Abstract
Development of improved RNA interference based strategies is of utmost clinical importance. While siRNA-mediated silencing of EphA2, an ovarian cancer oncogene, results in reduction of tumor growth, we present evidence that additional inhibition of EphA2 by a microRNA further ‘boosts’ its anti-tumor effects. We identified miR-520d-3p as a tumor suppressor upstream of EphA2, whose expression correlated with favorable outcomes in two independent patient cohorts comprising of 647 patients. Restoration of miR-520d-3p prominently decreased EphA2 protein levels, and suppressed tumor growth and migration/invasion both in vitro and in vivo. Dual inhibition of EphA2 in vivo using DOPC nano-liposomes loaded with miR-520d-3p and EphA2-siRNA showed synergistic anti-tumor efficiency and greater therapeutic efficacy than either monotherapy alone. This synergy is atleast in part due to miR-520d-3p targeting EphB2, another Eph receptor. Our data emphasize the feasibility of combined miRNA-siRNA therapy, and will have broad implications for innovative gene silencing therapies for cancer and other diseases.
Keywords: miR-520d-3p, EphA2, EphB2, ovarian cancer, RNA interference
INTRODUCTION
RNA interference (RNAi)-based therapeutics such as siRNA therapy is a novel approach that is currently under investigation to improve clinical trials and patient care for different cancers (1). However, early clinical trials testing siRNAs for cancer management have resulted in modest response and are yet to deliver on the full potential of this technology (2). One chief clinical concern with any targeted or siRNA-based therapy is their single-gene-management approach, which confines them to the ‘one-drug-one-target’ paradigm and renders them susceptible to resistance in due course. On the contrary, by the virtue of their ability to simultaneously target multiple protein-coding genes, microRNAs (miRNAs) have emerged to be promising novel intervention tools for cancer management (3). These small regulatory non-coding RNAs show widespread deregulation in many human cancers, and are thus associated with tumorigenesis and progression (4).
Epithelial ovarian cancer (OC) remains the most lethal form of gynecological malignancies, with the 5-year survival rate for patients less than 44% (5). In recent years, EphA2 has emerged as an important target for OC therapy (6). EphA2, a member of the Eph-receptor family, is a receptor tyrosine kinase that has been shown to be oncogenic in several human malignancies including ovarian (6, 7), breast (8), colorectal (9, 10), glioblastoma (11), pancreatic (12), esophageal (13), lung (14), melanoma (15), and prostate cancers (16) and promotes proliferation, migration, invasion and metastasis (17-19). EphA2 is overexpressed in >75% of OC patients, and its expression has been linked to increased tumor growth and angiogenesis and poor clinical outcome (7, 20, 21). Consistent with these findings, it was shown that delivery of EphA2 siRNA to OC tumors using neutral nano-liposomes potently inhibits EphA2 expression and suppresses tumor growth and prolongs survival in orthotopic mouse models of OC, emphasizing that EphA2 is a strong oncogenic target for OC therapy (22-24). However, given the incomplete target silencing with siRNA alone, we questioned whether addition of a miRNA that targets the same pathway would further enhance the efficacy of EphA2 inhibition and tumor suppression.
Based on this, we hypothesized that combination of miRNA and siRNA-based treatment could afford improved dual inhibition of a target protein as well as concurrent modulation of other oncogenic members of the same pathway. In this study, we test the hypothesis that EphA2 siRNA will present greater anti-tumor potency when combined with a specific miRNA targeting the Eph pathway. For this purpose, we identified a clinically relevant miRNA, miR-520d-3p that is an independent prognostic marker for epithelial OC patients using The Cancer Genome Atlas (TCGA) and MD Anderson Cancer Center (MDACC) datasets. We demonstrate that dual targeting of EphA2 using siRNA-EphA2 and miR-520d-3p exhibits synergistic inhibition of EphA2 and significantly augments tumor regression compared to either monotherapy alone. These findings provide proof-of-principle for the clinical application of a previously unrecognized approach combining miRNA and siRNA therapy for targeting a common oncogenic pathway in OC.
RESULTS
miR-520d-3p is an independent prognostic factor in serous ovarian cancer
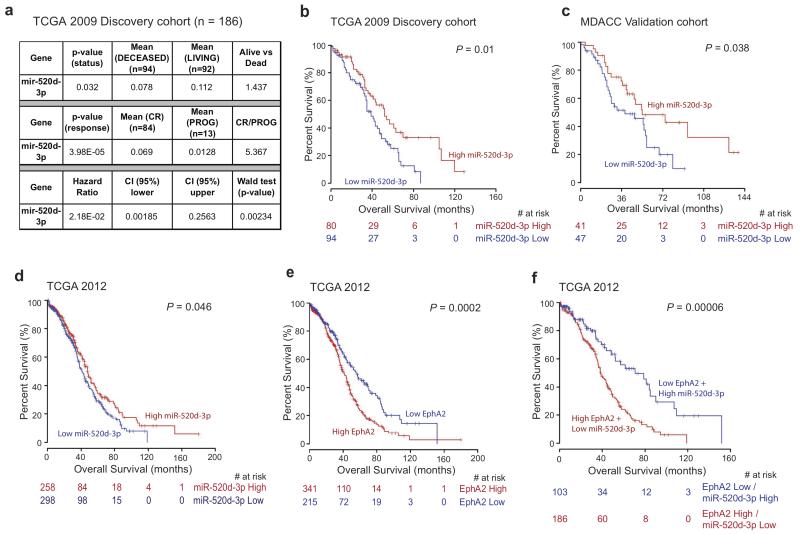
To detect novel miRNAs associated with clinical outcome, we utilized the data available at the beginning of our study from the 2009 TCGA (25) dataset for OC, comprising of 186 patients whose survival status was available (recorded as living, n = 92 or deceased, n = 94). Response to therapy was known for 118 of them, and was recorded as complete response (CR = 84), partial response (PR = 19), progressive disease (PROG = 13), or stable disease (SD = 2). Using analysis of variance (ANOVA), we identified 80 miRNAs that were significantly associated with longer overall survival (OS) (when comparing alive vs. deceased) and 75 miRNAs that correlated with good response to therapy (when comparing CR vs. PROG). A total of 14 miRNAs were found to be common between the two lists (Supp. Table 1). We also performed additional univariate Cox regression analysis on the discovery cohort with miRNA expression levels as continuous variables (data not shown). Next, we used multiple miRNA target prediction programs (RNA22, TargetScan, miRanda, microT and PicTar) to determine if any of these 14 miRNAs were predicted to target EphA2, an important oncogenic target in ovarian cancer. Interestingly, we identified miR-520d-3p (also called miR-520d) to be predicted to target EphA2, as well as statistically correlate with better survival and prognosis in OC patients (Fig. 1a, univariate and multivariate analysis in Table 1). High miR-520d-3p had a hazard ratio of 0.0218 (95% Confidence Interval = 0.00185-0.2563, Wald test P = 0.000234) (Fig. 1a). Subjects with high miR-520d-3p expression (cut-off = 0.54) had a significantly longer survival time (median 52 months) compared to patients with low miR-520d-3p expression (median 39 months) (P = 0.01; Fig. 1b). Instead, miR-520d-5p (also called miR-520d*), which is produced from the same precursor miRNA (pre-miR) and is considerably less expressed in OC cell lines (Supp. Fig. 1), does not correlate with any of these clinical parameters and is also not predicted to target EphA2 (data not shown); thus further supporting the biological exclusivity and relevance of miR-520d-3p::EphA2 interaction.
Figure 1. miR-520d-3p is an independent positive prognostic factor in OC.
(a) Analysis of variance (ANOVA) statistics identifying miR-520d-3p to be important predictor of overall survival (alive vs. deceased) and response to therapy (complete response vs. progressive disease), and cox proportional hazard model showing hazard ratio of miR-520d-3p using the 2009 TCGA database (n = 186). (b, c) Kaplan-Meier curves representing the percent overall survival in patients with OC based on miR-520d-3p median expression levels in TCGA 2009 database (n = 186) (b) and in MDACC cohort (n = 91) (c). (d, e, f) Kaplan-Meier curves representing the percent overall survival of 556 OC patients from TCGA 2012 dataset based on miR-520d-3p median expression alone (d) or EphA2 median expression alone (e) or after combined EphA2 and miR-520d-3p expression levels (f). The patients were grouped into percentiles according to median mRNA/miRNA expression. The Log-rank test was employed to determine the significance between mRNA/miRNA expression and overall survival. The colored numbers (red or blue) below the curves represent patients at risk at the specified time points.
Table 1. Univariate and multivariate analysis of overall survival and progression free survival results of 556 patients from TCGA dataset (a) and 91 OC patients from MDACC (b, c) - Data Portal (https://tcga-data.nci.nih.gov/tcga).
| a. OS (n = 556) | |||||
|---|---|---|---|---|---|
| Variable | Univariate analysis | Multivariate analysis | |||
| HR (95%CI) | P value | HR (95%CI) | P value | ||
| EphA2 | Low vs. High | 0.62 (0.48-0.8) | 0.0002 | 0.78 (0.7-1.01) | 0.0575 |
| EphB2 | Low vs. High | 0.69 (0.53-0.9) | 0.0051 | 0.93 (0.73-1.2) | 0.5972 |
| miR-520d-3p | Low vs. High | 1.29 (1.02-1.63) | 0.03 | 1.32 (1.03-1.69) | 0.0297 |
| DICER1 | Low vs. High | 1.43 (1.11-1.82) | 0.0032 | 0.94 (0.73-1.21) | 0.6271 |
|
Cytoreduction
operation |
(Residual disease vs. no residual disease) |
0.94 (0.62-1.43) | 0.77 | 0.92 (0.6-1.41) | 0.6935 |
| b. OS (n = 91) | |||||
| MiR-520d-3p | Low vs. High | 1.873 (1.026-3.420) | 0.041 | 3.168 (1.654- 6.066) |
0.0005 |
| Recurrence | Positive vs. Negative |
29.967(1.587-565.723) | 0.023 | ||
| Dicer_di | Low vs. High | 2.518(1.396-4.542) | 0.002 | 2.551(1.306-4.984) | 0.0061 |
| Drosha_di | Low vs. High | 2.594(1.431-4.705) | 0.002 | 2.471(1.220-5.003) | 0.0119 |
| c. PFS (n = 91) | |||||
| MiR-520d-3p | Low vs. High | 2.183 (1.208-3.944) | 0.01 | 2.871 (1.563- 5.275) |
0.0006 |
|
Cytoreduction
operation |
Suboptimal vs. optimal |
1.638 (0.864-3.102) | 0.007 | ||
| Node metastasis | Positive vs. Negative |
2.458 (1.091-50658) | 0.03 | ||
| Dicer_di | Low vs. High | 2.396 (1.319-4.355) | 0.004 | 3.224 (1.7444- 5.959) |
0.00018 |
| Drosha_di | Low vs. High | 2.244 (1.241-4.060) | 0.008 | ||
To validate our findings, we confirmed miR-520d-3p as a favorable prognostic factor in an independent cohort of OC samples collected from MDACC (n = 91; Clinical data in Supp. Table 2). The patients with high miR-520d-3p expression correlated with longer survival time (median 54 months) as compared to those with low miR-520d-3p levels (median 38 months) (P = 0.038; Fig. 1c, Table 1). High miR-520d-3p expression was also a favorable predictor of progression-free survival (PFS) in these patient samples (P = 0.0016; Supp. Fig. 2, Table 1). As expected, miR-520d-5p was not found to correlate with either OS or PFS in this dataset (data not shown). miR-520d-3p was also confirmed to be prognostic for OC patients in the updated 2012 TCGA dataset (n = 556, including the 186 patients initially analyzed, recorded as living, n = 265 or deceased, n = 291, P = 0.046; Fig. 1d). These findings suggest that miR-520d-3p is a favorable prognostic factor for OC independent of other clinicopathological parameters.
We further sought to determine whether combined expression of miR-520d-3p and EphA2 would serve as a better prognostic set for outcome of OC patients. In agreement with previous reports, EphA2 is differentially expressed in high-grade OC (cut-off = 0.386, P = 0.0014; data not shown) and high EphA2 expression levels correlated with shorter overall survival (median survival of 41 months compared to 56.5 months in patients with low expression, P = 0.0002; Fig. 1e). However, combined expression of EphA2 and miR-520d-3p significantly improved the separation curves, and patients showing EphA2(high)/miR-520d-3p(low) had significantly shorter survival (median 38.2 months) compared to those with EphA2(low)/miR-520d-3p(high) (median 70.8 months) (P = 0.00006; Fig. 1f). These findings further validate the importance of miR-520d-3p in OC, which led us to investigate its specific cellular and biological functions and its association with EphA2.
EphA2 is a direct functional target of miR-520d-3p
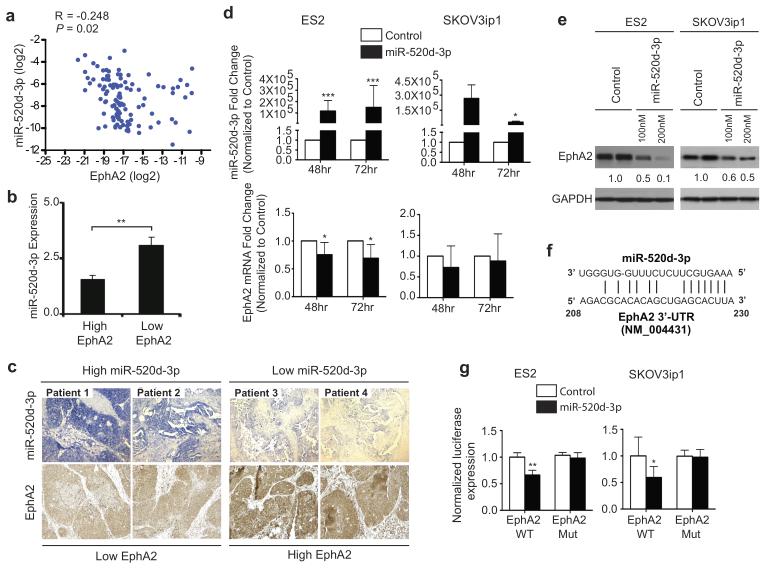
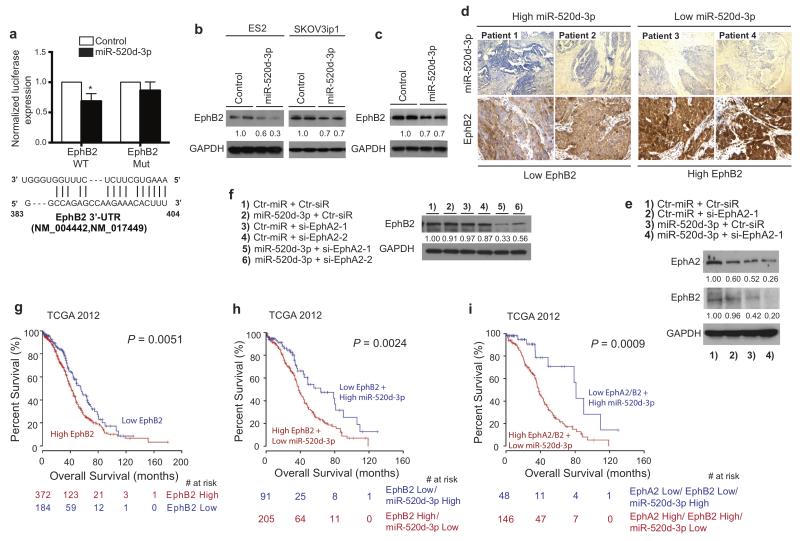
To determine whether EphA2 is indeed a direct target of miR-520d-3p, we first examined the correlation between miR-520d-3p and EphA2 mRNA expression in the 91 MDACC OC patient dataset. We found statistically significant inverse correlation between miR-520d-3p and EphA2 expression in these patient samples, but at a low strength (Spearman correlation coefficient (R) < −0.248; P = 0.02; Fig. 2a). To further analyze this relationship, we immunostained ovarian cancer patient samples for miR-520d-3p and EphA2. Immunostaining confirmed that tumors with high miR-520d-3p expression showed weak EphA2 staining, while tumors with low miR-520d-3p expression showed strong EphA2 staining (Fig. 2b and 2c).
Figure 2. EphA2 is a direct and functional target of miR-520d-3p.
(a) Scatter plot showing negative correlation between EphA2 mRNA (normalized to 18S) and miR-520d-3p (normalized to U6) in MDACC patient set using Spearman’s correlation analysis (R < −0.248; P = 0.02). (b) Quantification of EphA2 and miR-520d-3p immunostaining from four patients showing negative correlation in OC tumors. (c) Representative images of the immunostaining in panel (b). (d) qRT-PCR analysis showing transient overexpression of miR-520d-3p in ES2 and SKOV3ip1 cells (upper panel) results in downregulation of EphA2 mRNA after 48 and 72 hours (lower panel). (e) Immunoblotting of EphA2 and GAPDH in ES2 and SKOV3ip1 cells transfected with miR-520d-3p (100nM or 200nM) or a scrambled control. (f) Representative diagram of the conserved binding site of miR-520d-3p in the 3’-UTR of EphA2 mRNA. (g) Luciferase activity of a reporter construct fused to wild-type or mutant EphA2 3′-UTR in ES2 and SKOV3ip1 cells with ectopic miR-520d-3p expression. Control, cells transfected with a scrambled miRNA control. Data are average of three independent experiments. Statistical significance was determined by unpaired, two-tailed Student’s t test. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 Data are mean ± s.d.
To further study the relationship between miR-520d-3p and EphA2, we ectopically expressed miR-520d-3p in ES2 and SKOV3ip1, two cell lines with low miR-520d-3p expression. Compared to the negative control, transient miR-520d-3p expression reduced EphA2 mRNA levels in ES2 cells (Fig. 2d), while EphA2 protein levels were decreased in both cell lines (Fig. 2e). Conversely, miR-520d-5p transfection did not influence EphA2 protein levels in SKOV3ip1 cells (Supp. Fig. 3). Using multiple target prediction programs, we identified a conserved miR-520d-3p binding site in the 3′-untranslated region (3′-UTR) of EphA2 mRNA (Fig. 2f). In both ES2 and SKOV3ip1 cells, ectopic expression of miR-520d-3p significantly reduced the activity of a luciferase reporter fused to the wild-type EphA2 3′-UTR. Deletion mutations in the miR-520d-3p interacting seed region rescued the luciferase activity, thus confirming a direct interaction between EphA2 3′-UTR and miR-520d-3p (Fig. 2g). Taken together, these data suggest that EphA2 is a direct functional target of miR-520d-3p and its expression is regulated by miR-520d-3p in OC.
miR-520d-3p expression inhibits migration, invasion and tumor growth
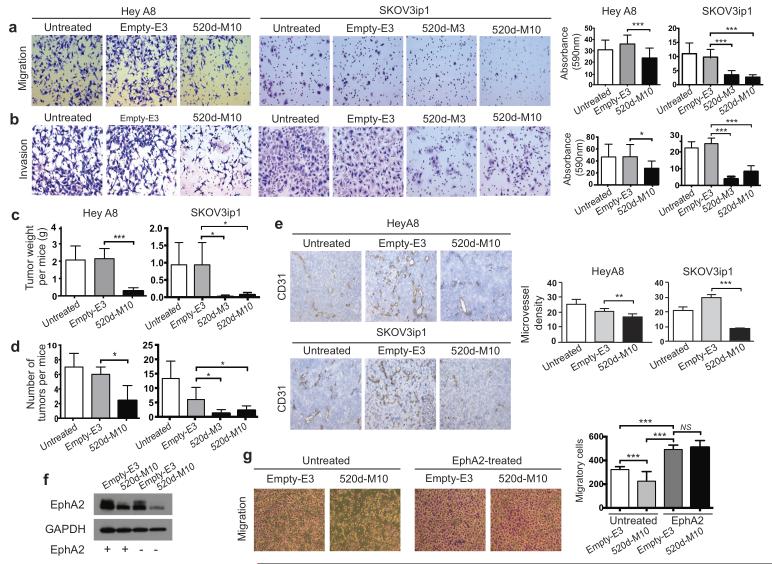
To study the in vitro and in vivo function of miR-520d-3p, we stably overexpressed miR-520d-3p in SKOV3ip1 and HeyA8 (chosen since HeyA8 has been better characterized in vivo in OC as compared to ES2) cells. We observed a marked reduction in EphA2 protein levels after miR-520d-3p overexpression (data not shown). Based on the miR-520d-3p and EphA2 expression levels, multiple clones from both cell line models were selected for performing further functional studies (Supp. Fig. 4). Ectopic expression of miR-520d-3p, both transiently and stably, in SKOV3ip1 cells significantly decreased cell proliferation (Supp. Fig. 5). To understand the role of miR-520d-3p in tumor progression, we studied the in vitro migratory and invasive capacity of miR-520d-3p-overexpressing clones. Tumor cell migration was significantly reduced in miR-520d-3p overexpressing stable clones (HeyA8-520d-M10 – P < 0.001, SKOV3ip1-520d-M3 – P < 0.001 and SKOV3ip1-520d-M10 – P < 0.001) when compared to empty controls (HeyA8-E3 and SKOV3ip1-E3 respectively) (Fig. 3a). Similarly, cell invasion decreased significantly in HeyA8-520d-M10 (P < 0.05), and in SKOV3-520d-M3 (P < 0.001) and SKOV3-520d-M10 (P < 0.001) when compared to empty clones (Fig. 3b). Thus, restoration of miR-520d-3p was able to substantially reduce cell migration and invasion in OC models.
Figure 3. miR-520d-3p expression inhibits migration, invasion and tumor growth.
(a, b) Representative images (at 100×) showing effect of miR-520d-3p stable overexpression on migration (a) and invasion (b) of HeyA8 and SKOV3ip1 cells using Transwell migration and Matrigel invasion assays (Left panel). Absorbance was measured at 590nm after 24 hours. The data from one representative experiment are shown in right panel. Experiment was performed in triplicates at three independent times. (c, d) Total tumor weight (c) and number of metastatic tumor nodules (d) in mice (n = 10 per group) with intra peritoneal injection of miR-520d-3p-transduced or control-transduced or parental untreated HeyA8 (33 days) or SKOV3ip1 (46 days) cells after implantation. (e) Representative images of CD31 staining (at 100×) to identify endothelial cells in untreated, control miR-transfected and miR-520d-3p-transfected HeyA8 and SKOV3ip1 tumors. Quantification of CD31 staining is shown in right panel. A lumen with positive CD31 staining was counted as a single microvessel. Data are average of three independent experiments. (f) Immunoblotting for EphA2 and GAPDH in control or EphA2-transfected HeyA8 empty-E3 or miR-520d-3p-overexpressing-M10 clones. (g) Representative images (at 40×) showing migration of untreated control or EphA2-overexpressing HeyA8 empty-E3 or miR-520d-3p-overexpressing-M10 clones. Quantification of migratory cells counted is shown in right panel. Experiment was repeated in duplicates at three independent times. Statistical significance was determined by unpaired, two-tailed Student’s t test when compared to empty clones for all analyses. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 Data are mean ± s.d.
We next injected the empty and miR-520d-3p overexpressing clones of both cell lines into the peritoneal cavity of a murine orthotopic metastasis model (1×106cells/mouse, n = 10 per group) that mimics the pattern of tumor spread in patients with advanced OC (26-28). Mice were sacrificed and necropsied after 33 days in HeyA8 clones and 46 days in SKOV3ip1 clones. Increased expression of miR-520d-3p was confirmed by qRT-PCR in five mice per group (Supp. Fig. 6), and a corresponding decrease in EphA2 protein levels was observed in the same mice (Supp. Fig. 6). In both models, mice bearing miR-520d-3p-overexpressing tumors had a significant reduction in aggregate intraperitoneal metastatic burden (HeyA8-520d-M10 – P < 0.001, and SKOV3ip1-520d-M3 – P < 0.05 and 520d-M10 – P < 0.05) as compared with empty vectors (Fig. 3c). This also corresponded with a significant decrease in intraperitoneal tumor nodules in these mice (HeyA8-520d-M10 – P < 0.05 compared to E3, and SKOV3ip1-520d-M3 – P < 0.05 and 520d-M10 – P < 0.05 compared to E3 respectively) (Fig. 3d). Since increased EphA2 expression has been associated with enhanced angiogenesis (20), we immunostained tissues from empty and miR-overexpressing tumors for CD31, a marker of endothelial cells (20). As expected, microvessel density (MVD) decreased significantly in miR-520d-3p tumors when compared to empty tumors (HeyA8-520d-M10 – P < 0.01, and SKOV3ip1-520d-M10 – P < 0.001) (Fig. 3e). Thus, restoration of miR-520d-3p effectively inhibits cell migration, invasion and angiogenesis both in vitro and in vivo in OC.
To assess if this function of miR-520d-3p is mediated by EphA2 activity, we asked if re-introduction of miR-520d-3p-resistant-EphA2 could rescue the tumorigenic phenotype of OC cell lines. Empty and miR-520d-3p overexpressing HeyA8 stable clones (E3 and 520d-M10) were transiently transfected with EphA2-ORF plasmid, which lacks the 3′-UTR and hence miR-520d-3p binding sites. Immunoblot assays confirmed the overexpression of EphA2 in these cells (Fig. 3f). EphA2-transfected cells showed significant increase in migration compared to untransfected controls (Fig 3g). Further, EphA2-transfected 520d-M10 stable cells showed no change in their migratory ability compared to EphA2-transfected Empty-E3 controls (Fig 3g). These results suggest that activity of miR-520d-3p in OC is dependent on EphA2 downregulation.
Synergistic effect of combined miR-520d-3p and EphA2-siRNA therapy
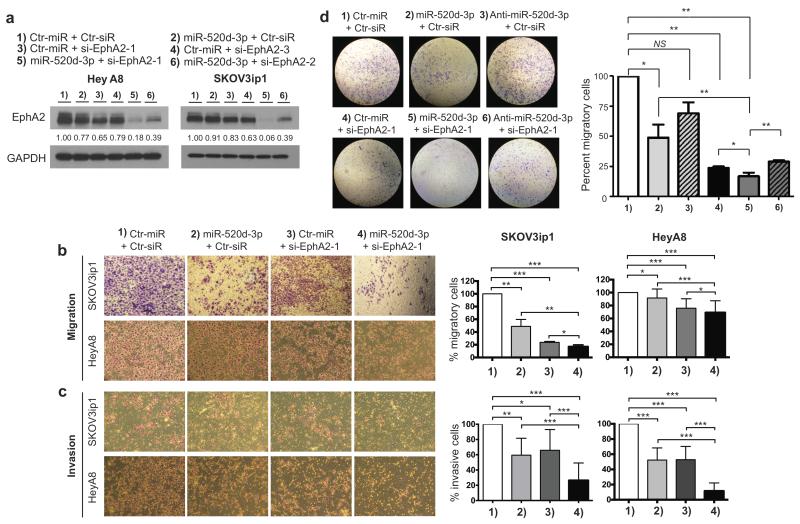
Since EphA2 has been previously shown to be a targetable protein in OC (6, 22-24), we sought to experimentally evaluate whether dual inhibition of EphA2 by siRNA and miRNA showed synergistic anti-tumor efficacy. For this purpose, we designed four different siRNAs targeting EphA2 and confirmed their ability to knockdown EphA2 (Supp. Fig. 7). Based on their efficiency, si-EphA2-1 (currently under consideration for human clinical trials) and si-EphA2-2 (highest efficiency in EphA2 knockdown) were selected for further analysis. Combination of each siRNA with miR-520d-3p lead to a remarkable reduction in EphA2 protein levels in both HeyA8 and SKOV3ip1 cells (Fig. 4a). Since combination of si-EphA2-1 and miR-520d-3p showed the highest efficiency in EphA2 knockdown, this combination was used to perform additional functional studies. Overexpression of miR-520d-3p in the combination studies in both cell lines was confirmed by qRT-PCR (Supp. Fig. 8). These data show that co-treatment with miR-520d-3p and EphA2-siRNA significantly enhanced EphA2 knockdown in both HeyA8 and SKOV3ip1 cells.
Figure 4. Combination of miR-520d-3p and siRNA-EphA2 treatment shows enhanced EphA2 inhibition and anti-tumor efficiency in vitro.
(a) Immunoblotting of EphA2 and GAPDH in HeyA8 and SKOV3ip1 cells after treatment with miR-520d-3p or different EphA2-targeting siRNAs or a combination of both (1 – 6). (b, c) Representative images showing effect of different combination treatments (1 – 4) on SKOV3ip1 and HeyA8 migration (b) and invasion (c) using Transwell migration assay (left panel). Cells were counted in 10 random fields per well at 40× after 6 hrs for migration and 24 hrs for invasion and the percent migratory or percent invasive cells were calculated compared to control treatment. A representative experiment is shown in right panel. Experiment was performed in duplicates at three independent times. (d) Representative images showing effect of rescue treatment with anti-miR-520d-3p in different combinations (1 – 6) on SKOV3ip1 migration using Transwell migration assay (left panel). Absorbance was measured at 590nm after 24 hours and the percent migratory cells were calculated compared to control treatment. The data from one representative experiment are shown in right panel. Experiment was performed in triplicates at three independent times. Statistical significance was determined by unpaired, two-tailed Student’s t test when compared to empty clones for all analyses. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 Data are mean ± s.d.
To further refine these correlations and to determine the functional consequences of this combination, we performed in vitro cell proliferation, migration and invasion analysis after miR-520d-3p/si-EphA2-1 treatment in HeyA8 and SKOV3ip1 cells. MTT analysis demonstrated a dose-dependent decrease in cell viability after individual monotherapies; however, combination therapy further decreased the cell viability in both cell lines (Supp. Fig. 9). The combination Index (CI) obtained after performing Isobologram analysis using the CompuSyn software showed synergistic cytotoxicity between the two agents (Supp. Fig. 9). Similarly, in migration and invasion analysis, treatment with individual therapy significantly decreased the migratory and invasive capabilities of both cell lines in comparison to control treatment (Fig. 4b, c), However, combination of both treatments further enhanced the inhibition of cell migration and invasion (Fig. 4b, c). To evaluate if this inhibition is synergistic, we adopted the method shown in (29, 30) as described in Materials and Methods. Using this analysis, we determined that the total anti-migratory effect of combined treatment was partially additive and not synergistic (Fig. 4b). However, the combination therapy showed synergistic inhibition of cell invasion in both cell lines (Ratio of expected:observed Fa is 2.3 for HeyA8 and 1.5 for SKOV3ip1 cells respectively) (Fig. 4c). Next, to validate the role of miR-520d-3p in combination therapy, we performed a rescue experiment after treatment with anti-miR-520d-3p. As shown in Fig. 4d, treatment with anti-miR-520d-3p along with miR-520d-3p/si-EphA2-1 treatment successfully rescued the migratory phenotype of SKOV3ip1 cells. Further, treatment with anti-miR-520d-3p was able to restore EphA2 protein levels in a dose-dependent manner (Supp. Fig. 10). These results together emphasize the functional significance of miR-520d-3p and support enhanced efficiency of miR-520d-3p/si-EphA2-1 combination in vitro.
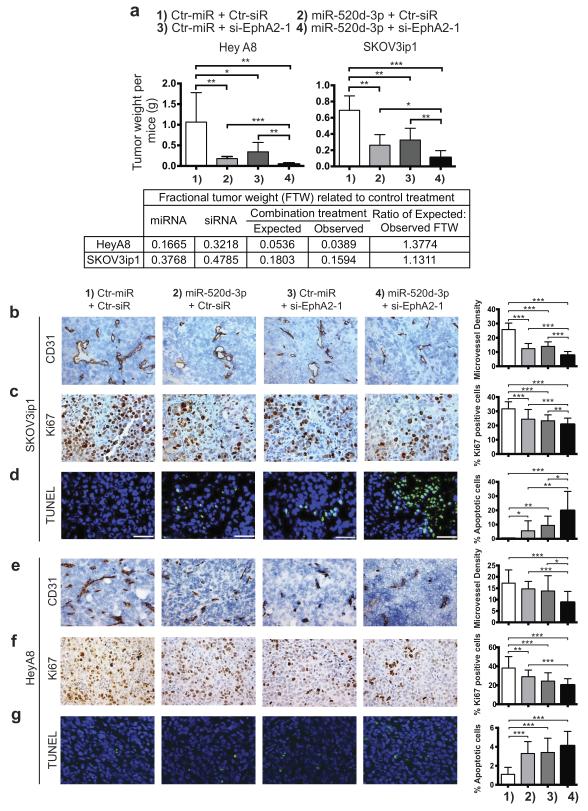
Correspondingly, in vivo administration of miR-520d-3p/si-EphA2-1 combination induced potent synergy resulting in substantial inhibition of tumor growth when compared to individual treatments. As described in Materials and Methods, we administered si-EphA2-1-DOPC (150 μg siRNA/kg) and miR-520d-3p-DOPC (200 μg miRNA/kg), twice weekly for two weeks, into the peritoneal cavity of orthotopic tumor bearing mice. The EphA2 combinatorial target approach proved remarkably effective, demonstrating a significant reduction in tumor weight compared to miR-520d-3p-DOPC or si-EphA2-1-DOPC monotherapies in both HeyA8 and SKOV3ip1 tumor bearing mice (Fig. 5a). The reduction in tumor weight after combined therapy was synergistic for both HeyA8 (Ratio of expected:observed Fa = 1.37) and SKOV3ip1 (Ratio of expected:observed Fa = 1.13) tumor bearing mice. We next examined the effects of dual therapy on angiogenesis, proliferation and apoptosis in the in vivo SKOV3ip1 (Fig. 5b, c, d) and HeyA8 (Fig. 5e, f, g) models. CD31 immunostaining showed that compared to control, MVD decreased significantly after miR-520d-3p-DOPC (P < 0.001) as well as si-EphA2-1-DOPC (P < 0.001) treatment. However, combination of these treatments resulted in further reduction (P < 0.001) in CD31 positive cells (Fig. 5b). To determine the effect on cell proliferation, Ki67 staining was assessed for all treatment groups. While monotherapy with miR-520d-3p-DOPC and si-EphA2-1-DOPC individually decreased cell proliferation (P < 0.001 for both), combined treatment resulted in improved reduction (P < 0.001) in tumor cell proliferation (Fig. 5c). We next assessed the degree of apoptosis for all treatments using the TUNEL assay (Fig. 5d). miR-520d-3p-DOPC or si-EphA2-1-DOPC treatment resulted in modest increase in apoptosis compared to the control treatment (5-fold and 10-fold respectively, P < 0.05 for miR-520d-3p-DOPC and P < 0.01 for si-EphA2-1-DOPC), while the combination treatment resulted in substantially higher apoptosis compared to the control treatment (20-fold, P < 0.001). A similar effect on tumor angiogenesis, proliferation and apoptosis was also confirmed in the HeyA8 model (Fig. 5e, f, g, Supp. Fig. 11). Detailed analysis of the data indicated that while combined treatment afforded improved tumor inhibition, its action was not synergistic, but additive. However, it is important to consider that even though miR-520d-3p/si-EphA2-1 combination showed additive effect in inhibiting in vivo tumor angiogenesis, proliferation and apoptosis (Fig. 5 b-g), it displayed a synergistic reduction in total tumor weight after dual therapy (Fig. 5a). Putting together the in vitro and in vivo combination therapy data, these results together confirm the improved therapeutic efficiency and anti-tumor activity of combined miRNA and siRNA therapy as compared to individual therapies in OC.
Figure 5. Co-treatment with miR-520d-3p and siRNA-EphA2 shows potent synergy and improved therapeutic efficiency in vivo.
(a) Total tumor weight after various combination treatments (1 – 4) of HeyA8 (left panel) and SKOV3ip1 (right panel) tumors. The lower panel shows calculation to demonstrate synergism as described in Materials and Methods. (b, c, d) Effect of combined miR-520d-3p + siEphA2-1 treatment on angiogenesis, proliferation and apoptosis in SKOV3ip1 cells. Representative images of CD31 (b) Ki67 (c) and TUNEL (d) immunostaining following various combination treatments (1 – 4) are shown (images were acquired at 100×). Quantification of immunostaining in (b, c, d) are shown in the right panels. (e, f, g) Effect of combined miR-520d-3p + siEphA2-1 treatment on angiogenesis, proliferation and apoptosis in HeyA8 cells. Representative images of CD31 (e), Ki67 (f) and TUNEL (g) immunostaining following various combination treatments (1 – 4) are shown (images were acquired at 100×). Quantification of immunostaining in (e, f, g) are shown in the right panels. Statistical significance was determined by unpaired, two-tailed Student’s t test when compared to empty clones for all analyses. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 Data are mean ± s.d.
miR-520d-3p also targets EphB2, which is an independent prognostic marker for OC
We hypothesized that the superior tumor suppression following combination treatment is possibly due to miR-520d-3p targeting multiple Eph pathway oncogenes in addition of EphA2. To identify these additional targets, we combined microarray gene expression with miRNA target prediction analyses to look for transcripts that are downregulated in stable clones of both cell lines and also predicted as miR-520d-3p targets. We identified three members of the Eph-receptor family: EphA2 (as expected by our previous results), EphA8 and EphB2 (Supp. Fig. 12). Using luciferase reporter analysis, we confirmed that miR-520d-3p targets EphB2 at a conserved seed region in its 3′-UTR (Fig. 6a). Consistent with the reporter assay, endogenous expression of EphB2 protein was reduced by ectopic miR-520d-3p overexpression in both ES2 and SKOV3ip1 cells (Fig. 6b), as well as in miR-520d-3p-overexpressing SKOV3ip1 stable clones (Fig. 6c). Though EphA8 was not confirmed as a direct target, it might still be a potential indirect target. Further analysis of EphB2 and miR-520d-3p levels by immunostaining revealed that OC tumors exhibit an inverse expression pattern between miR-520d-3p and EphB2 levels. Tumors with high miR-520d-3p expression showed weak EphB2 staining, while tumors with low miR-520d-3p expression showed strong EphB2 staining (Fig. 6d), further confirming EphB2 as an additional target of miR-520d-3p.
Figure 6. EphB2 is a direct and functional target of miR-520d-3p, and a prognostic factor for patients with OC.
(a) Luciferase activity of a reporter construct fused to wild-type or mutant EphB2 3′-UTR in MCF7 cells with ectopic miR-520d-3p expression (upper panel). Control, cells transfected with a scrambled control. Data are average of four independent experiments. Representative diagram of miR-520d-3p binding site on EphB2 mRNA (lower panel). (b) Immunoblotting of EphB2 and GAPDH in ES2 and SKOV3ip1 cells transfected with miR-520d-3p or a scrambled control. (c) Immunoblotting of EphB2 and GAPDH in miR-520d-3p overexpressing SKOV3ip1 stable clones. (d) Representative images of the immunostaining for EphA2 and miR-520d-3p from four patients showing negative correlation in OC tumors. (e) Immunoblotting of EphA2, EphB2 and GAPDH in SKOV3ip1 cells after various combination treatments (1 – 4). (f) Immunoblotting of EphB2 and GAPDH in SKOV3ip1 cells after treatment with miR-520d-3p or different EphA2-targeting siRNAs or a combination of both (1 – 6). (g, h, i) Kaplan-Meier curves representing the percent overall survival of 556 patients from TCGA 2012 dataset based on EphB2 median expression (f), combined EphB2 and miR-520d-3p expression levels (g) or combined EphA2, EphB2 and miR-520d-3p expression levels (h). The colored numbers (red or blue) below the curves represent patients at risk at the specified time points. Statistical significance was determined by unpaired, two-tailed Student’s t test. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 Data are mean ± s.d.
To investigate the functional significance of EphB2::miR-520d-3p interaction in OC, we sought to determine whether EphB2 played a role in the tumor suppression observed after miR-520d-3p/si-EphA2-1 dual-targeting of EphA2. We discovered that EphB2 protein levels were maximally reduced in combined miR-520d-3p/si-EphA2-1 treated SKOV3ip1 cells (Fig. 6e). While EphA2-targeting siRNAs (si-EphA2-1 and si-EphA2-2) alone did not affect EphB2 levels, combined treatment with miR-520d-3p and si-EphA2 greatly reduced EphB2 levels (Fig. 6f). Based on these data, we concluded that miR-520d-3p targets multiple genes in the Eph pathway, and when combined with standard single-target siRNA therapy, it contributed to superior tumor regression by potentially targeting Eph pathway signaling among other possible targets.
Further analyzing the TCGA dataset, we found that patients with EphB2-high tumors had significantly shorter overall survival (median 41.1 months) than those with EphB2-low tumors (median 55.2 months) (P = 0.0051; Fig. 6g). Addition of miR-520d-3p expression status to EphB2 expression further improved the stratification of the patient’s overall survival. Patients showing EphB2(high)/miR-520d-3p(low) had a significantly shorter survival and poorer prognosis (median 38.2 months) compared to patients with EphB2(low)/miR-520-3p(high) (median 64 months) (P = 0.0024; Fig. 6h). We next looked at combined EphA2, EphB2 and miR-520d-3p expression status in the TCGA dataset. Remarkably, this signature showed a further improved separation with patients having EphA2(low)/ EphB2(low)/miR-520d-3p(high) survived significantly longer (median 81.1 months) than patients with EphA2(high)/ EphB2(high)/miR-520d-3p(low) (median 38 months) (P = 0.0009; Fig. 6i). These data suggest the potential use of EphA2/EphB2/miR-520d-3p gene expression in prognostic stratification of patients with OC and highlight the rationale and therapeutic potential of using miRNAs to target multiple oncogenic pathways simultaneously.
DISCUSSION
In this study, we exploited large-scale cancer genomic databases and bioinformatic approaches to discover novel therapeutic applications of RNAi in ovarian cancer. We report that inhibition of EphA2 by an EphA2-targeting-siRNA and miR-520d-3p led to substantial augmentation in therapeutic efficiency, and potently suppressed tumor progression and metastasis in vitro and in vivo. This is the first study depicting that dual inhibition of a specific oncogene by its single-targeting-siRNA and a multi-targeting-miRNA can be utilized to obtain improved therapeutic efficiency. We named this ‘boosting effect’ of miRNAs and the mechanism of this improved activity is partly explained by the ability of miR-520d-3p to transcriptionally repress two Eph-receptors, EphA2 and EphB2, both highly upregulated in cancers (21, 31). This could provide an important clinical advantage since targeting of a single Eph receptor might raise the possibility of mutual compensation by other Eph receptors due to their overlapping functional roles, several common downstream targets and structural homology (32). Our data, however, does not rule out that miR-520-3p might target additional genes involved in multiple cancer-associated pathways, and these targets might assist in its anti-tumor phenotype. In summary, the data collectively supports application of combined miR-520d-3p/si-EphA2-1 therapy for clinical trials in OC patients.
Our experimental findings are also clinically relevant as the principle presented in this paper can be successfully applied to other tumor suppressive miRNAs in different human cancers. This approach is useful for concurrent targeting of distinct molecular defects in canonical cancer-associated pathways. One potential complexity with this approach could be the apparent saturation of the RISC (RNA-induced silencing complex) assembly essential for RNAi occurrence. While no loss of efficiency was observed at the concentrations used in our study, further dose-response studies need to be performed to determine the optimal pharmacological therapeutic window for this combination.
We also identified that miR-520d-3p serves as an effective and independent predictor of outcome in OC. While one previous study reported the role of miR-520d-3p in non-obstructive azoospermia (33), our study is the first to show a role of miR-520d-3p in cancer. We identified that high miR-520d-3p expression is linked to better clinical outcome and longer overall and relapse-free survival. While miR-520d-3p can serve as a useful prognostic marker in OC, our study highlights its tumor suppressive function both in vitro and in vivo. We further demonstrated that miR-520d-3p directly inhibits expression of EphA2 and EphB2, two key receptors of the Eph-pathway. Constitutive activation of the Eph-pathway by overexpression of Eph-receptors has been linked to tumor aggressiveness in multiple human cancers, modulating tumor microenvironment and contributing to tumor growth, invasiveness, angiogenesis, metastasis and resistance (34). Thus, therapeutic restoration of miR-520d-3p expression or function using miRNA mimics could be a useful approach for OC treatment.
Another key finding of this study is the identification of a novel gene-expression signature comprising of EphA2(low)/EphB2(low)/miR-520d-3p(high) that can predict favorable OC prognosis with powerful accuracy. High expression of EphA2 and EphB2 has been previously shown to be associated with clinical aggressiveness, shorter survival and poorer prognosis in OC (21, 31). Our study reveals that integration of these into a multi-gene signature markedly improved its prognostic power, and this signature, alone or in combination with other molecular markers, may improve outcome prediction and stratification of OC patients if prospectively validated.
Finally, while genomic approaches are identifying many new potential therapeutic targets, the targeted treatments currently available (e.g., small molecule inhibitors or antibodies) are still impractical or impossible due to a number of factors including large structure (e.g., proteins such as p130Cas), kinase-independent functions, and multiple structural domains with independent functions. Several small molecule inhibitors lack specificity and can be associated with intolerable side effects. Similarly, while monoclonal antibodies have shown promise against specific targets such as VEGF, their use is limited to either ligands or surface receptors. The development of combined miR-520d-3p and EphA2-targeting-siRNA therapy allows for therapeutic targeting of the Eph pathway and other proteins that would otherwise not be “drugable” in OC. The findings reported in our study emphasize on the development and application of novel RNAi-based therapeutics to improve the efficacy of targeted therapy and holds potential for improved management of OC patients.
METHODS
Clinical samples
A total of 91 patient samples with OC used in this study were collected from the Gynecologic Oncology tumor bank at MDACC. All samples were collected according to the institutional policies and obtained following patient’s informed consent. Data were de-identified prior to any analyses using standard procedures. The clinicopathological features of the OC are detailed in Supp. Table 1. The 2009 (n = 186) and 2012 (n = 556) TCGA datasets were downloaded from data portal at https://tcga-data.nci.nih.gov/tcga.
Cell culture
All the epithelial ovarian cancer cell lines used in this study (HeyA8, SKOV3ip1 and ES2) were purchased from the American Type Culture Collection (ATCC) and cultured under the conditions specified by the manufacturer. All cell lines were validated by the Characterized Cell Line Core at The University of Texas MD Anderson Cancer Center using STR DNA fingerprinting.
RNA extraction and real-time RT-PCR
Total RNA was isolated from tissues and cell lines as previously described (35) for both microRNA and EphA2 mRNA expression analyses. For quantification of miRNA levels, total RNA was reverse transcribed with miRNA-specific primers using TaqMan microRNA reverse transcription kit (Applied Biosystems, Foster City, CA), and then real-time PCR was carried out using TaqMan MicroRNA Assay Kit (Applied Biosystems) according to manufacturer’s instructions. For EphA2 mRNA expression study, RNA was reverse transcribed using SuperScript III (Invitrogen) with random hexamers and real-time PCR analysis was carried out with iQ SYBR Green Supermix (Bio-Rad) using primer sequences as previously described (22) according to the manufacturer’s protocol. 18S and U6 were used as normalizing controls for mRNA and miR quantification, respectively. The 2-ΔΔCt method was used to calculate the relative abundance compared with empty controls.
Plasmids and siRNAs
The interaction sites for miR-520d-3p were predicted on EphA2 (NM_004431.3) and EphB2 (NM_004442, NM_017449) 3′UTRs and the sequences were PCR amplified from human genomic DNA using primers: EphA2-F: 5′-CGTCTAGAGGCCACTGGGGACTTTATTT-3′, EphA2-R: 5′-CGTCTAGACCAGCTCACGAATGTTTGAC-3′, EphB2-F: 5′-GGCGGGAAATACAAGGAATA-3′ and EphB2-R: 5′-ATTTTCCCAGAGGGGTTCTC-3′. PCR products were cloned into the Xba1-site of pGL3-control vector (Promega, Madison, WI, USA), to obtain pGL3-EphA2-WT and pGL3-EphB2-WT luciferase constructs. Mutant constructs, pGL3-EphA2-mut and pGL3-EphB2-mut, were prepared by deleting the entire miR-520d-3p binding seed region in their 3′ UTRs using Quick change II site-directed mutagenesis kit (Stratagene). To establish stable cell lines with miR-520d-3p overexpression, we used the pcDNA3.1(+) vector system (Invitrogen). We first amplified a human genomic span that contains miR-520d-3p by using the following primers: pF: 5′ TCTAGAGAATTCTCAACAAGAAACCCAGAGTG 3′, and pR: 5′ TCTAGACTCGAGCAAAACAGAACCCCACCATC 3′. We then cloned this PCR product in pcDNA3.1(+) using EcoRI and XhoI restriction enzymes. The nonspecific, non-targeting siRNA used in this study is 5′-UUCUCCGAACGUGUCACGU-3′, while EphA2-targeting siRNAs used are: si-EphA2-1: 5′-UGACAUGCCGAUCUACAUG-3′, si-EphA2-2: 5′-CCAUCAAGAUGCAGCAGUA-3′, si-EphA2-3: 5′-CGUAUCUUCAUUGAGCUCA-3′ and si-EphA2-4: 5′-CAGAGAAGCAGCGAGUGGA-3′.
Luciferase reporter assay
Luciferase reporter assay to confirm miRNA targets were as previously described (35).
Immunoblotting
Immunoblotting staining of EphA2, EphB2 and GAPDH were performed on cell lines and stable clones as previously described (35).
Immunohistochemistry
Immunohistochemical staining of EphA2, Ki67 and CD31 were performed on tumor samples from patients and from mice as described previously (22).
In situ hybridization
The FFPE tissue sections were first digested with 5 μg/mL proteinase K for 20 minutes at RT, and were then loaded onto Ventana Discovery Ultra system (Ventana Medical Systems, Inc, Tucson, AZ) for in situ hybridization. The tissue slides were incubated with double-DIG labeled LNA probe for miR-520d-3p (Exiqon) for 2 hrs at 55° C. The digoxigenins were detected with a polyclonal anti-DIG antibody and Alkaline Phosphatase conjugated second antibody (Ventana) using NBT-BCIP as the substrate.
In vitro transfections
Transfections with miRNA, anti-miRNA or siRNA were performed as previously described (35). Briefly, cells were transfected with 100nM or 200nM of specified RNA using Lipofectamine 2000 (Invitrogen) for specified number of hours and cells were then used for further analysis.
In vitro Migration and Invasion assays
Cell migration assay was performed using 6.5-mm diameter Transwell chambers with 8.0 μm porous membrane (Corning Incorporated), and cell invasion assay was performed using BioCat growth-factor reduced Matrigel invasion chambers (BD Bioscience) according to the protocol previously described (35) on parental untreated, Empty vector clones and miR-520d-3p overexpressing clones of HeyA8 and SKOV3ip1 cells. Each experiment was performed in at least triplicate, and repeated three times. For in vitro combination analysis, 24-well trans-well plates with 8 μm pore size chambers were coated with 0.1% gelatin (migration) or defined matrix (invasion) separating the upper and lower wells. HeyA8 or SKOV3ip1 cells were transfected with miRNA or siRNA. Twenty four hours later, 6 × 104 cells were resuspended in 100 μl of serum-free media and added to upper wells and 5% serum-containing media was added to he lower wells. The culture system was incubated (migration, 6 hrs; invasion, 24 hrs) at 37 °C. Membranes were fixed, stained, and counted (10 random fields under 40×) using light microscopy. Experiments were done in triplicate and in three independent experiments.
Xenograft models of ovarian cancer
Female athymic nude mice (NCr-nu) were purchased from the National Cancer Institute, Frederick Cancer Research and Development Center (Frederick, MD) and maintained in specific pathogen-free conditions. The animals were cared for according to guidelines set forth by the American Association for Assessment and Accreditation of Laboratory Animal Care and the US Public Health Service Policy on Human Care and Use of Laboratory Animals. All mouse studies were approved and supervised by the MD Anderson Cancer Center Institutional Animal Care and Use Committee. To produce orthotopic tumors, mice were injected into the peritoneal cavity with 1 × 106 parental untreated, empty vector clones or miR-520d-3p overexpressing clones of HeyA8 and SKOV3ip1 cells (n = 10 mice per group). The cells were treated with trypsin, washed and resuspended in Hank’s balanced salt solution (Gibco, Carlsbad, CA) at a concentration of 5 × 106 cells/ml. About 33 days in HeyA8 clones and 46 days in SKOV3ip1 clones after cell injection, all mice were sacrificed and necropsy was performed. The individual tumor nodules were isolated from the supporting tissue and counted. The total tumor weight was also measured. Tissue samples were fixed in formalin for paraffin embedding, and frozen in optimal cutting temperature (OCT) media for preparation of frozen slides, or snap frozen for mRNA as described above.
Liposomal siRNA and miRNA preparation
For in vivo delivery, siRNA was incorporated into dioleoyl-snglycero-3-phosphocholine (DOPC). DOPC and siRNA were mixed in the presence of excess tertiary butanol at a ratio of 1:10 (w/w) siRNA/DOPC (22). Before in vivo administration, the preparation was hydrated with normal 0.9% saline (100 μL per mouse) for i.v. or i.p.injection.
In vivo treatment with miR520d-3p-DOPC and si-EphA2-1-DOPC
miR520d-3p, control miRNA, siRNA-EphA2 and control siRNA were purchased from Sigma. These miRNAs and siRNAs were conjugated with DOPC as described above. The appropriate dosage for microRNA treatment was determined by performing dose-response analysis consisting of three different doses (200μg/kg, 400μg/kg, 600μg/kg) of miR520d-3p and 200μg/kg of control miRNA (data not shown). For in vivo combination analysis, female athymic nude mice (NCr-nu) were injected into the peritoneal cavity with 1 × 106 HeyA8 or SKOV3ip1 cells. Mice were divided into 4 groups (n = 10 per group): (a) Control miRNA + siRNA, (b) miR-520d-3p-DOPC + control siRNA, (c) Control miRNA + si-EphA2-1-DOPC, and (d) miR-520d-3p-DOPC + si-EphA2-1-DOPC. One week after injection, each miRNA was administered thrice weekly at 200μg/kg body weight, and each siRNA was given twice weekly at 200μg/kg body weight. Treatment was continued until control mice became moribund (33 days in HeyA8 cells and 46 days in SKOV3ip1 cells), and the last treatment was done 48 hours (HeyA8) and 24 hours (SKOV3ip1) before sacrificing them. At the time of sacrifice, mouse weight, tumor weight, number of nodules and distribution of tumors were recorded.
Calculations for synergism
Isobologram analysis was performed using the CompuSyn software program (ComboSyn, Inc., USA) (36). Briefly, cells were treated with different concentrations of miR-520d-3p alone, si-EphA2-1 alone or combination of miR-520d-3p + si-EphA2-1 in 1:1 ratio for 72 hrs followed by MTT analysis to determine percent cell viability and a combination index (CI) was calculated using the Chou-Talalay method (37). A CI of < 1.0 indicates synergism, a CI of 1 indicates additive activity and a CI > 1.0 indicates antagonism. For single dose experiments, the potential synergy between two antitumor agents was evaluated as described by (29, 30). Using this method, the Fractional activity (Fa) for miR-520d-3p alone or si-EphA2-1 alone or combined treatment is calculated compared to the control, and the ratio of ‘Expected Fa’ to ‘Observed Fa’ is determined for the combination of the two agents. A ratio of > 1 indicates a synergistic effect, and a ratio of < 1 indicates less than additive effect.
TCGA data analyses
The input data were downloaded from data portal at https://tcga-data.nci.nih.gov/tcga. Data have been imported on BRB-ArrayTools Version: 3.7.2 and average values of the replicate spots of each miRNA were background subtracted, normalized, and subjected to further analysis. Normalization was performed by using per chip median normalization method and the median array as referenced. Class comparison analysis using t test identified miRNAs that were differentially expressed (P < 0.001). Class prediction algorithms in BRB array tools were used to determine whether microRNA microarray expression patterns could accurately differentiate between classes (P < 0.001).
Statistical analysis
Survival analyses were performed in R (version 2.14.2) and SPSS 16.0. The patients were grouped into percentiles according to mRNA/miRNA expression. We checked for a relation with the survival by choosing a cut-off to optimally split the samples into two groups. Optimally was defined as significant separation in OS or PFS using the best P values for both TCGA and MDACC datasets. The Log-rank test was employed to determine the association between mRNA/miRNA expression and OS and PFS respectively. The Kaplan-Meyer method was used to generate survival curves. We computed a significance level for each miRNA and clinicobiological factor based on a univariate Cox proportional hazard regression model. For multivariate analysis, a full Cox proportional hazards model was fitted. The relationship between miRNA expression and experimental groups (transfection group vs. control) was assessed using Student’s t test. Data are represented as mean ± standard deviation (s.d.). Statistical analysis was performed using SPSS 16.0. All tests were two-sided, and an effect was considered to be statistically significant at P < 0.05.
Supplementary Material
SIGNIFICANCE.
This study addresses a new concept of RNA inhibition therapy by combining miRNA and siRNA in nano-liposomal particles to target oncogenic pathways altered in ovarian cancer. Combined targeting of Eph pathway using EphA2-targeting-siRNA and tumor suppressor miR-520d-3p exhibits remarkable therapeutic synergy and enhanced tumor suppression in vitro and in vivo compared to either monotherapy alone.
ACKNOWLEDGEMENTS AND GRANT SUPPORT
Dr. George Calin is the Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. He is supported as a Fellow at The University of Texas MD Anderson Research Trust, as a University of Texas System Regents Research Scholar, and by the CLL Global Research Foundation. Work in Dr. Calin’s laboratory is supported in part by the NIH/NCI (CA135444), a Department of Defense Breast Cancer Idea Award, Developmental Research Awards in Breast Cancer, Ovarian Cancer, Brain Cancer, Prostate Cancer, Multiple Myeloma, and Leukemia SPOREs, an MDACC Sister Institution Network Fund grant in colorectal cancer, the Laura and John Arnold Foundation, the RGK Foundation, and the Estate of C. G. Johnson, JR. M. Y. S. is a Rosalie B. Hite fellow. Portions of this work were supported by the NIH (UH2 TR000943-01, CA 109298, P50 CA083639, P50 CA098258, CA128797, RC2GM092599, CA151668), the Ovarian Cancer Research Fund, Inc. (Program Project Development Grant), the DOD (OC120547, OC073399, W81XWH-10-1-0158, BC085265), NSC-96-3111-B, the Zarrow Foundation, the Marcus Foundation, and the Betty Anne Asche Murray Distinguished Professorship. Short tandem repeat DNA fingerprinting was performed by the Cancer Center Support grant-funded Characterized Cell Line Core, NCI number CA16672.
Abbreviations
- RNAi
RNA-interference
- miRNA
microRNA
- siRNA
silencing RNA
- OC
Ovarian cancer
- TCGA
The Cancer Genome Atlas
- MDACC
MD Anderson Cancer Center
- pre-miR
precursor miRNA
- OS
Overall survival
- PFS
Progression-free survival
- 3′-UTR
3′-Untranslated Region
- mRNA
messenger RNA
- MVD
Microvessel density
- CI
Combination Index
- DOPC
dioleoyl-snglycero-3-phosphocholine
- RISC
RNA-induced silencing complex
- qRT-PCR
quantitative reverse transcription PCR
- ATCC
American Type Culture Collection
Footnotes
Conflict of Interest: All authors declare no conflict of interest.
REFERENCES
- 1.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chemistry & Biology. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nature Reviews Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.American Cancer Society . Cancer Facts & Figures 2012. American Cancer Society; Atlanta: [Google Scholar]
- 6.Landen CN, Kinch MS, Sood AK. EphA2 as a target for ovarian cancer therapy. Expert Opinion On Therapeutic Targets. 2005;9:1179–87. doi: 10.1517/14728222.9.6.1179. [DOI] [PubMed] [Google Scholar]
- 7.Lu C, Shahzad MM, Wang H, Landen CN, Kim SW, Allen J, et al. EphA2 overexpression promotes ovarian cancer growth. Cancer Biology & Therapy. 2008;7:1098–103. doi: 10.4161/cbt.7.7.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox BP, Kandpal RP. Invasiveness of breast carcinoma cells and transcript profile: Eph receptors and ephrin ligands as molecular markers of potential diagnostic and prognostic application. Biochemical & Biophysical Research Communications. 2004;318:882–92. doi: 10.1016/j.bbrc.2004.04.102. [DOI] [PubMed] [Google Scholar]
- 9.Herath NI, Boyd AW. The role of Eph receptors and ephrin ligands in colorectal cancer. International Journal of Cancer. 2010;126:2003–11. doi: 10.1002/ijc.25147. [DOI] [PubMed] [Google Scholar]
- 10.Kataoka H, Igarashi H, Kanamori M, Ihara M, Wang JD, Wang YJ, et al. Correlation of EPHA2 overexpression with high microvessel count in human primary colorectal cancer. Cancer Science. 2004;95:136–41. doi: 10.1111/j.1349-7006.2004.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Wang L, Gu JW, Li B, Liu WP, Wang YG, et al. Up-regulation of EphA2 and down-regulation of EphrinA1 are associated with the aggressive phenotype and poor prognosis of malignant glioma. Tumour Biology. 2010;31:477–88. doi: 10.1007/s13277-010-0060-6. [DOI] [PubMed] [Google Scholar]
- 12.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. EphA2: a determinant of malignant cellular behavior and a potential therapeutic target in pancreatic adenocarcinoma. Oncogene. 2004;23:1448–56. doi: 10.1038/sj.onc.1207247. [DOI] [PubMed] [Google Scholar]
- 13.Nemoto T, Ohashi K, Akashi T, Johnson JD, Hirokawa K. Overexpression of protein tyrosine kinases in human esophageal cancer. Pathobiology. 1997;65:195–203. doi: 10.1159/000164123. [DOI] [PubMed] [Google Scholar]
- 14.Brannan JM, Dong W, Prudkin L, Behrens C, Lotan R, Bekele BN, et al. Expression of the receptor tyrosine kinase EphA2 is increased in smokers and predicts poor survival in non-small cell lung cancer. Clinical Cancer Research. 2009;15:4423–30. doi: 10.1158/1078-0432.CCR-09-0473. [DOI] [PubMed] [Google Scholar]
- 15.Easty DJ, Bennett DC. Protein tyrosine kinases in malignant melanoma. Melanoma Research. 2000;10:401–11. doi: 10.1097/00008390-200010000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Kamat AA, Coffey D, Merritt WM, Nugent E, Urbauer D, Lin YG, et al. EphA2 overexpression is associated with lack of hormone receptor expression and poor outcome in endometrial cancer. Cancer. 2009;115:2684–92. doi: 10.1002/cncr.24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu M, Miller KD, Gokmen-Polar Y, Jeng MH, Kinch MS. EphA2 overexpression decreases estrogen dependence and tamoxifen sensitivity. Cancer Research. 2003;63:3425–9. [PubMed] [Google Scholar]
- 18.Andres AC, Reid HH, Zurcher G, Blaschke RJ, Albrecht D, Ziemiecki A. Expression of two novel eph-related receptor protein tyrosine kinases in mammary gland development and carcinogenesis. Oncogene. 1994;9:1461–7. [PubMed] [Google Scholar]
- 19.Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Research. 2001;61:2301–6. [PubMed] [Google Scholar]
- 20.Lin YG, Han LY, Kamat AA, Merritt WM, Landen CN, Deavers MT, et al. EphA2 overexpression is associated with angiogenesis in ovarian cancer. Cancer. 2007;109:332–40. doi: 10.1002/cncr.22415. [DOI] [PubMed] [Google Scholar]
- 21.Thaker PH, Deavers M, Celestino J, Thornton A, Fletcher MS, Landen CN, et al. EphA2 expression is associated with aggressive features in ovarian carcinoma. Clinical Cancer Research. 2004;10:5145–50. doi: 10.1158/1078-0432.CCR-03-0589. [DOI] [PubMed] [Google Scholar]
- 22.Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, Sood AK. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Research. 2005;65:6910–8. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 23.Shahzad MM, Lu C, Lee JW, Stone RL, Mitra R, Mangala LS, et al. Dual targeting of EphA2 and FAK in ovarian carcinoma. Cancer Biology & Therapy. 2009;8:1027–34. doi: 10.4161/cbt.8.11.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JW, Han HD, Shahzad MM, Kim SW, Mangala LS, Nick AM, et al. EphA2 immunoconjugate as molecularly targeted chemotherapy for ovarian carcinoma. Journal of the National Cancer Institute. 2009;101:1193–205. doi: 10.1093/jnci/djp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clinical Cancer Research. 2003;9:4227–39. [PubMed] [Google Scholar]
- 27.Yoneda J, Kuniyasu H, Crispens MA, Price JE, Bucana CD, Fidler IJ. Expression of angiogenesis-related genes and progression of human ovarian carcinomas in nude mice. Journal of the National Cancer Institute. 1998;90:447–54. doi: 10.1093/jnci/90.6.447. [DOI] [PubMed] [Google Scholar]
- 28.Tedjarati S, Baker CH, Apte S, Huang S, Wolf JK, Killion JJ, Fidler IJ. Synergistic therapy of human ovarian carcinoma implanted orthotopically in nude mice by optimal biological dose of pegylated interferon alpha combined with paclitaxel. Clinical Cancer Research. 2002;8:2413–22. [PubMed] [Google Scholar]
- 29.Yokoyama Y, Dhanabal M, Griffioen AW, Sukhatme VP, Ramakrishnan S. Synergy between angiostatin and endostatin: inhibition of ovarian cancer growth. Cancer Research. 2000;60:2190–6. [PubMed] [Google Scholar]
- 30.Takahashi N, Haba A, Matsuno F, Seon BK. Antiangiogenic therapy of established tumors in human skin/severe combined immunodeficiency mouse chimeras by anti-endoglin (CD105) monoclonal antibodies, and synergy between anti-endoglin antibody and cyclophosphamide. Cancer Research. 2001;61:7846–54. [PubMed] [Google Scholar]
- 31.Wu Q, Suo Z, Kristensen GB, Baekelandt M, Nesland JM. The prognostic impact of EphB2/B4 expression on patients with advanced ovarian carcinoma. Gynecologic Oncology. 2006;102:15–21. doi: 10.1016/j.ygyno.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 32.Kullander K, Klein R. Mechanisms and functions of Eph and ephrin signalling. Nature Reviews Molecular Cell Biology. 2002;3:475–86. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- 33.Lian J, Zhang X, Tian H, Liang N, Wang Y, Liang C, et al. Altered microRNA expression in patients with non-obstructive azoospermia. Reproductive Biology & Endocrinology. 2009;7:13–20. doi: 10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nature Reviews Cancer. 2010;10:165–80. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almeida MI, Nicoloso MS, Zeng L, Ivan C, Spizzo R, Gafa R, et al. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142:886–96. doi: 10.1053/j.gastro.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou TC, Martin N. CompuSyn for Drug Combinations: PC Software and User’s Guide: A Computer Program for Quantitation of Synergism and Antagonism in Drug Combinations, and the Determination of IC50 and ED50 and LD50 Values. ComboSyn; Paramus (NJ): 2005. Available at: www.combosyn.com. [Google Scholar]
- 37.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in Enzyme Regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.