pups were weaned and infected three successive days with S. mutans UA159. Oral infection of the pups was confirmed at age 25 days by plating on Mitis Salivarius Agar plus bacitracin agar (MSB; Sigma-Aldrich, St. Louis, MO). The pups were then randomly placed into three groups of 13 animals. From this point, the molar teeth of the animals were treated topically by means of a camel hair brush twice daily, as follows: NV (800 μg/mL), 250 ppm fluoride, and vehicle control (20% ethanol, v/v). Fluoride at 225–250 ppm is commonly used in most mouth rinses (Zero, 2006) and has been used as a positive control in caries studies using rodent models (Koo et al., 2005, 2010b; Murata et al. 2010; Branco-de-Almeida et al. 2011; Falsetta et al., 2012). Each group of 13 animals was provided with diet 2000 and 5% sucrose water ad libitum (Bowen et al. 1988). The animals were weighed weekly and their behavior and physical appearance was noted daily. The experiment proceeded for 5 weeks. At the end, the animals were euthanized by CO2 asphyxiation. The lower left jaw was aseptically dissected, suspended in 5.0 mL sterile saline solution (0.9 %, w/v) and sonicated (three 10 s pulses at 5 s intervals, at 30 W; Vibracell, Sonics and Material Inc). The suspension was plated on MSB to estimate the S. mutans UA159 populations and on blood agar to determine the total cultivable microorganisms (Bowen et al. 1988). Smooth and sulcal caries and their severities were evaluated according to Larson’s modification of Keyes’ system (Larson 1981). The determination of the caries score was blinded by codification of the jaws and performed by one calibrated examiner.
Statistical Analysis
For in vitro studies, exploratory data analysis was performed to determine the most appropriate statistical test; the assumptions of equality of variances and normal distribution of errors were also checked. The data were then analyzed using ANOVA, and the F test was used to identify any difference between the groups. When significant differences were detected, a pairwise comparison was made between all the groups using Tukey-Kramer HSD method to adjust for multiple comparisons. For the animal study, an analysis of outcome measures was done with transformed values by LOG10 of the measures in order to stabilize variances as detailed in Raubertas et al. (1999). The data were then subjected to ANOVA in the Tukey-Kramer Honest Standard Deviation (HSD) test for all pairs. The statistical software BioEst version 5.0 was used to perform the analyses. The level of significance was set at 5%.
Results
Chemical analysis of bioactive fraction
Analysis revealed that the bioactive fraction was composed of primarily two compounds (Figure 1). Compound 1 was identified as 2′,4′-dihydroxy-7-methoxyisoflavone (neovestitol), corresponding to a 60.4% of relative percentage of NV, while compound 2 was identified as 2′,7-diidroxi-4′ metoxiisoflavan (vestitol), correlating with a 27.6 % of relative percentage of NV.
Figure 1.
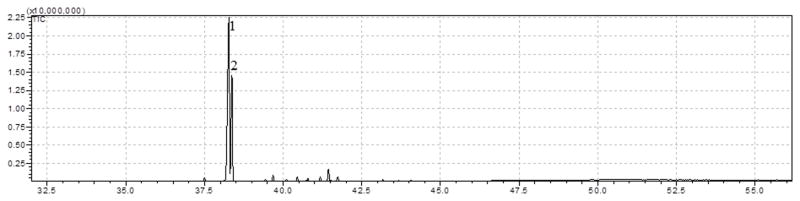
Chemical profile of NV through GC-MS. Compound number 1 was identified as Neovestitol (60.4 % of NV) and compound number 2 was identified as Vestitol (27.6 % of NV).
Biofilm analysis
A preliminary study determined the most effective concentration of NV (800 μg/ml) based on biochemical data of the treated biofilms (Supplemental Figure 3). Topical applications of NV (800 μg/ml) significantly reduced the dry-weight and, particularly, the total amount of soluble and insoluble extracellular polysaccharides (35–40% vs vehicle-control; P < 0.05). NV treatments did not reduce the total number of viable bacterial cells within treated-biofilms (vs. vehicle-control, P > 0.05).
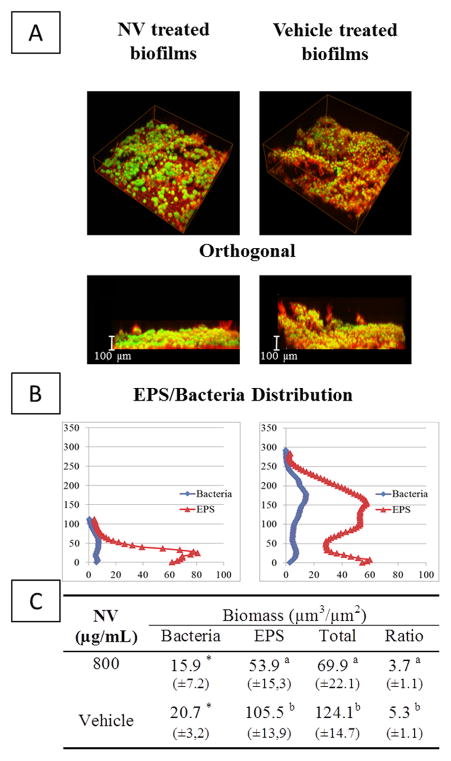
A significant reduction in the amount of exopolysaccharide prompted us to examine whether the assembly and 3D architecture of the biofilm was affected by NV treatment. Confocal microscopy and computational analyses of NV-treated biofilms revealed major changes in the EPS:bacteria ratio and the spatial organization of the bacterial cells and EPS when compared to vehicle-treated biofilms at 67 h (Figure 2). EPS biomass was markedly reduced (48.9%), while the bacterial biomass was less dramatically affected (23.1%), altogether resulting in a significant reduction in the EPS:bacteria ratio within biofilms (Figure 2). The NV-treated biofilms exhibited less EPS across the biofilm depth than those treated with vehicle-control (Figure 2), especially in the outer layers (above 40 μm). The biofilm changes at earlier developmental phase (43 h) were negligible (data not shown), indicating a greater effect of NV against further biofilm accumulation/maturation.
Figure 2.
Confocal imaging analysis of S. mutans biofilm following topical treatments with NV and vehicle-control. (A) Representative 3D rendered images of S. mutans biofilm post-treatments at 67 h. The cells are depicted in green (SYTO 9), while the EPS matrix is depicted in red (Alexa Fluor 647 dextran); (B) EPS/Bacteria distribution across the biofilm architecture (% of disc’s area covered by EPS/Bacteria versus biofilm height); (C) Quantitative analysis of EPS and bacteria biomass by COMSTAT. Statistical analyses was performed using ANOVA, with Tukey-Kramer test without any transformation or (*)ANOVA, with Kruskal-Wallis test
The data are congruent with biochemical assays (Suppl. Fig. 3), which indicate that brief exposure of the topically applied agent is capable of disrupting EPS accumulation without significant bactericidal activity. Thus, we examined whether NV affects the activity of glucosyltranferases (Gtfs) and/or the expression of their encoding genes.
Influences of NV on EPS synthesis by Gtfs and on gtfBCD expression
The effects of NV on the enzymatic activity of glucosyltransferases (Gtfs) in solution or adsorbed to saliva-coated hydroxyapatite (sHA) surfaces are shown in Figure 3. NV inhibited the activity of GtfB, GtfC, and GtfD in the solution phase by 60% (vs. vehicle-control, P<0.05). Surface-adsorbed GtfD was also significantly inhibited by NV (~45% inhibition vs. vehicle-control, P<0.05). However, it was less effective against surface-GtfB (18% inhibition) while GtfC was not significantly inhibited (P>0.05). Inhibitors are often less active (or without effect) when the Gtfs are adsorbed to a surface, likely due to conformational changes in the immobilized enzyme (Bowen and Koo, 2011). The exact reason why NV is more effective against surface adsorbed GtfD than other Gtfs is unclear, and warrants further investigation.
Figure 3.
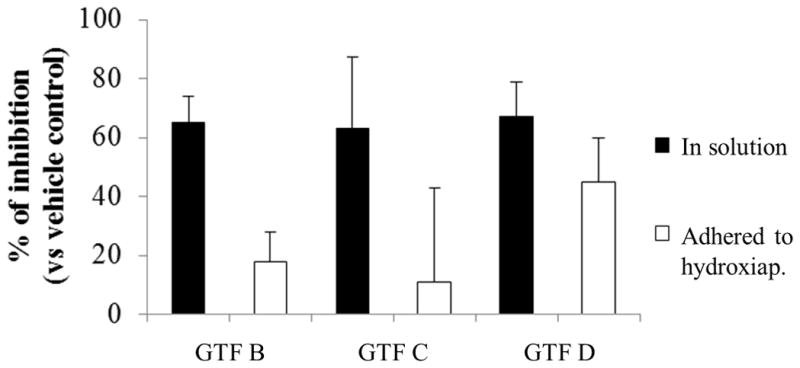
Glucosyltransferase activity assays. The percent of inhibition was calculated considering the vehicle-control as 100% Gtf activity. Data are expressed as means ± standard deviations of triplicates from at least two separate experiments. Values marked with an asterisk are significantly different from vehicle-control (P<0.05, Tukey’s test).
We also examined the impact of NV on gtfBCD expression by S. mutans within biofilms post treatment with NV. The gene expression profile was determined 1h after the topical treatment at 46 h of biofilm development. This time-point was selected based on our previous studies on the dynamics of the S. mutans transcriptome response during biofilm development and after the topical application of antibiofilm agents (Klein et al., 2010; Falsetta et al., 2012). The results of qRT-PCR assays are shown in Figure 4. Among the gtf genes, only gtfD expression was slightly reduced in NV-treated biofilms (vs. vehicle-control). It appears that one possible mechanism of action by which NV reduces EPS-matrix assembly and biofilm accumulation is primarily inhibiting GtfD activity (and at a lesser extent GtfB), while having minimal effects on gtfBCD gene expression. We investigated whether NV has additional antibiofilm properties by analyzing the global effect of NV on S. mutans transcriptome via microarrays.
Figure 4.
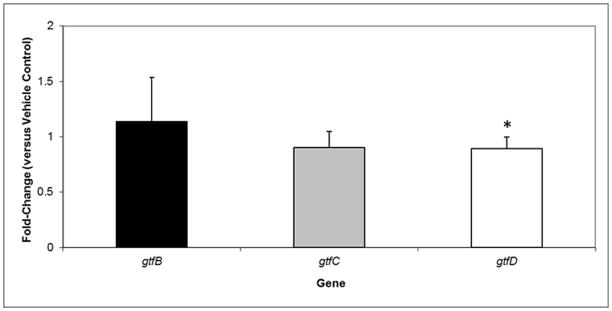
qRT-PCR analysis of gtfBCD expression by S. mutans within biofilms following topical treatments with NV and vehicle-control at 45 h time-point. These values were compared to those from vehicle-treated biofilms (corresponding to an arbitrary value of 1) to determine the change (n-fold) in gene expression. Data are expressed as means ± standard deviations of triplicates from at least three separate experiments. Values marked with an asterisk are significantly different from the value for the vehicle-treated biofilms (P<0.05, Tukey’s test).
Other putative biofilm disruption mechanisms
A total of 59 genes were differentially regulated in response to treatment (Figure 5). The majority of the genes identified were down-regulated compared to the vehicle control (52 genes), although a small number were induced in response to treatment (7 genes). A previously study developed a “Gene of Interest” (GOI) classification system (Falsetta et al., 2012) was used to categorize these genes into the following functional groups: EPS, glycolytic pathway, stress (oxidative), regulators, hypothetical, other (genes with functions that are not relevant to biofilm growth and/or infection) (Figure 5). In addition, the expression of genes related to IPS and biofilm formation/adhesion categories was unaffected by NV treatment.
Figure 5.
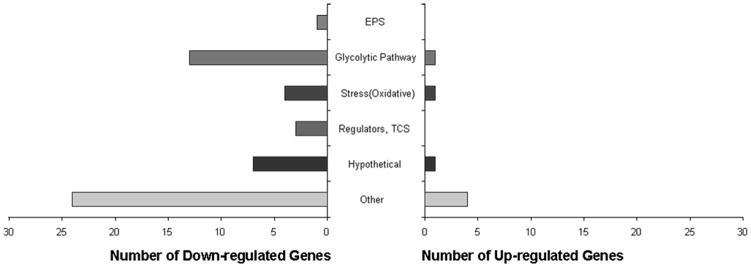
Microarray data, in which genes were organized in functional groups (EPS, glycolytic pathway, stress, regulators, hypothetical and other), from single species biofilm treated with NV and vehicle.
The greatest number of genes identified in this study belonged to the “Other” category (Figure 5) and are subsequently less attractive candidates for study, as they lack identified roles in biofilm formation and/or cariogenesis. However, this does not eliminate the possibility that they could have novel functions in biofilm formation that remain to be elucidated. The next most abundant and relevant categories were stress, specifically oxidative stress-responsive genes and glycolytic pathway genes. Several genes were selected for validation with RT-qPCR, including the following: copY, copA, copZ and sloA. All of these were significantly repressed in response to NV treatment in 46 h biofilms (Figure 6). It is apparent that NV treatments not only impair EPS synthesis but also affect the expression of specific traits associated with stress survival and bacterial fitness, which may affect the assembly of cariogenic biofilms in vivo.
Figure 6.
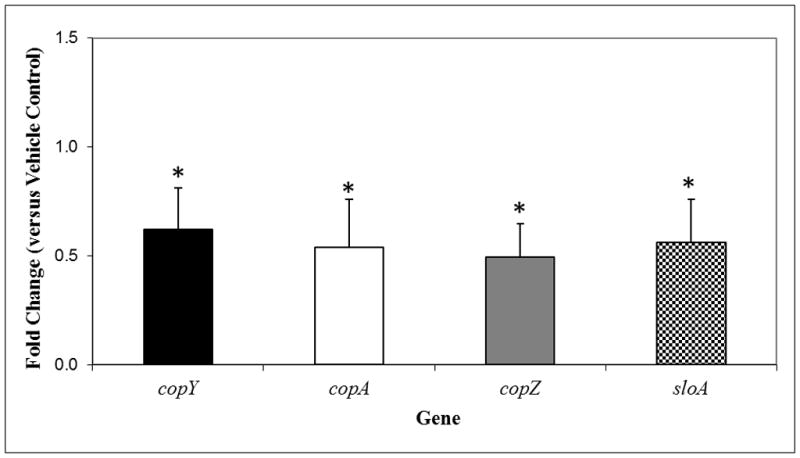
qRT-PCRanalysis (microarray validation) of copY, copA, copZ and sloA genes expression by S. mutans within biofilms following topical treatments with NV and vehicle-control. These values were compared to those from vehicle-treated biofilms (corresponding to an arbitrary value of 1) to determine the change (n-fold) in gene expression. Data are expressed as means ± standard deviations of triplicates from at least three separate experiments. Values marked with an asterisk are significantly different from the value for the vehicle-treated biofilms (P<0.05, Tukey’s test).
In vivo caries study
The animals were treated topically using a similar regimen as used in vitro. Fluoride treatment was included as a positive control because of its clinically proven anticaries effects. During the course of the animal study, the rats remained in good health, exhibited normal behavior, and gained weight as expected. The average weight gain for individual groups of rats (all the three groups) was not significantly different from one another (P > 0.05, data not shown).
Table 1 shows the total cultivable microflora and S. mutans population recovered from the plaque samples of infected animals, as determined by CFU counting. There was no significant difference in the total microbial and S. mutans populations among the treatment groups.
Table 1.
Influences of treatments on viable populations of S. mutans and total cultivable aerotolerant cells in plaque-biofilms in vivo using a rodent dental caries model. Statistical analysis was performed using an ANOVA, using Tukey—Kramer transformed with LOG10
| Treatments | Microorganisms | |
|---|---|---|
| Total | S. mutans | |
| NV | 0.5E+05 (±0.7E+05) | 0.2E+05 (±0.4E+05) |
| Fluoride | 0.7E+05 (±1.1E+05) | 0.2E+05 (±0.4E+05) |
| Vehicle | 3.6E+05 (±3.7E+05) | 2.9E+05 (±3.0E+05) |
The effects of each of the treatments on the incidence and severity of smooth and sulcal surface caries are shown in Table 2. NV treatments effectively reduced dental caries development in vivo, and results were comparable with those observed with fluoride (positive control). The animals treated with NV or fluoride displayed significantly less total smooth and sulcal surface carious lesions than those treated with the vehicle control (P<0.05). Furthermore, both NV and fluoride treatment significantly reduced the severity of smooth and sulcal surface caries when compared to the vehicle-control group in all levels of dentin damage: Ds (slight dentinal caries), Dm (moderate) and Dx (extensive).
Table 2.
Influences of treatments on development of dental caries in vivo (Keyes’ score).
| Treatments | Smooth-surface caries | Sulcal Caries | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| E | Ds | Dm | Dx# | E | Ds | Dm | Dx# | |
| NV | 38.8b (±7.9) | 29.9b (±8.4) | 7.3b (±5.3) | 0.8 (±0.6) | 40.5 (±4.4) | 29.8b (±5.1) | 10.0b (±6.0) | 2.3b (±2.7) |
| Fluoride | 38.6b (±8.8) | 29.5b (±8.0) | 6.9b (±5.5) | 1.6 (±2.5) | 40.7 (±4.2) | 30.3a,b (±5.5) | 8.0b (±4.1) | 1.3b (±1.9) |
| Vehicle | 49.0a (±12.9) | 41.1a (±14.2) | 14.1a (±8.6) | 3.2 (±3.2) | 44.6 (±5.5) | 35.8a (±6.1) | 18.4a (±7.3) | 6.8a (±5.4) |
E, enamel caries; Ds, slight dentinal caries; Dm, moderate dentinal caries; Dx, extensive dentinal caries. Values followed by the same letters are not significantly different from each other (P < 0.05). ANOVA, using Tukey—Kramer without any transformation and # ANOVA, using Tukey—Kramer transformed with LOG10
Discussion
Natural products are proven rich sources of bioactive compounds (Newman and Cragg, 2012, Butler 2005, Koehn and Carter 2005). However, further characterization of bioactivity and effectiveness of such agents is warranted to assess the true value of natural products in preventing oral diseases, especially in studies that use clinically relevant treatment regimens and in vivo models (Jeon et al., 2011). This work demonstrates the antibiofilm and anticaries properties of a highly purified neovestitol-vestitol containing fraction (NV) (Figure 1) isolated from Brazilian red propolis (Alencar et al. 2007; Righi et al. 2011; Piccinelli et al. 2011).
We used a combination of biochemical, confocal fluorescence imaging and gene expression analysis to characterize the biological actions of topical applications of NV on biofilm accumulation by S. mutans. This approach demonstrated that NV treatments reduce biofilm accumulation and the content of EPS across the biofilm architecture. We have identified at least two possible antibiofilm mechanisms: 1) inhibition of the enzymatic activity of GtfD (both in solution and surface phases), and 2) repression of specific genes associated with S. mutans stress survival/fitness. Topical applications of NV did not display bactericidal effects in vitro and in the animal’s plaque in vivo, despite showing antibacterial activity against planktonic cells (data not shown). This observation is not surprising given the recalcitrant nature of biofilms to antimicrobial agents (vs. planktonic phase) (Ceri et al., 1999; Jeon et al., 2011).
Inhibition of the activity of GtfD by NV has many implications for biofilm accumulation and expression of virulence. GtfD contributes to EPS rich-matrix development by producing soluble glucans which serve as a primer for insoluble EPS synthesis by GtfB, enhancing its enzymatic activity (Venkitaraman et al, 1995). GtfB activity is required for the assembly of an insoluble EPS-matrix within biofilms and for the production of EPS on bacterial surfaces, which ultimately modulates microcolony development (Koo et al., 2010; Xiao et al., 2012). It is apparent that there is positive cooperativity between GtfB and GtfD in the establishment of the matrix and biofilm formation (Ooshima et al, 2001). Furthermore, GtfD-derived glucans serves as a reserve polysaccharide that can be metabolized by S. mutans and other oral bacteria, enhancing the extent of acidification within the biofilm (Bowen and Koo, 2011).
Our observations may explain, at least in part, the reason that NV treatments affect the EPS/bacteria ratio and the EPS content in the outer layers of the biofilms despite weak GtfB or GtfC inhibition. These effects are relevant in the context of biofilm virulence, because S. mutans cells defective in assembling the EPS-rich matrix are compromised in their ability to create and maintain acidic microenvironments across the biofilm and at the surface of attachment, despite normal bacterial acidogenesis (Xiao et al., 2012). Thus, NV could be a promising interfering agent against optimal development of the EPS-matrix, which could attenuate both the accumulation and virulence of cariogenic biofilms.
Although NV treatment displayed negligible effects on gtfBCD expression, it significantly repressed genes associated with S. mutans stress survival and fitness, including sloA and copYAZ. Specifically, sloABCR encodes a manganese/iron transport system and sloA is required for virulence and oxidative stress tolerance by S. mutans (Paik et al., 2003; Rolerson et al., 2006). Furthermore, deletions in the sloR regulatory sequence that represses transcription of the operon induce S. mutans cariogenicity (Spatafora et al., 2001). The cop operon on the other hand is presumed to be involved in copper transport which is important for critical enzymatic and metabolic reactions (Vats and Lee, 2001). In particular, CopY may play a role in biofilm detachment by S. gordonii (Mitrakul et al., 2004); whether it modulates biofilm development by S. mutans needs further elucidation. The repression of these genes by NV treatments could reduce the ability of S. mutans to cope with environmental stress. Altogether, the biological effects of NV treatments against S. mutans biofilms could have direct implications in the pathogenesis of dental caries.
The in vivo data show that topical applications of NV impaired S. mutans ability to develop carious lesions in a rodent model of the disease. Both the incidence and severity of dental caries was significantly reduced by NV (vs. vehicle-control, P<0.05). Consistent with our in vitro data, NV treatments did not affect the number of S. mutans viable cells or total microbial populations in the plaque. Thus, it would appear that NV reduces caries onset by mediating the expression of S. mutans virulence.
Remarkably, NV treatment was as effective in vivo as fluoride, which is considered the gold standard in caries prevention (Clarkson, 2000). Fluoride may not adequately address the infectious elements of the disease despite subtle effects on glucan synthesis and on S. mutans acidogenesis/acid-tolerance (Jeon et al., 2011). Rather, it has a role in physico-chemical interference of caries development by reducing demineralization and enhancing remineralization of early lesions (Dawes and ten Cate, 1990). Thus, it is possible that NV could complement or even enhance fluoride effectiveness when used in combination. This is in line with overall enhancement of the cariostatic effects of fluoride found in previous studies involving other natural compounds (Koo et al. 2005, Falsetta et al., 2012). Hence, future studies should test NV or neovestitol (major compound of NV) in combination with fluoride.
Collectively, our data demonstrate that NV is a promising antibiofilm agent that could be useful for the development of alternative or adjunctive anticaries therapies. Additional studies are needed to further characterize the molecular mechanisms of action of NV as well as its toxicity/safety prior to clinical efficacy studies.
Supplementary Material
Acknowledgments
The authors are grateful to Mr. Alessandro Esteves for providing the Brazilian red propolis samples. This research was supported by FAPESP (#2008/58492-8), CNPq (CNPq 200174/2009-6) and NIH (NIDCR DE016139 and DE18023).
References
- Alencar SM, Oldoni TL, Castro ML, Cabral IS, Costa-Neto CM, Cury JA, Rosalen PL, Ikegaki M. Chemical composition and biological activity of a new type of Brazilian propolis: red propolis. J Ethnopharmacol. 2007;113(2):278–283. doi: 10.1016/j.jep.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Baelum V, van Palenstein Helderman W, Hugoson A, Yee R, Fejerskov O. A global perspective on changes in the burden of caries and periodontitis: implications for dentistry. J Oral Rehabil. 2007;34(12):872–906. doi: 10.1111/j.1365-2842.2007.01799.x. discussion 940. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45(1):69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Madison KM, Pearson SK. Influence of desalivation in rats on incidence of caries in intact cagemates. J Dent Res. 1988;67(10):1316–1318. doi: 10.1177/00220345880670101401. [DOI] [PubMed] [Google Scholar]
- Branco-de-Almeida LS, Murata RM, Franco EM, dos Santos MH, de Alencar SM, Koo H, Rosalen PL. Effects of 7-epiclusianone on Streptococcus mutans and caries development in rats. Planta Med. 2011;77(1):40–45. doi: 10.1055/s-0030-1250121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno-Silva B, Alencar SM, Koo H, Ikegaki M, Silva GV, Napimoga MH, Rosalen PL. Anti-inflammatory and antimicrobial evaluation of neovestitol and vestitol isolated from brazilian red propolis. J Agric Food Chem. 2013;61(19):4546–4550. doi: 10.1021/jf305468f. [DOI] [PubMed] [Google Scholar]
- Butler MS. Natural products to drugs: natural product derived compounds in clinical trials. Nat Prod Rep. 2005;22(2):162–195. doi: 10.1039/b402985m. [DOI] [PubMed] [Google Scholar]
- Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37(6):1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson JJ, McLoughlin J. Role of fluoride in oral health promotion. Int Dent J. 2000;50(3):119–128. doi: 10.1111/j.1875-595x.2000.tb00552.x. [DOI] [PubMed] [Google Scholar]
- Cury JA, Koo H. Extraction and purification of total RNA from Streptococcus mutans biofilms. Anal Biochem. 2007;365(2):208–214. doi: 10.1016/j.ab.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Dawes C, ten Cate JM. International symposium on fluorides: mechanisms of action and recommendations for use. J Dent Res. 1990;69: 505–836. [Google Scholar]
- Dye BA, Fisher MA, Yellowitz JA, Fryar CD, Vargas CM. Receipt of dental care, dental status and workforce in U.S. nursing homes: 1997 National Nursing Home Survey. Spec Care Dentist. 2007;27(5):177–186. doi: 10.1111/j.1754-4505.2007.tb00343.x. [DOI] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Lemos JA, Silva BB, Agidi S, Scott-Anne KK, Koo H. Novel antibiofilm chemotherapy targets exopolysaccharide synthesis and stress tolerance in Streptococcus mutans to modulate virulence expression in vivo. Antimicrob Agents Chemother. 2012;56(12):6201–6211. doi: 10.1128/AAC.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146 (Pt 10):2395–407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Jeon JG, Rosalen PL, Falsetta ML, Koo H. Natural products in caries research: current (limited) knowledge, challenges and future perspective. Caries Res. 2011;45(3):243–263. doi: 10.1159/000327250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MI, Duarte S, Xiao J, Mitra S, Foster TH, Koo H. Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilm development. Appl Environ Microbiol. 2009;75(3):837–841. doi: 10.1128/AEM.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MI, DeBaz L, Agidi S, Lee H, Xie G, Lin AH, Hamaker BR, Lemos JA, Koo H. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS ONE. 2010;5(10):e13478. doi: 10.1371/journal.pone.0013478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4(3):206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010;192(12):3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Rosalen PL, Cury JA, Park YK, Bowen WH. Effects of compounds found in propolis on Streptococcus mutans growth and on glucosyltransferase activity. Antimicrob Agents Chemother. 2002;46(5):1302–1309. doi: 10.1128/AAC.46.5.1302-1309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Rosalen PL, Cury JA, Park YK, Ikegaki M, Sattler A. Effect of Apis mellifera propolis from two Brazilian regions on caries development in desalivated rats. Caries Res. 1999;33(5):393–400. doi: 10.1159/000016539. [DOI] [PubMed] [Google Scholar]
- Koo H, Seils J, Abranches J, Burne RA, Bowen WH, Quivey RG., Jr Influence of apigenin on gtf gene expression in Streptococcus mutans UA159. Antimicrob Agents Chemother. 2006;50(2):542–546. doi: 10.1128/AAC.50.2.542-546.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Schobel B, Scott-Anne K, Watson G, Bowen WH, Cury JA, Rosalen PL, Park YK. Apigenin and tt-farnesol with fluoride effects on S. mutans biofilms and dental caries. J Dent Res. 2005;84(11):1016–1020. doi: 10.1177/154405910508401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Duarte S, Murata RM, Scott-Anne K, Gregoire S, Watson GE, Singh AP, Vorsa N. Influence of cranberry proanthocyanidins on formation of biofilms by Streptococcus mutans on saliva-coated apatitic surface and on dental caries development in vivo. Caries Res. 2010;44(2):116–126. doi: 10.1159/000296306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RM. Merits end modifications of scoring rat dental caries by Keyes’ method. In: Tanzer JM, editor. Microbiology Abstracts. Washington: IRL Press; 1981. pp. 195–203. Animal Models in Cariology, Special Supplement. [Google Scholar]
- Lemos JA, Burne RA. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology. 2008;154(Pt 11):3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD. Plaque as a biofilm: pharmacological principles of drug delivery and action in the sub- and supragingival environment. Oral Dis. 2003;9(Suppl 1):16–22. doi: 10.1034/j.1601-0825.9.s1.4.x. [DOI] [PubMed] [Google Scholar]
- Mitrakul K, Loo CY, Hughes CV, Ganeshkumar N. Role of a Streptococcus gordonii copper-transport operon, copYAZ, in biofilm detachment. Oral Microbiol Immunol. 2004;19(6):395–402. doi: 10.1111/j.1399-302x.2004.00176.x. [DOI] [PubMed] [Google Scholar]
- Murata RM, Branco-de-Almeida LS, Franco EM, Yatsuda R, dos Santos MH, de Alencar SM, Koo H, Rosalen PL. Inhibition of Streptococcus mutans biofilm accumulation and development of dental caries in vivo by 7-epiclusianone and fluoride. Biofouling. 2010;26(7):865–872. doi: 10.1080/08927014.2010.527435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooshima T, Matsumura M, Hoshino T, Kawabata S, Sobue S, Fujiwara T. Contributions of three glycosyltransferases to sucrose-dependent adherence of Streptococcus mutans. J Dent Res. 2001;80(7):1672–1677. doi: 10.1177/00220345010800071401. [DOI] [PubMed] [Google Scholar]
- Paes Leme AF, Koo H, Bellato CM, Bedi G, Cury JA. The role of sucrose in cariogenic dental biofilm formation--new insight. J Dent Res. 2006;85(10):878–887. doi: 10.1177/154405910608501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S, Brown A, Munro CL, Cornelissen CN, Kitten T. The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J Bacteriol. 2003;185(20):5967–5975. doi: 10.1128/JB.185.20.5967-5975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinelli AL, Lotti C, Campone L, Cuesta-Rubio O, Campo Fernandez M, Rastrelli L. Cuban and Brazilian red propolis: botanical origin and comparative analysis by high-performance liquid chromatography-photodiode array detection/electrospray ionization tandem mass spectrometry. J Agric Food Chem. 2011;59(12):6484–6491. doi: 10.1021/jf201280z. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Kuhnert WL, Hahn K. Adaptation of oral streptococci to low pH. Adv Microb Physiol. 2000;42:239–274. doi: 10.1016/s0065-2911(00)42004-7. [DOI] [PubMed] [Google Scholar]
- Raubertas RF, Davis BA, Bowen WH, Pearson SK, Watson GE. Litter effects on caries in rats and implications for experimental design. Caries Res. 1999;33(2):164–169. doi: 10.1159/000016511. [DOI] [PubMed] [Google Scholar]
- Righi AA, Alves TR, Negri G, Marques LM, Breyer H, Salatino A. Brazilian red propolis: unreported substances, antioxidant and antimicrobial activities. J Sci Food Agric. 2011;91(13):2363–2370. doi: 10.1002/jsfa.4468. [DOI] [PubMed] [Google Scholar]
- Rolerson E, Swick A, Newlon L, Palmer C, Pan Y, Keeshan B, Spatafora G. The SloR/Dlg metalloregulator modulates Streptococcus mutans virulence gene expression. J Bacteriol. 2006;188(14):5033–5044. doi: 10.1128/JB.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KM, Bowen WH. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect Immun. 1992;60(1):284–295. doi: 10.1128/iai.60.1.284-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva BB, Rosalen PL, Cury JA, Ikegaki M, Souza VC, Esteves A, Alencar SM. Chemical composition and botanical origin of red propolis, a new type of brazilian propolis. Evid Based Complement Alternat Med. 2008;5(3):313–316. doi: 10.1093/ecam/nem059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora G, Moore M, Landgren S, Stonehouse E, Michalek S. Expression of Streptococcus mutans fimA is iron-responsive and regulated by a DtxR homologue. Microbiology. 2001;147:1599–1610. doi: 10.1099/00221287-147-6-1599. [DOI] [PubMed] [Google Scholar]
- Vats N, Lee SF. Characterization of a copper-transport operon, copYAZ, from Streptococcus mutans. Microbiology. 2001;147(Pt 3):653–662. doi: 10.1099/00221287-147-3-653. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR, Vacca-Smith AM, Kopec LK, Bowen WH. Characterization of glucosyltransferaseB, GtfC, and GtfD in solution and on the surface of hydroxyapatite. J Dent Res. 1995;74(10):1695–1701. doi: 10.1177/00220345950740101101. [DOI] [PubMed] [Google Scholar]
- Wunder D, Bowen WH. Action of agents on glucosyltransferases from Streptococcus mutans in solution and adsorbed to experimental pellicle. Arch Oral Biol. 1999;44(3):203–214. doi: 10.1016/s0003-9969(98)00129-0. [DOI] [PubMed] [Google Scholar]
- Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR, 3rd, Heydorn A, Koo H. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8(4):e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.