Abstract
MicroRNAs (miRNAs) are master regulators of gene expression. By degrading or blocking translation of messenger RNA targets, these non-coding RNAs can modulate the expression of more than half the protein-coding genes in mammalian genomes. MiRNAs play important regulatory roles in a variety of cellular functions and in several diseases, including cancer. Aberrant miRNA expression has been well characterized in cancer, with implications for progression and prognosis. Recently, the discovery of miRNAs in body fluids, such as serum and plasma, opens up the possibility of using them as noninvasive biomarkers of disease and therapy response. In this chapter, we discuss the use of circulating miRNAs as biomarkers of disease and therapy response and as diagnostic and prognostic markers in breast cancer. We also discuss the main issues related to establishing circulating miRNAs as biomarkers in cancer.
1 Introduction
MicroRNAs (miRNAs) are short (~22-nucleotide) non-coding RNAs that play important roles in post-transcriptional gene silencing of target messenger RNAs (mRNAs) [1]. MiRNAs are involved virtually in all biologic processes, including cell proliferation and apoptosis, development, differentiation, metabolism, immunity, neuronal patterning, stress response, aging, and cell-cycle control [1–5]. MiRNAs are strongly conserved among distantly related invertebrates, vertebrates, and plants [6], and more than 1,400 have been identified in humans [7]. It has been estimated that more than 50% of protein-coding genes are regulated by miRNAs in mammalian genomes [8, 9]. MiRNAs negatively regulate gene expression through mRNA cleavage in cases of perfect complementarity to the 3′-UTR of the target mRNA or through translational repression in cases of partial complementarity [10–12]. However, the results of recent studies demonstrate that miRNAs can also target the 5′-UTR of a target mRNA, both open reading frames and promoter regions [13, 14]. Recent studies have demonstrated that the open reading frames of many repeat-rich genes contain strikingly large numbers of particular miRNA target sites [15, 16].
Because a single miRNA can target hundreds of mRNAs, aberrant miRNA expression is capable of disrupting the expression of several mRNAs and proteins and is involved in the initiation of many diseases, such as cancer [17]. The first evidence of miRNAs’ involvement in cancer was found in a study on miR-15a and miR-16a, which are located on chromosome region 13q14, a region that is deleted in more than half of all B-cell chronic lymphocytic leukemia patients [18, 19]. Since then, several studies have detected aberrations in miRNA expression in virtually all cancer types [20–25]. In breast cancer, Iorio et al. [26] identified 29 miRNAs differentially expressed in 76 breast tumor and 34 normal tissue specimens. In addition, miR-30 expression is associated with biopathologic features such as estrogen receptor (ER) and progesterone receptor (PR) expression, and miR-213 and miR-203 expression is related to tumor stage. Mattie et al. [27] identified unique sets of miRNAs associated with breast cancers currently defined by their HER2/neu or ER/PR status.
With the exception of leukemias, for which malignant cells are easily available, tissues for profiling solid cancers are obtained either by biopsy or surgery. Therefore, studies that demonstrate the diagnostic and prognostic usefulness of circulating miRNAs in body fluids, such as serum and plasma, are of high interest. MiRNAs have also been detected in other body fluids, such as tears, breast milk, bronchial lavage, colostrum, and seminal, amniotic, pleural, peritoneal, and cerebrospinal fluids [28]. Specific compositions and concentrations are found in each body fluid type analyzed. These findings might be useful if a correlation exists between specific miRNA levels in body fluids and various disease states.
In this chapter, we discuss detecting circulating miRNAs in serum and plasma and their applicability as diagnostic and prognostic markers in breast cancer.
2 Circulating MicroRNAs in Breast Cancer
The first study that measured miRNA levels in serum was conducted by Lawrie et al. [29], who found that sera levels of miR-21 were associated with relapse-free survival in patients with diffuse large B-cell lymphoma. Since then, several studies have assessed the potential use of serum or plasma miRNAs as biomarkers in different types of cancers, such as prostate cancer [30], lung cancer, colorectal cancer [31, 32], ovarian cancer [33], renal cell carcinoma [34], squamous cell carcinoma of the tongue [35], and glioblastoma [36].
In breast cancer, some studies have assessed the use of circulating miRNAs as biomarkers to differentiate normal from diseased states and monitor response to therapy. In one of the first studies, Zhu et al. [37] demonstrated that PR-positive tumors had higher miR-155 expression levels than did negative tumors in serum specimens from 21 women with and without breast cancer. In another study, Heneghan et al. [38] identified cancer-specific miRNAs that were significantly altered in the circulation of 148 breast cancer patients and that increased systemic miR-195 levels in breast cancer patients were reflected in breast tumors. Furthermore, the authors found that circulating levels of miR-195 and let-7a decreased in cancer patients after tumor resection and that specific circulating miRNAs were correlated with certain clinicopathologic variables, namely nodal status and ER status.
In one study, serum miRNA levels were highly correlated with breast tumor tissue types. miR-21, miR-106a, and miR-155 were significantly overexpressed in tumor specimens compared with in normal controls, whereas miR-126, miR-199a, and miR-335 were significantly underexpressed. Furthermore, the relative expression levels of miR-21, miR-126, miR-155, miR-199a, and miR-335 were closely associated with breast cancer histologic tumor grades and sex hormone receptor expression status [39]. Asaga et al. [40] demonstrated that circulating miR-21 concentrations could be used to distinguish breast cancer patients from healthy women and further distinguish patients with distant metastases from those with locoregional disease. Expression levels of circulating miR-10b, miR-34a, and miR-155 were used to discriminate 59 breast cancer patients from 29 healthy individuals [41]. Zhao et al. [42] identified 26 differentially expressed miRNAs in 20 breast cancer patients compared with 20 healthy donors. In this study, let-7c and miR-589 were significantly downregulated and upregulated in breast cancer patients, respectively. Using a deep sequencing technique, Wu et al. [43] found significantly higher miR-29a and miR-21 in the serum of breast cancer patients compared to controls. Another avenue of current research is the identification of miRNAs in circulating tumor cells (CTCs) in the peripheral blood. Sieuwerts et al. [44] identified 10 miRNAs that were more abundantly expressed in 32 patients with CTCs than in 9 patients with no detectable CTCs and healthy blood donors. Technical issues regarding CTC isolation must be addressed to establish CTCs’ relevance in clinical use, but they represent a promising approach because they are shed from the primary tumor or its metastases.
Some studies have identified differentially expressed circulating miRNAs in breast cancer patients and controls; few cases of overlapping expression have been reported. The main reason for these disparities is the lack of a standardized and robust method, with universal parameters, for detecting tumor-specific miRNA in body fluids. Nonetheless, studies of the circulating miRNAs associated with breast cancer have been limited to date. Therefore, more extensive studies are needed to establish circulating miRNA as noninvasive biomarkers and predictors of therapy response. Furthermore, not only technical aspects compromise the establishment of circulating miRNAs as biomarkers in breast cancer and also in other types of cancer; to date the source of circulating miRNAs is not clear. Therefore, more studies are necessary to clarify whether circulating miRNAs detected in body fluids are tumor-specific or the product of dead cells, either from tumor or healthy tissues.
In the following section, we discuss the main findings regarding the origin of circulating miRNAs, as well as recently published data that may explain the stability of circulating miRNAs in the bloodstream.
3 Stability and Origin of Circulating MicroRNAs
The potential of circulating miRNAs as cancer biomarkers relies mainly on their high stability and their capacity to reflect tumor status and predict therapy response. Many studies have systematically demonstrated that circulating miRNAs remain stable after being subjected to severe conditions that would normally degrade RNAs, such as boiling, low or high pH levels, extended storage, and ten freeze–thaw cycles [45, 46]. This remarkable stability is partly explained by miRNAs’ association with protein complexes and the presence of these small RNAs in circulating microvesicles called exosomes.
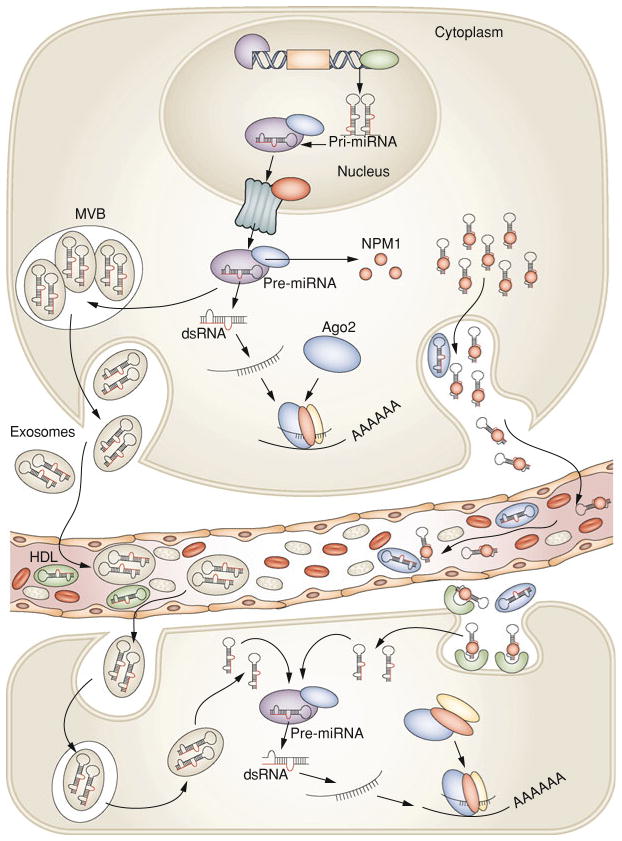
Recently, Arroyo et al. [47] found that most circulating miRNAs in plasma are cofractionated with Argonaute2 (Ago2), suggesting that circulating Ago2 complexes are the mechanisms responsible for plasma miRNA stability. Ago2 is part of an RNA-induced silencing complex and is the key effector protein of miRNA-mediated silencing. The results of Arroyo et al. study suggest that vesicle-associated versus Ago2 complex-associated miRNAs originate from different cell types and reflect cell type-specific miRNA expression or release mechanisms. However, only 10% of circulating miRNAs were vesicle associated in plasma. These findings were confirmed by Turchinovich et al. [48], who also demonstrated that extracellular miRNA is ultra filtrated with Ago2 and that most miRNA in plasma and cell culture remains in the supernatant after ultracentrifugation at 110,000 g, indicating a non-vesicular origin for extracellular miRNA. Furthermore, non-miRNA species such as U6 RNA, RNU24, RNU43, RNU44, RNU48, RNU6B, and mRNAs, which are not associated with Ago proteins, are absent in the extracellular environment or present in only low amounts. Other proteins may be associated with circulating miRNAs because some were present in the supernatant after Ago2 immunoprecipitation [47]. Recent findings demonstrate that the RNA-binding protein nucleophosmin 1 plays a role in the exportation, packaging, and protection of extracellular miRNAs [36]. Another mechanism that may involve the nSMase2 pathway was discovered, demonstrating that high-density lipoprotein transports circulating miRNAs and can alter gene expression by transferring miRNAs to recipient cells [49]. Nevertheless, miRNAs may also be involved in autocrine and paracrine miRNA signaling through exosomes. Of note, studies that have demonstrated that most circulating miRNAs are exosome-free only used samples from healthy donors or culture media [47, 48]. The two populations of circulating miRNAs (i.e., extracellular and exosomal) were not assessed or compared between healthy donor and cancer patients in these studies. Importantly, the results indicate that cancer patients have elevated levels of tumor-derived exosomes in plasma compared with those in healthy donors [50]. Furthermore, exosomes containing miRNAs were found not only in blood [51] but also in other body fluid types, such as saliva [52]. In Fig. 1, we depicted a hypothetical way in which circulating miRNAs are generated in the bloodstream. Nonetheless, further studies are necessary to explain the origin of these small RNAs in body fluids.
Fig. 1.
After being transcribed in the nucleus, pre-miRNA molecules can be processed further by dicer in the cytoplasm. In addition, based on recent findings [36, 47, 49] there are at least two ways that pre-miRNAs can be packaged and transported using exosomes and MVBs or other (not fully explored) pathways together with RNA-binding proteins. After fusion with the plasma membrane, MVBs release exosomes into the circulating compartments and bloodstream. Likewise, pre-miRNA inside the donor cell can be stably exported in conjunction with RNA-binding proteins, such as NPM1, and Ago2, or by HDL. Circulating miRNAs enter the bloodstream and are taken up by the recipient cells by endocytosis or, hypothetically, by binding to receptors present at the recipient cellular membrane capable of recognizing RNA-binding proteins. More studies are necessary to elucidate how miRNAs are loaded into exosomes and how they can be internalized by recipient cells. Exosomal miRNAs are processed by the same machinery used in miRNA biogenesis and thus have widespread consequences within the cell by inhibiting the expression of target protein-coding genes. MVBs multivesicular bodies, NPM1, nucleophosmin 1, Ago2 Argonaute2, HDL high-density lipoprotein. Figure modified with permission from Cortez etal. [58]
Interestingly, one group of researchers demonstrated the existence of tumor-derived exosomes [53] and an miRNA signature for circulating ovarian cancer exosomes [54]. This miRNA signature was significantly correlated with primary tumor miRNA expression in cancer patients compared with in benign disease patients and was not identified in normal controls. A similarity between miRNA signatures in circulating exosomal miRNAs and originating tumor cells was also found in lung adenocarcinoma [55], with a significant difference in exosomal miRNA levels between cancer patients and controls. Therefore, exosomes may be a newly discovered mechanism by which donor cells can communicate and influence the gene expression of recipient cells. More studies are needed to elucidate its importance in cancer progression [56]. Indeed, one study confirmed these findings and demonstrated that exosomes released by glioblastoma cells containing mRNA, miRNAs, and angiogenic proteins, such as epidermal growth factor receptor vIII, are taken up by normal recipient cells, such as brain micro-vascular endothelial cells [50]. Another study showed that exosomal miRNAs are associated with maintenance of dormant breast cancer cells in bone marrow stroma, which is related to recurrence and poor prognosis in breast cancer. In this study, Lim et al. [57] found that miRNAs play a role in breast cancer cell quiescence by demonstrating their passage through gap junctional intercellular communication and stroma-derived exosomes between breast cancer quiescent cells and bone marrow stroma.
Because Ago2/miRNA complexes are extremely stable and found in the cellular cytoplasm, some researchers have hypothesized that extracellular circulating miRNAs originate from dead cells [41, 48]. Indeed, miRNAs in body fluids can originate from apoptotic and necrotic cells of tumors and other sources, such as blood cells, the liver, the lungs, the kidneys, and other organs in which extensive contact between cells and the blood plasma occurs [48]. This hypothesis suggests that caution should be applied in using extracellular circulating miRNAs as biomarkers because cancer-specific miRNAs can be masked by circulating miRNAs from healthy tissues. It is necessary to clarify whether differential expression between tumors and normal tissues is affected solely by the tumor or by the affected organ or system. The real origin of circulating miRNAs, the mechanisms by which miRNAs are generated in the bloodstream, and the biologic effects of these molecules at distant sites are unknown and require further study.
4 Conclusions
Successful breast cancer treatment relies on early disease detection. Because aberrant miRNA expression is an early event in tumorigenesis, circulating miRNAs may represent noninvasive biomarkers in breast cancer. Nonetheless, studies in large populations are needed and some aspects of experimental reliability must be assessed before circulating miRNAs can be used as biomarkers. Likewise, given that most current approaches to cancer screening are invasive and unable to detect early stage disease, it is important to determine when tumor-related circulating miRNAs are detectable in the bloodstream during disease evolution. Moreover, important issues need to be addressed to establish circulating miRNAs as biomarkers for cancer.
First, larger prospective clinical trials are needed to validate these results because most published studies have small sample sizes and lack long-term outcome data. Second, because common upregulated miRNAs in body fluids are shared by several cancer types, especially those with common origins, further studies are necessary to establish a well-characterized panel of miRNAs specific to each tumor type. The use of known biomarkers as cancer antigens, along with miRNAs, can also increase cancer detection specificity and sensitivity. Third, more studies are necessary to determine which circulating miRNAs indicate early or advanced cancer stage, response to treatment, and patient outcome. Fourth, a robust method for tumor-specific miRNA detection in body fluids with universal parameters is needed. Finally, to use miRNAs as biomarkers in cancer, it is important to determine the source of tumor-specific miRNAs in body fluids and establish a signature capable of differentiating diseased from healthy states.
Acknowledgments
G.A.C. is supported as a fellow by The University of Texas MD Anderson Research Trust, as a University of Texas System Regents research scholar, and by the CLL Global Research Foundation. Work in Dr. Calin’s laboratory is supported in part by the National Institutes of Health; a Department of Defense Breast Cancer Idea Award; Developmental Research Awards in Breast Cancer; MD Anderson’s Ovarian Cancer, Brain Cancer, and Leukemia SPORE grants; a CTT/3I-TD grant; a 2009 Seena Magowitz—Pancreatic Cancer Action Network AACR Pilot Grant; and MD Anderson’s Cancer Center Support Grant CA016672. J.W.Welsh is supported in part by Paul Calabreski K-12 grant. We thank Ann Sutton for her help in editing this manuscript.
Contributor Information
Maria Angelica Cortez, Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, 1881 East Road, Houston, TX 77054, USA. Department of Experimental Radiation Oncology, Unit 66, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
James William Welsh, Department of Experimental Radiation Oncology, Unit 97, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Boulevard, Houston, TX 77030, USA.
George Adrian Calin, Email: gcalin@mdanderson.org, Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, 1881 East Road, Houston, TX 77054, USA.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124(5):877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 5.Kato M, Slack FJ. MicroRNAs: small molecules with big roles—C. elegans to human cancer. Biol Cell. 2008;100(2):71–81. doi: 10.1042/BC20070078. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–676. doi: 10.1016/s0092-8674(03)00428-8. Erratum in: Cell. 2003 Jul 25;114(2):269. [DOI] [PubMed] [Google Scholar]
- 7.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Friedman JM, Jones PA. MicroRNAs: critical mediators of differentiation, development and disease. Swiss Med Wkly. 2009;139(33–34):466–472. doi: 10.4414/smw.2009.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. Erratum in: Nat Rev Genet. 2004;5(8):631. [DOI] [PubMed] [Google Scholar]
- 11.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- 13.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnall-Levin M, Rissland OS, Johnston WK, Perrimon N, Bartel DP, Berger B. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res. 2011;21(9):1395–1403. doi: 10.1101/gr.121210.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chin LJ, Slack FJ. A truth serum for cancer-microRNAs have major potential as cancer biomarkers. Cell Res. 2008;18(10):983–984. doi: 10.1038/cr.2008.290. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 21.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 22.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 25.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 27.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, Ohyashiki K, Kuroda M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4(5):e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 32.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 33.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112(1):55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 34.Feng G, Li G, Gentil-Perret A, Tostain J, Genin C. Elevated serum-circulating RNA in patients with conventional renal cell cancer. Anticancer Res. 2008;28(1A):321–326. [PubMed] [Google Scholar]
- 35.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14(9):2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 36.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251(3):499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 39.Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119(3):586–593. doi: 10.1016/j.ygyno.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 40.Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 41.Roth C, Rack B, Müller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12(6):R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5(10):e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Q, Lu Z, Li H, Lu J, Guo L, Ge Q. Next-generation sequencing of microRNAs for breast cancer detection. J Biomed Biotechnol. 2011;2011:597145. doi: 10.1155/2011/597145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sieuwerts AM, Mostert B, Bolt-de Vries J, Peeters D, de Jongh FE, Stouthard JM, Dirix LY, van Dam PA, Van Galen A, de Weerd V, Kraan J, van der Spoel P, Ramírez-Moreno R, van Deurzen CH, Smid M, Yu JX, Jiang J, Wang Y, Gratama JW, Sleijfer S, Foekens JA, Martens JW. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin Cancer Res. 2011;17(11):3600–3618. doi: 10.1158/1078-0432.CCR-11-0255. [DOI] [PubMed] [Google Scholar]
- 45.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 46.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabrò E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108(9):3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. Erratum in: Gynecol Oncol. 2010 Jan;116(1):153. [DOI] [PubMed] [Google Scholar]
- 51.Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 53.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2010;3(11):e3694. doi: 10.1371/annotation/b15ca816-7b62-4474-a568-6b60b8959742. Erratum in: PLoS One. 2010;5(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16(1):34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor DD, Homesley HD, Doellgast GJ. Binding of specific peroxidase-labeled antibody to placental-type phosphatase on tumor-derived membrane fragments. Cancer Res. 1980;40(11):4064–4069. [PubMed] [Google Scholar]
- 56.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA, Greco SJ, Bryan M, Patel PS, Rameshwar P. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 2011;71(5):1550–1560. doi: 10.1158/0008-5472.CAN-10-2372. [DOI] [PubMed] [Google Scholar]
- 58.Cortez MA, et al. Nat Rev Clin Oncol. 2011;8(8):467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]