Summary
The tomato HIGH PIGMENT-2 gene encodes an orthologue of the Arabidopsis nuclear protein DE-ETIOLATED 1 (DET1). From genetic analyses it has been proposed that DET1 is a negative regulator of light signal transduction, and recent results indicate that it may control light-regulated gene expression at the level of chromatin remodelling. To gain further understanding about the function of DET1 during plant development, we generated a range of overexpression constructs and introduced them into tomato. Unexpectedly, we only observed phenotypes characteristic of DET1 inactivation, i.e. hyper-responsiveness to light. Molecular analysis indicated in all cases that these phenotypes were a result of suppression of endogenous DET1 expression, due to post-transcriptional gene silencing. DET1 silencing was often lethal when it occurred at relatively early stages of plant development, whereas light hyper-responsive phenotypes were obtained when silencing occurred later on. The appearance of phenotypes correlated with the generation of siRNAs but not DNA hypermethylation, and was most efficient when using constructs with mutations in the DET1 coding sequence or with constructs containing only the 3′-terminal portion of the gene. These results indicate an important function for DET1 throughout plant development and demonstrate that silencing of DET1 in fruits results in increased carotenoids, which may have biotechnological potential.
Keywords: DET1, DNA methylation, fruit ripening, photomorphogenesis, post-transcriptional gene silencing, siRNA, tomato
Introduction
Throughout the plant life cycle, light is a critical environmental signal that affects nearly every aspect of development. Plants must be able to monitor the intensity, quality, and direction of light signals, which is achieved using a series of photoreceptors that are classified into three known groups: the red/far-red light-absorbing phytochromes, and the blue/UV-A light-absorbing cryptochromes and phototropins (Quail, 2002).
In Arabidopsis much is now known about photoreceptor function and signal transduction (Schäfer and Bowler, 2002). Many putative components of photoreceptor signalling pathways have been identified using mutant screens. Some of these mutants appear to be mutated in general components of the light signalling machinery, whereas others are specific to individual photoreceptors (Fankhauser and Bowler, 2004).
An important class of Arabidopsis mutants display light responses in darkness and are known as constitutive photomorphogenic (cop) or de-etiolated (det) mutants (Schäfer and Bowler, 2002). A total of 10 such loci have now been described. Identification of the genetic lesions in these mutants has led to the elucidation of important mechanisms controlling photomorphogenic responses, such as polyubiquitin-mediated proteolysis of positively acting signalling intermediates, e.g. the transcription factor HY5 (Serino and Deng, 2003).
Tomato (Lycopersicon esculentum) has become a model system complementary to Arabidopsis for studying light responses, allowing general conclusions to be drawn about the roles played by individual photoreceptors in higher plants (Kendrick et al., 1997). Analysis of photoreceptor function in tomato also allows study of their roles during fruit development, which is more difficult in Arabidopsis. It has been known for 50 years that phytochromes can regulate the fruit ripening process (Piringer and Heinze, 1954). More recent studies have confirmed that red and far-red light can penetrate the epidermis and pericarp of both immature and mature tomato fruits, and that phytochromes are indeed present in fruit tissues (Alba et al., 2000). Other studies have shown that cryptochromes also play an important role in fruit ripening (Weller et al., 2001).
In tomato, no constitutive photomorphogenic mutants have ever been found. However, mutants with an exaggerated light responsiveness have been identified such as the high pigment mutants hp-1 and hp-2 (Kendrick et al., 1997). Light-grown hp mutants display high levels of anthocyanins, are shorter and darker than wild-type plants and have dark green immature fruits, so it is likely that they are mutated in genes encoding important negative regulators of photoreceptor-linked signal transduction pathways. Cloning of the HP-2 gene indeed revealed that it encodes the tomato homologue of Arabidopsis DET1 (Mustilli et al., 1999). This was nonetheless surprising, given that Arabidopsis det1 mutants display constitutive photomorphogenesis (Chory et al., 1989), whereas hp-2 mutants do not (Mustilli et al., 1999). However, it is not known whether hp-2 mutant alleles represent null alleles, and consequently the effect of DET1 inactivation in tomato is not known.
DET1 is a nuclear protein (Mustilli et al., 1999; Pepper et al., 1994), and recent findings suggest that the functional form of DET1 is within an approximately 350 kDa complex that also contains the plant homologue of UV-damaged DNA binding protein 1 (DDB1) (Schroeder et al., 2002). Interestingly, the hp-1 mutant is now known to be mutated in DDB1 (Lieberman et al., 2004; Liu et al., 2004). Furthermore, Benvenuto et al. (2002) have shown that DET1 binds to hypo-acetylated amino-terminal tails of the core histone H2B and have proposed that DET1 may be involved in chromatin remodelling around photoregulated genes (Schäfer and Bowler, 2002). For example, DET1 may limit access of positive regulatory factors to the promoters of light responsive genes.
In this report we analyse the phenotypes of transgenic tomato plants in which DET1 gene expression has been modulated. We have generated a number of independent transgenic lines containing different DET1 constructs and summarize the range of phenotypes observed. Surprisingly, in all cases phenotypes were a consequence of the induction of post-transcriptional gene silencing (PTGS) of the DET1 gene. Our data support the notion that DET1 is an important regulator of photomorphogenesis that plays a role during the entire life cycle of the plant.
Results
Transgenic plants containing tomato DET1 (TDET1) constructs show a range of light hyper-responsive phenotypes
To investigate the function and importance of DET1 in tomato, several transgenic lines were generated containing different forms of the TDET1 gene. Table 1 summarizes these constructs, the number of lines generated and the number of lines showing clear phenotypes. A total of 11 constructs were utilized, which contained three different promoters (CaMV 35S and enhanced 35S (e35S) promoters for constitutive transgene expression (Benfey and Chua, 1990) and the E8 promoter for fruit-specific expression (Deikman et al., 1992)), with different TDET1 transgene constructs (including sense, antisense, hp-2 and hp-2j mutant alleles, 5′-terminal and 3′-terminal), and two different genotypes (Money Maker (MM) and T56) (see Experimental procedures). Full-length TDET1 expressed from either the 35S or the e35S promoter was expected to generate light-insensitive phenotypes, whereas 5′-terminal and 3′-terminal constructs were made in an attempt to identify dominant negative phenotypes. Antisense constructs were expected to repress TDET1 gene expression and consequently to generate light hyper-responsive phenotypes. However, in all cases where phenotypes were visible they were invariably characteristic of exaggerated light sensitivity, i.e. shorter bushy plants and dark green immature fruits, reminiscent of high pigment mutants (Kendrick et al., 1997) (Figure 1).
Table 1.
Details of constructs used and numbers of lines generated
| Construct | Genotype | No. lines | No. lines with hp-like phenotypes | Severity of phenotypesa |
|---|---|---|---|---|
| 35S::TDET1 | MM | 5 | 0 | – |
| 35S::TDET1-5′ | MM | 3 | 2 | + |
| 35S::TDET1-3′ | MM | 2 | 0 | – |
| 35S::TDET1-AS | MM | 7 | 0 | – |
| 35S::hp2 | MM | 3 | 3 | +++ |
| T56 | 5 | 3 | +++ | |
| 35S::hp2j | MM | 5 | 0 | – |
| T56 | 13 | 5 | +++ | |
| e35S::TDET1 | T56 | 45 | 14 | + |
| e35S::TDET1-5′ | T56 | 15 | 6 | ++ |
| e35S::TDET1-3′ | T56 | 8 | 4 | +++ |
| E8::TDET1 | MM | 17 | 0 | – |
| E8::TDET1-AS | MM | 2 | 0 | – |
| T56 | 19 | 0 | – |
+ and ++ denote severity of blotchy phenotypes, +++ denotes hp phenotype.
Figure 1.
Phenotypes of transgenic plants containing different TDET1 constructs. In all cases [except in (h) and (j)], photographs were taken from MM plants containing the 35S::hp2 construct. Identical phenotypes were also observed with other constructs (see Table 1), albeit with different severity (see text).
(a) Wild-type phenotype fruits.
(b) Blotchy fruits.
(c) hp-like dark green immature fruits.
(d) Wild-type plant phenotype.
(e) Intermediate light hyper-responsive plant phenotype.
(f) Severe light hyper-responsive young plant phenotype.
(g) Severe light hyper-responsive old plant phenotype.
(h) Phenotype of MM genotype transformed with 35S::TDET1-3′ construct.
(i) Blotchy fruit phenotype in MM genotype.
(j) Blotchy fruit phenotype in T56 genotype.
From the data in Table 1, several observations can be made: (i) All sense-oriented TDET1 constructs produced phenotypes in at least one genetic background when expressed from a 35S-based construct. (ii) Phenotypes were observed in both Money Maker and T56 genotypes at similar frequencies. (iii) Expression of transgenes using an e35S promoter was slightly more effective in generating phenotypes than was the 35S promoter. (iv) Plants containing antisense TDET1 constructs displayed no obvious phenotypes. (v) Constructs containing the fruit-specific E8 promoter produced no phenotypes.
Beyond this simplified summary, we however observed notable differences, including the severity of phenotypes (i.e. more dwarf statures and darker green immature fruits) generated by different constructs (Table 1). In particular, transgenes containing the hp-2 and hp-2j TDET1 mutant alleles produced more dramatic phenotypes compared with the TDET1 wild-type transgene, and the truncated 3′-terminal construct tended to produce plants with more severe phenotypes than the full length or truncated 5′-terminal constructs. These phenotypes are illustrated in Figure 1 and are more thoroughly described below.
Range of phenotypes in transgenic plants
Figure 1 shows the range of light hyper-responsive phenotypes observed in transgenic plants containing different TDET1 constructs. In many cases fruit phenotypes ranged from normal wild type to intermediate blotchy (normal fruits with dark green patches) to uniform dark green high pigment (hp)-like phenotypes (Figure 1a–c). As mentioned earlier the full-length TDET1 construct produced only the weak blotchy phenotype, whereas the truncated and mutant constructs often generated the more severe hp phenotypes. Similarly, whole plant phenotypes varied from wild type to intermediate dark green to very dark green bushy plants (Figure 1d–f).
Both the 3′-terminal and the hp-2 and hp-2j mutant allele constructs produced the most severe whole plant phenotypes, whereas the 5′-terminal construct only gave the intermediate dark green phenotype. In most cases the phenotypes were not visible in seedlings but appeared during the later development of the plants, after at least one month. In some cases, the plants produced wild-type fruits and only subsequently did blotchy and hp phenotypes appear. In other cases the plants generated only blotchy or hp fruits. In a few cases, most notably with the 3′-terminal and mutant allele constructs, the phenotypes were more severe and appeared earliest during plant development compared with plants containing the other constructs. These plants were dark green and often died before producing fruits (Figure 1g).
Phenotypes caused by the 3′-terminal construct were slightly different than with the other constructs. These plants were severely dwarfed and in the apex of the plant the leaves were arranged much closer together than normal and became progressively yellowish rather than dark green (Figure 1h). Finally blotchy phenotypes varied between the Money Maker and T56 genotypes. In Money Maker the fruits showed dark green blotches (Figure 1i), whereas in T56 the dark green patches appeared as more precisely defined sectors (Figure 1j).
Fruits from transgenic plants produce high levels of pigments
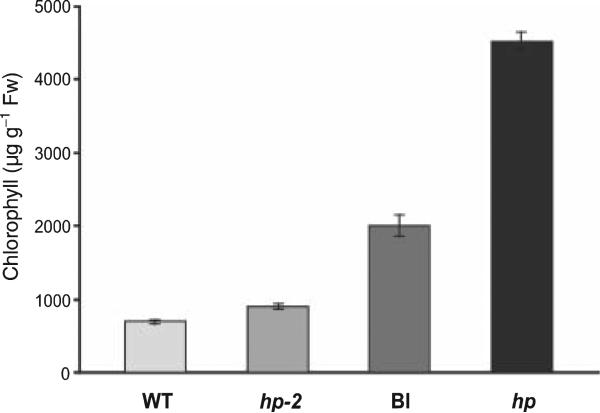
Pigment analysis of immature fruits showing phenotypes confirmed that they contain high levels of chlorophyll (Figure 2). There was a clear difference in pigment content between wild type, blotchy and dark green fruits, and these latter fruits contained more than five times the chlorophyll levels of wild-type fruits and four times more than hp-2 mutant fruits (Figure 2). The intermediate blotchy fruits contained approximately three times more chlorophyll than wild-type fruits. This was further confirmed by Northern blot analysis on these fruits, which showed upregulation of CAB gene expression in hp-phenotype fruits from transgenic plants (see Figure 3b).
Figure 2.
Chlorophyll analysis of blotchy (Bl) and hp phenotype fruits from T56 plants transformed with 35S::hp2. Fruits from T56 (WT) and hp-2 mutants (MM background) were used for comparison. Green immature fruits were harvested from five plants per genotype. Standard error bars are shown (n = 5).
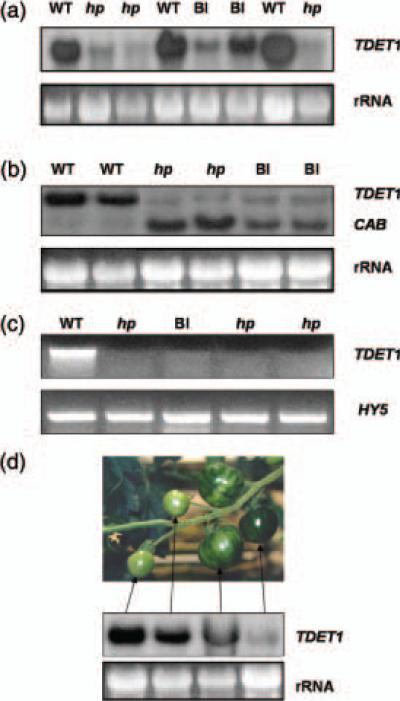
Figure 3.
Molecular analysis of transgenic plants showing a range of light hyper-responsive phenotypes.
(a) Northern blot analysis of TDET1 mRNA levels in leaves of transgenic plants with different phenotypes.
(b) Northern blot analysis of TDET1 and CAB mRNA levels in fruits of transgenic plants with different phenotypes.
(c) RT-PCR analysis of fruits from transgenic plants showing different phenotypes. Full-length TDET1 was synthesized using specific primers and HY5 primers were used as a control for cDNA synthesis and loading.
(d) Northern blot analysis of TDET1 mRNA levels in fruits from the same truss. Severity of light hyper-responsive phenotypes correlated negatively with expression levels of TDET1.
In (a), (b) and (d) 28S rRNA is shown as loading control. MM plants containing the 35S::hp2 construct were used for all analyses. Wild-type (WT), Blotchy (Bl) and high pigment (hp) fruit phenotypes.
We also examined the levels of lycopene and β-carotene in mature red ripe fruits (Table 2). Levels of lycopene were increased more than twofold in hp phenotype fruits compared with wild-type fruits, and levels of β-carotene were increased fivefold. Table 2 also shows that the brix values and sugar content of the more pigmented transgenic fruits were slightly lower than wild-type fruits. Furthermore, the total yield of fruits from plants displaying hp-like phenotypes was significantly reduced, which is characteristic of hp-2 mutants (Mustilli et al., 1999).
Table 2.
Pigment analysis of fruits from transgenic lines with hp phenotypesa
| Plant material | Fruit yield per plant (g) | Brix (g 100 g–1) | Sugars (g 100 g–1) | Lycopene (p.p.m.) | β-carotene (p.p.m.) | Vitamin A (IU g–1)b |
|---|---|---|---|---|---|---|
| Wild type T56 | 224 ± 9 | 4.73 ± 0.06 | 2.58 ± 0.08 | 85.7 ± 4.3 | 3.03 ± 0.41 | 5.04 ± 0.68 |
| Transgenic T56 with hp phenotype | 151 ± 29 | 4.60 ± 0.08 | 2.30 ± 0.25 | 186.8 ± 28.4 | 15.62 ± 5.69 | 26.03 ± 9.49 |
All red ripe fruits were harvested from individual T2 generation plants, weighed, and analysed as in Experimental procedures. Wild-type material was derived from non-transformed T56 genotype (six plants), whereas plants with hp phenotypes wereT2 generation T56 plants containing either the 35S::hp2j or e35S::TDETI-3′ constructs (five plants). Plants were grown in Woodland, CA, during the Spring of 2002. Data are ±standard errors.
International units of vitamin A per gram.
Molecular analysis of transgenic plants
Southern blot analyses were used to confirm the presence of the transgenes, and lines containing single copy inserts were chosen for further molecular analysis (data not shown). RNA from leaves and fruits of transgenic plants displaying either wild-type or light hyper-responsive phenotypes was extracted and TDET1 mRNA levels were examined by Northern blot hybridization. Endogenous TDET1 mRNA is undetectable by this method because the gene is expressed at extremely low levels (G. R. Davuluri and C. Bowler, unpublished results). Consequently, any observed expression should be derived from the TDET1 transgene. Interestingly, severity of the light hyper-responsive phenotypes was negatively correlated with transgene expression levels (Figure 3a,b).
Because the endogenous TDET1 gene is normally expressed at only very low levels we performed RT-PCR analysis with specific primers to determine whether TDET1 expression levels in transgenic material displaying light hyper-responsive phenotypes were below the levels normally found in wild-type plants. This analysis revealed that TDET1 expression levels were indeed below normal levels and were often completely undetectable (Figure 3c). Furthermore, a negative correlation between expression levels and strength of phenotype was again found in this analysis. These observations therefore demonstrate that loss of endogenous TDET1 gene expression results in severe light hyper-responsive phenotypes, thereby confirming a previous hypothesis that loss-of-function mutations in the TDET1 gene are responsible for the hp-2 mutation (Mustilli et al., 1999), thus implying the importance of TDET1 for controlling light responses in tomato. Furthermore, these data indicate that the phenotypes observed were probably caused by suppression of endogenous TDET1 expression rather than by overexpression of the gene. We therefore investigated the underlying mechanism and used these plants for further understanding the importance of DET1 in tomato development.
Based upon phenotypic observations of transgenic plants, suppression of TDET1 expression appeared to occur at a certain moment and then to spread to neighbouring regions, or even throughout the whole plant (see above). TDET1 expression levels were therefore examined in RNA extracted from different regions of the same plant. One such example is illustrated in Figure 3(d), in which TDET1 expression is shown in fruits from a truss which displayed phenotypes ranging from wild type to fully hp. This analysis revealed that hp fruits had extremely low levels of TDET1 mRNA, that wild-type phenotype fruits had high levels of TDET1 transgene mRNA, and that blotchy fruits had intermediate levels (Figure 3d). Furthermore, in such cases the oldest fruits were visually dark green, and a gradient of phenotypic severity was often observed in fruits at different distances. This, as well as the observed sudden death of healthy plants in some cases (Figure 1g), inferred the systemic spread of a silencing signal from one area of the plant to another. Such a phenomenon has been found to occur during post-transcriptional gene silencing (PTGS) (Vaucheret and Fagard, 2001).
It was curious that no phenotypes characteristic of DET1 overexpression, i.e. light insensitivity, were ever observed, even with constructs designed to overexpress the full-length wild-type gene. This could be because the constructs were not functional, i.e. incapable of generating a functional protein, or because overexpression was lethal. To test this, the 35S::TDET1 construct was transformed into the hp-2 mutant. The fruit phenotypes of these plants demonstrated that the transgene could indeed complement the phenotypes of hp-2 mutants, confirming that it was functional (Figure 4). Nonetheless, in some cases complementation was only partial because fruits sometimes displayed sectors with normal pigmentation on a dark green background (Figure 4c). These observations were again reminiscent of TDET1 gene silencing.
Figure 4.

Complementation of hp-2 mutant phenotype with 35S::TDET1 construct.
(a) hp-2 mutant fruits.
(b) Fruits from a hp-2 mutant plant containing the 35S::TDET1 construct.
(c) Blotchy fruits from a hp-2 mutant plant containing the 35S::TDET1 construct, indicative of only partial complementation.
TDET1 silencing is not inherited but is acquired epigenetically from generation to generation
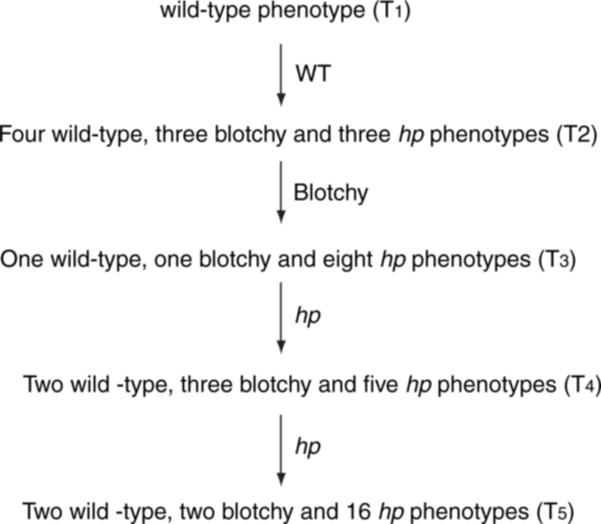
Analysis of MM and T56 transgenic plants from up to five generations showed that phenotypes often reappeared but that they were not inherited normally, e.g. seeds from a plant producing hp fruits gave rise to plants with both wild-type and hp phenotypes that segregated in a non-Mendelian fashion. For example, out of 10 plants sown in the T2 generation from a WT T1 plant, three produced hp fruits, three blotchy and four were phenotypically wild type (Figure 5). In the next generation, we obtained eight hp, one blotchy and one WT plant from seeds derived from a blotchy phenotype T2 plant. Furthermore, penetration of the hp phenotype increased in successive generations and sometimes reached 100% by the fifth generation (data not shown).
Figure 5.
Phenotypes of plants in successive generations.
The example shown is from an MM transformant containing a single copy of the 35S::hp2 construct. Phenotypes were scored in five successive generations. All plants were kanamycin-resistant and contained the TDET1 transgene (verified by PCR).
Following germination, seedlings invariably displayed wild-type phenotypes and the onset of the appearance of light hyper-responsive characteristics occurred during the course of development of the plant, typically after at least 1 month. Because loss of DET1 activity results in easily observable phenotypes in seedlings, e.g. shorter hypocotyls and increased anthocyanin content (Mustilli et al., 1999), these data indicate that the light hyper-responsive phenotype of TDET1-suppressed plants is not inherited but that it must be acquired each time by successive generations. The phenomenon therefore has an underlying epigenetic basis.
Light hyper-responsive phenotypes in transgenic plants are caused by PTGS of the TDET1 gene
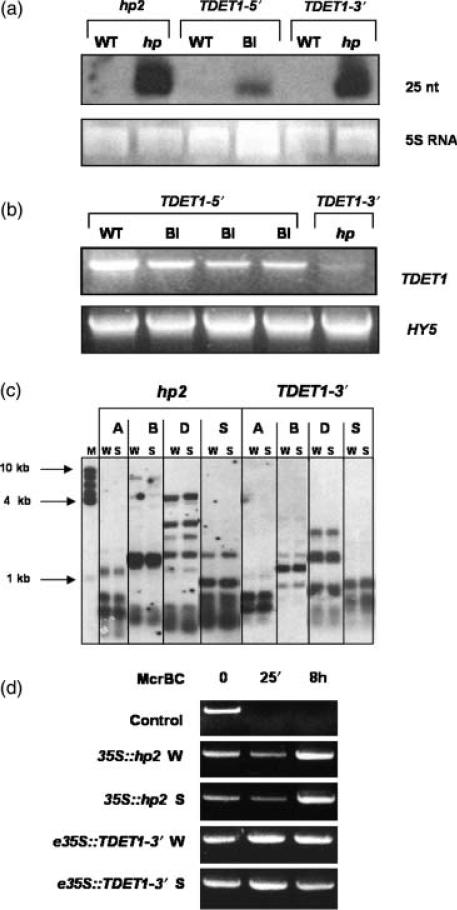
Systemic spread of gene silencing and its epigenetic inheritance are characteristic features of PTGS (Vaucheret and Fagard, 2001). A diagnostic molecular characteristic of PTGS is the appearance of short interfering RNAs (siRNAs) of between 21 and 25 nucleotides (Hamilton and Baulcombe, 1999). To detect these we isolated low molecular weight RNA from leaves and fruits of transgenic plants, ran it on denaturing polyacrylamide gels, blotted onto nylon membranes, and hybridized with an in vitro-transcribed TDET1 antisense probe. Figure 6 shows that we were indeed able to detect TDET1-derived siRNAs in plant material displaying hp phenotypes. The molecular size of the siRNAs was 25 nucleotides. Furthermore, the amounts of TDET1-derived siRNAs were directly proportional to the severity of the light hyper-responsive phenotypes and in particular were present at the highest levels in plants containing TDET1-3′ or the hp-2 mutant allele construct (Figure 6a). Interestingly, the TDET1-3′ construct was much more efficient at siRNA generation than the TDET1-5′ construct (Figure 6a), a result which was confirmed by examining eight transgenic plants from several independent lines containing each construct (data not shown). Conversely, semi-quantitative RT-PCR analysis of TDET1 mRNA levels showed that material from TDET1-5′ transgenic plants contained slightly more full-length TDET1 mRNA than did TDET1-3′-containing plants (Figure 6b). Levels were nonetheless lower than in plants with wild-type phenotypes. These results therefore demonstrate that the 3′-terminal construct was more efficient in generating siRNAs than was the 5′-terminal construct, which also correlated with the severity of the observed phenotypes (i.e. strong hp phenotypes were never observed in plants containing the TDET1-5′ constructs). The same result was observed with the hp-2 and hp-2j constructs with respect to full-length wild-type TDET1 constructs (data not shown).
Figure 6.
Molecular analysis of TDET1 gene silencing.
(a) siRNA analysis of transgenic lines overexpressing different TDET1 constructs. In vitro-synthesized TDET1 was used as probe. 5S rRNA is shown as loading control.
(b) RT-PCR analysis of transgenic lines containing e35S::TDET1 5′ and e35S:: TDET1 3′ constructs. Full-length TDET1 was synthesized using specific TDET1 primers and HY5 primers were used as control for cDNA synthesis and loading.
(c) Methylation analysis of genomic DNA from transgenic plants containing 35S::hp2 and e35S::TDET1 3′ constructs. Genomic DNA from wild type (W) and fully silenced (S) plants was digested with Alu1 (A), BglII/EcoRI (B), Dde1 (D) and Sau3A1 (S). Full-length TDET1 was used as probe. 1 kb size markers are indicated.
(d) McrPCR analysis of genomic DNA from transgenic plants containing 35S::hp2 and e35S::TDET1 3′ constructs. Genomic DNA from wild type (W) and fully silenced (S) plants was digested with McrBC at the indicated times and the TDET1 transgene subsequently amplified by PCR. A methylated control DNA is also shown.
35S::hp2 plants were MM genotype, whereas the e35S::TDET1-3′ plants were T56 genotype. The DNA used for methylation analysis was derived from plants from T4 generation, and results obtained were identical to those found in the T3 generation (data not shown).
Finally we tested whether silencing of the TDET1 gene was also caused by methylation of the TDET1 coding sequence, which is another characteristic of PTGS (Vaucheret and Fagard, 2001). We tested a range of methylation-sensitive restriction enzymes on genomic DNA extracted from plants containing either hp-2 or TDET1-3′ constructs. By Southern blot analysis we found no difference between silenced and wild-type plants, indicating that neither the TDET1 endogene nor the transgene were methylated (Figure 6c).
As a more efficient method to examine DNA methylation we used the McrPCR technique, which has been developed recently for high-throughput methylation analyses (Lippman et al., 2003). McrBC is an endonuclease which only cleaves DNA containing methylcytosine residues on one or both strands, so the successful amplification of DNA after digestion with McrBC indicates the lack of methylation. Genomic DNA (2 μg) from both wild-type and silenced plants was digested with McrBC for different times and 50 ng of genomic DNA was then used to amplify the TDET1 gene. Figure 6(d) shows the successful amplification of the TDET1 transgene from both wild-type and silenced plants, indicative of the absence of methylation. Similar analyses were performed using intron-derived primers to amplify the TDET1 endogene and we again did not find any evidence for methylation (data not shown). These results show that the observed light hyper-responsive phenotypes were correlated with DET1 mRNA degradation and siRNA generation but not through DNA hypermethylation.
Discussion
Although DET1 has been studied in both Arabidopsis and tomato (Mustilli et al., 1999; Pepper et al., 1994), its function is only partially understood (Benvenuto et al., 2002; Schroeder et al., 2002). In an attempt to obtain new insights into the role of this protein in a developmental context in tomato, we have generated several transgenic tomato plants containing a range of TDET1 constructs under the control of three different promoters. When phenotypes were visible they were invariably characteristic of exaggerated light sensitivity. Molecular examination of these plants indicated in all cases that the phenotypes were a result of suppression of endogenous TDET1 expression rather than the overexpression of wild-type or dominant negative versions of TDET1. Subsequent analyses showed that these phenotypes were caused by PTGS of TDET1.
Reiteration of light hyper-responsive high pigment phenotypes by suppression of TDET1 expression is consistent with the hypothesis that hp-2-mutant phenotypes are caused by reductions rather than alterations in TDET1 activity (Mustilli et al., 1999). This was further confirmed by complementation of the hp-2 mutant with a wild-type TDET1 gene (Figure 4). The importance of DET1 function at all stages of development was evident from the severity of phenotypes generated by suppression of its expression (Figure 1). On the contrary, plants overexpressing high levels of the transgene had wild-type phenotypes in all cases studied. These findings indicate that plants are very sensitive to reductions in DET1 levels but not to increased levels, perhaps because DET1 levels are already saturated. Similarly, in Arabidopsis overexpression of DET1 did not appear to confer any altered phenotypes (Schroeder et al., 2002).
Whereas DET1 silencing was often lethal when it occurred at a relatively early stage during development (Figure 1g), this was never the case in fruits. Even in cases of extreme silencing in fruits, the fruits remained healthy in spite of their extreme pigmentation. This finding could indicate that DET1 activity regulates essential processes in the vegetative parts of the plant but not in the fruits. In tomato fruits it is known that phytochromes can regulate pigment accumulation (Alba et al., 2000), although how it does so is unclear. They may simply regulate the expression of genes encoding pigment biosynthetic enzymes (Giuliano et al., 1993) or they may regulate plastid compartment size within the cells of the fruit (Cookson et al., 2003). The transgenic plants generated in this study could be useful for examining these possibilities.
Overexpression of either mutant (hp-2 and hp-2j) or truncated versions of the TDET1 gene generated the strongest silencing of TDET1, and caused the most severe developmental defects. Hence, it appears that expression of aberrant transgenes was much more efficient at inducing PTGS than were wild-type sequences. The fact that a sequence with a single point mutation (hp-2j) generated such severe phenotypes may be an indication of the impressive capacity of the plant cell's surveillance system to detect abnormal RNAs (Vaucheret and Fagard, 2001). Alternatively, the weaker phenotypes generated in e35S::TDET1 transgenic plants may be a result of the production of some viable DET1 protein. The severity of phenotypes in plants containing the 5′-terminal and 3′-terminal constructs was also different, in that the 3′-terminal constructs were able to generate much stronger phenotypes than the 5′-terminal constructs. Accordingly, severity of phenotype was clearly correlated with the amounts of 25 nt siRNAs, indicating that production of dsRNAs was more efficient from 3′ sequences of the TDET1 gene. Similar results were reported by Han and Grierson (2002), who found higher amounts of siRNAs derived from the 3′-terminus of a silenced polygalacturonase gene. These authors concluded that siRNAs are synthesized preferentially from the 3′-region of a transgene.
Because loss of DET1 activity results in dark green phenotypes, these plants are useful to follow the systemic spread of silencing within a plant. This was most clear in the fruits, and one such example is shown in Figure 3(d). When silencing was triggered within the mature fruits of one truss, a silencing signal appeared to travel to other fruits of the same truss, particularly the younger ones, and a clear gradient of phenotypes was observed, which was also confirmed by RT-PCR (data not shown). In many cases the fruits were not completely dark green, but appeared blotchy and remained so for several weeks and often up to complete maturity. On the contrary, in the vegetative parts of the plant, silencing appeared to spread to neighbouring parts much more quickly, generally towards the younger upper leaves. This may be because the systemic silencing signal travelled faster through phloem than from one cell to another. An alternative explanation could be that because tomato fruits are sink organs, signals tend not to be released efficiently from them. It has been proposed that phloem-mediated transmission of PTGS may be dependent upon the generation of 25 nt siRNAs, whereas long-range cell-to-cell transmission may require only 21 nt siRNAs (Hamilton et al., 2002; Himber et al., 2003). Assuming that long-range cell-to-cell transmission is responsible for our blotchy fruit phenotypes, the fact that we only observed siRNAs of the 25 nt size class does not support this hypothesis.
The silencing phenotype was not inherited from generation to generation, but had to be acquired in each successive generation (Figure 5). The wild-type phenotypes of progeny seedlings derived from silenced plants suggest that silencing was relieved during sexual reproduction and that siRNAs did not enter the seeds. A previous study of PTGS in tobacco reported similar observations (Mitsuhara et al., 2002).
Interestingly, our results indicate that PTGS of TDET1 acts only at the level of RNA degradation and not at the genomic level, because we were unable to find any evidence for hypermethylation of either the transgene or the endogene (Figure 6). This indicates that PTGS does not require DNA methylation per se, which had been previously inferred from other studies (Wang and Waterhouse, 2000). The lack of any evidence for DNA hypermethylation is nonetheless curious considering the increased penetrance of the silencing phenotype in successive generations (Figure 5), and demonstrates a lack of correlation between 25 nt siRNA production and DNA hypermethylation (Himber et al., 2003).
Finally, these results may have implications for the genetic improvement of plant nutritional quality. Tomatoes are known to be a rich source of health-promoting carotenoids and vitamins and are a multi-billion dollar global industry. In TDET1-silenced fruits, lycopene and β-carotene levels were increased dramatically. These results indicate that manipulation of TDET1 levels has promising biotechnological potential. However, the reduced yields and collateral negative effects on the vegetative parts of the plant must first be eliminated. Use of a fruit-specific promoter in combination with a TDET1 silencing construct may provide an ideal strategy, although the fruit-specific E8 promoter used in this study (Deikman et al., 1992) was clearly ineffective (Table 1). Alternative promoters or a more efficient silencing construct (e.g. containing inverted repeats; Wesley et al., 2001) may therefore be necessary.
In conclusion, the results presented here clearly support the idea that DET1 is an important negative regulator of light signalling that is required throughout the plant life cycle. Our results also indicate that DET1 may function differently during plant and fruit development, being essential in the former but not in the latter. Finally, it appears that the role of DET1 may be to ensure quantitatively appropriate responses to incoming light signals and that DET1 levels in wild-type cells are already sufficient for this, although DET1 is expressed at extremely low levels. The mechanistic significance of these findings in the context of the proposed role of DET1 in chromatin remodelling (Benvenuto et al., 2002) will be a major challenge to address in future research.
Experimental procedures
Plasmid construction
All expression cassettes were made in plasmid pBI121 (Bevan, 1984), which is a binary vector commonly used for Agrobacterium tumefaciens-mediated transformation of plant tissues (McCormick, 1991; Yoder et al., 1988). This vector contains NPT II as a marker gene encoding the neomycin phosphotransferase II gene for kanamycin resistance driven by the CaMV 35S promoter (Benfey and Chua, 1990). An additional 35S promoter is located upstream of a cloning site for the insertion of genes of interest. The full-length tomato DET1 gene (TDET1) (Mustilli et al., 1999) was first cloned in pBS-KS to generate suitable restriction sites and was subsequently cloned in pBI121 as a transcriptional fusion between the 35S promoter and the nos terminator as a BamH1-Sac1 fragment. This construct was denoted 35S::TDET1. Similarly, the hp-2 and hp-2j mutant versions of TDET1 (Mustilli et al., 1999) were cloned first in pBS-KS and then in pBI121. These constructs were denoted 35S::hp2 and 35S::hp2j. Two other constructs were designed to express N-terminal and C-terminal truncated versions of TDET1. The 5′ portion of the TDET1 gene up to nucleotide 1254 was cloned in pBI121 after first adding a stop codon by PCR at the end of the sequence. This construct was denoted 35S::TDET1-5′. A 3′ portion of TDET1 was cloned, starting from nucleotide 750, after adding a start codon at the 5′ end. This construct was denoted 35S::TDET1-3′. The primers used for generating the 5′-terminal construct were: GCG GCG AGC TCT TAA AAT GGT CGC TGA ACA G and TTA AAA ATG GTC GTC GAA CAG (to add the stop codon for 35S::TDET1-5′). The primers used for the 3′-terminal construct were: ATG TTC CAC CTT TTG AGG TTG GTG (to add the start codon for 35S::TDET1-3′) and GCG GCG AAT CCA TGT TCC ACC TTT TGA GGT TGG TG. The TDET1 gene was also inserted in the antisense orientation to generate 35S::TDET1-AS construct. In addition to the 35S-based antisense construct, we also generated a construct with the fruit-specific E8 promoter (Deikman et al., 1992). A 1.2 kb E8 promoter fragment was cloned in pBI121 between HindIII and BamH1 sites using 5′-GGGGAAGCTTTTTCACGAAATCGGCCCTTA-3′ and 5′-CCCGGATCCTTCTTTTGCACTGTGAATGATTAG-3′, and the full length TDET1 coding sequence was fused in either sense or antisense orientations downstream of the promoter as BamH1-Sac1 fragments. These constructs were denoted pE8::TDET1 and pE8:: TDET1-AS respectively. For stronger expression, the e35S promoter, which contains a duplicated 35S promoter sequence without the leader sequence was used. These constructs were made in an analogous fashion. All the constructs were introduced into A. tumefaciens strains LBA4404 or ABI for tomato plant transformation. A summary of constructs and results obtained is shown in Table 1.
Plant transformation
To generate transgenic tomato plants, cotyledons from 1 to 2-week-old seedlings were used for protocols as described by McCormick (1991) and Yoder et al. (1988). As wild-type backgrounds we used Money Maker and T56 genotypes. T56 is a commercial processing tomato variety. The Money Maker genotype was transformed as described by McCormick (1991) whereas T56 was transformed essentially according to Yoder et al. (1988). For T56, tomato seeds were sterilized using 2% commercial bleach and germinated on MSSV medium (Fillatti et al., 1987). Four to 7-day-old cotyledons were excised and placed onto freshly prepared tobacco feeder plates, prepared by decanting 1–2 ml of tobacco cells in suspension culture onto 2Z medium (Thomas and Pratt, 1981). After 48 h the cotyledons were immersed for 5 min in an overnight culture of A. tumefaciens strain ABI diluted to an A600 of 0.1. After 24 h the explants were plated onto 2Z medium containing 350 mg l–1 carbenicillin and 100 mg l–1 kanamycin sulphate. Excised shoots were placed onto rooting medium containing 50 mg l–1 kanamycin. Primary transformants were screened for the presence of the transgene by PCR and were transferred to soil. Adult plants were grown under shaded greenhouse conditions (25°C day/17°C night). It was observed that regenerated callus from transformation with some of the constructs was dark green, and that some of the regenerated plants were darker green and produced dark green immature fruit phenotypes (see text for details).
Molecular characterization of plants
DNA was isolated from young leaves using the CTAB method (Dellaporta et al., 1983). For methylation analysis, 10 μg of DNA was digested overnight with Alu1, BglII + EcoRI, Dde1, and Sau3A1 restriction enzymes. The digested DNA was run on 1.0% agarose gels and blotted to Hybond N+ membranes (Amersham Pharmacia Biotech, Little Chalfont, UK) overnight using 20X SSC. Filters were hybridized with random-primed full-length TDET1 probes labelled with α-32P-dCTP. Hybridizations were carried out overnight at 60°C in Church and Gilbert buffer (7% SDS, 0.5 m NaPO4 pH 7.2 and 1 mm EDTA) (Church and Gilbert, 1984). The washes were carried out using 0.1% SDS and 1X SSC at the same temperature twice for 10 min each.
For isolation of RNA tomato leaves or pericarp of immature fruits (0.2 g) were ground to a fine powder in liquid nitrogen and subsequently extracted using the hot phenol method (DellaPenna et al., 1986). RNA (10 μg) was loaded onto formaldehyde gels electrophoresed and blotted to Hybond N+ membranes using 10X SSC overnight. Hybridization was carried out overnight at 60°C using an α-32P-labelled TDET1 probe with Church and Gilbert buffer. The washes were performed in 0.1% SDS and 1X SSC twice at 60°C for 5 min each. Blots were stripped using hot SDS (1%) and reprobed with an α-32P-labelled CAB6 gene probe.
For isolating low molecular weight RNA, we used the protocols described by Hamilton and Baulcombe (1999) with small modifications. Low molecular weight RNA was separated from high molecular weight RNA using 10% PEG8000 and 0.5 m NaCl; 50 μg of low molecular weight RNA was run on 15% polyacrylamide gels containing 7 m urea and electoblotted onto Hybond NX membranes. In vitro-transcribed α-32P-labelled full-length TDET1 anti-sense RNA was used as a probe and hybridizations were carried out at 60°C in Church and Gilbert buffer for 24 h. The blots were washed with 0.1% SDS and 20 mm NaPO4 twice for 10 min each.
For RT-PCR analysis, poly A+ mRNA was isolated using Dynal mRNA extraction kits and protocols recommended by the manufacturers (Dynal Inc., Oslo, Norway). poly A+ RNA (100 ng) was used to synthesize first-strand cDNA using the Invitrogen Superscript First Strand Synthesis kit (Invitrogen Corporation, Carlsbad, CA, USA). From this 2 μl of first-strand cDNA was used for TDET1 amplification using specific primers 5′-GTATGATTCACTAGTTTAATGCTGCTGAAAG-3′ and 5′-CCCATACTAGTCGTCTTGGCACTCTATCAAG-3′. As a control for normalization, the HY5 gene (Oyama et al., 1997) was amplified using specific primers 5′-ATGCAAGAGCAAGCGACGGTTCTAT-3′ and 5′-GTCCACGTGTCCTTCCCTCCTTCA-3′.
For McrPCR, 2 μg of genomic DNA extracted from leaves of transgenic plants was digested with 30 units of McrBC enzyme as suggested by the supplier (New England Biolabs, Beverly, MA, USA). DNA was incubated at 37°C for 25 min and 8 h. As a control, premethylated DNA from the supplier of the enzyme was used. After digestion the enzyme was heat inactivated by incubating at 65°C for 20 min. DNA (50 ng) was then used to amplify the TDET1 transgene using primers 5′-GTATGATTCACTAGTTTAATGCTGCTGAAAG-3′ and 5′-CCCATACTAGTCGTCTTGGCACTCTATCAAG-3′ for the full-length transgene and 5′-GAAAGCAGCCGTTGCT-3′ and 5′-CCCATACTAGTCGTCTTGGCACTCTATC-3′ for the TDET1-3′ construct. After 25 PCR cycles the reaction was analysed on 1% agarose gels.
Biochemical analysis of fruits
For chlorophyll extraction, approximately 0.05 g of immature fruit peel was weighed and put in 5 ml DMSO. The tubes were incubated at 65°C for 48 h in the dark. Chlorophyll content was determined by measuring absorbance at 649 and 665 nm and concentrations were calculated using the equations for ethanol published by Lichtenthaler and Wellburn (1983). The analysis was repeated thrice, in each case with five fruits from each phenotype.
Fully red ripe greenhouse fruits were harvested between the 15 and 20 day post-breaker stage for biochemical analysis. Carotenoids were extracted from tomato puree using 15 ml of acetone/methanol (2:1) and 4 ml hexane. Following addition of approximately 21 ml cold saline water, samples were shaken vigorously and then centrifuged into separate phases. An aliquot of the hexane phase was taken and analysed on an HP 1050 system configured with a diode array detector. The carotenoids were separated on a Whatman Partisil 5 ODS-3 column using a solvent mix of 81.7% acetonitrile, 9.6% methanol, 5.4% isopropyl alcohol and 3.3% MQ water. Lycopene was detected at 504 nm and β-carotene at 450 nm. The Brix (Soluble Solids) value was measured on a Bellingham & Stanley RFM-91 refractometer (Jepson Bolton & Co. Ltd., Watford, UK) using filtered tomato puree. Sugars were extracted from tomato puree using ethanol (final concentration 80%). Following centrifugation, an aliquot of the extract was removed and analysed on an HP 1050 system configured with a refractometer. The sugars were separated on a Whatman Partisil PAC column using a solvent mix of 87% acetonitrile and 13% water.
Acknowledgements
This work was partially supported by funding from the European Union (contract QLK5-CT-2000-00357), the Italian Ministry for Research and Education (FIRB contract RBNE01CFKB) and the Italian Ministry of Agriculture and Forestry (contract EcoPom).
References
- Alba R, Cordonnier-Pratt MM, Pratt LH. Fruit-localized phytochromes regulate lycopene accumulation independently of ethylene production in tomato. Plant Physiol. 2000;123:363–370. doi: 10.1104/pp.123.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Chua N-H. The cauliflower mosaic virus 35S promoter: combinational regulation of transcription in plants. Science. 1990;250:959–966. doi: 10.1126/science.250.4983.959. [DOI] [PubMed] [Google Scholar]
- Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C. The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr. Biol. 2002;12:1529–1534. doi: 10.1016/s0960-9822(02)01105-3. [DOI] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:871–872. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Protocols for nucleic acids hybridizations. Proc. Natl Acad. Sci. USA. 1984;81:1991–1992. [Google Scholar]
- Cookson PJ, Kiano JW, Shipton CA, Fraser PD, Romer S, Schuch W, Bramley PM, Pyke KA. Increase in cell elongation, plastid compartment size and phytoene synthase activity underlie the phenotype of the high pigment-1 mutant of tomato. Planta. 2003;217:896–903. doi: 10.1007/s00425-003-1065-9. [DOI] [PubMed] [Google Scholar]
- Deikman J, Klone R, Fischer RL. Organization of ripening and ethylene regulatory regions in a fruit-specific promoter from tomato (Lycopersicon esculentum). Plant Physiol. 1992;100:2013–2017. doi: 10.1104/pp.100.4.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Alexander DC, Bennett AB. Molecular cloning of tomato fruit polygalacturonase: analysis of polygalacturonase mRNA levels during ripening. Proc. Natl Acad. Sci. USA. 1986;83:6420–6424. doi: 10.1073/pnas.83.17.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA mini-preparation: version II. Plant Mol. Biol. Rep. 1983;1:19–21. [Google Scholar]
- Fankhauser C, Bowler C. Biochemical and molecular analysis of signaling components. In: Schäfer E, Nagy F, editors. Photomorphogenesis in Plants and Bacteria. Kluwer Academic Publishers; The Netherlands: 2004. In press. [Google Scholar]
- Fillatti JJ, Kiser J, Rose R, Comai L. Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Bio/Technology. 1987;5:726–730. [Google Scholar]
- Giuliano G, Bartley GE, Scolnik PA. Regulation of carotenoid biosynthesis during tomato development. Plant Cell. 1993;5:379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A novel species of small antisense RNA in post-transcriptional gene silencing. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Grierson D. Relationship between small antisense RNAs and aberrant RNAs associated with sense transgene mediated gene silencing in tomato. Plant J. 2002;29:509–519. doi: 10.1046/j.1365-313x.2002.01236.x. [DOI] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kerckhoffs LHJ, van Tuinen A, Koornneef M. Photomorphogenic mutants of tomato. Plant Cell Environ. 1997;20:746–751. [Google Scholar]
- Lichtenthaler HK, Wellburn AR. Determinations of total carotenoids and chlorophyll a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983;11:591–592. [Google Scholar]
- Lieberman M, Segev O, Gilboa N, Lalazar A, Levin I. The tomato homolog of the gene encoding UV-damaged DNA binding protein 1 (DDB1) underlined as the gene that causes the high pigment-1 mutant phenotype. Theor. Appl. Genet. 2004;108:1574–1581. doi: 10.1007/s00122-004-1584-1. [DOI] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biology. 2003;1:420–428. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J. Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl Acad. Sci. USA. 2004;101:9897–9902. doi: 10.1073/pnas.0400935101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. Transformation of tomato with Agrobacterium tumefaciens. Plant Tissue Culture Manual. 1991;B6:1–9. [Google Scholar]
- Mitsuhara I, Shirasawa-Seo N, Iwai T, Nakamura S, Honkura R, Ohashi Y. Release from post-transcriptional gene silencing by cell proliferation in transgenic tobacco plants: possible mechanism for noninheritance of the silencing. Genetics. 2002;160:343–352. doi: 10.1093/genetics/160.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C. Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell. 1999;11:145–157. doi: 10.1105/tpc.11.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- Piringer AA, Heinze PH. Effect of light on the formation of a pigment in the tomato fruit cuticle. Plant Physiol. 1954;29:467–472. doi: 10.1104/pp.29.5.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. Photosensory perception and signalling in plant cells: new paradigms? Curr. Opin. Cell Biol. 2002;14:180–188. doi: 10.1016/s0955-0674(02)00309-5. [DOI] [PubMed] [Google Scholar]
- Schäfer E, Bowler C. Phytochrome-mediated photo-perception and signal transduction in higher plants. EMBO Rep. 2002;3:1042–1048. doi: 10.1093/embo-reports/kvf222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 2002;12:1462–1472. doi: 10.1016/s0960-9822(02)01106-5. [DOI] [PubMed] [Google Scholar]
- Serino G, Deng X-W. The COP9 signalosome: regulating plant development through the control of proteolysis. Annu. Rev. Plant Biol. 2003;54:165–182. doi: 10.1146/annurev.arplant.54.031902.134847. [DOI] [PubMed] [Google Scholar]
- Thomas BR, Pratt D. Efficient hybridization between Lycopersicon esculentum and L. Peruvianum via embryo callus. Theor. Appl. Genet. 1981;59:215–219. doi: 10.1007/BF00265495. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Fagard M. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 2001;17:29–35. doi: 10.1016/s0168-9525(00)02166-1. [DOI] [PubMed] [Google Scholar]
- Wang MB, Waterhouse PM. High-efficiency silencing of a beta-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation. Plant Mol. Biol. 2000;43:67–82. doi: 10.1023/a:1006490331303. [DOI] [PubMed] [Google Scholar]
- Weller JL, Perrotta G, Schreuder ME, van Tuinen A, Koornneef M, Giuliano G, Kendrick RE. Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochromes A, B1 and B2. Plant J. 2001;25:427–440. doi: 10.1046/j.1365-313x.2001.00978.x. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Yoder JI, Palys J, Alpert K, Lassner M. Ac transposition in transgenic tomato plants. Mol. Gen. Genet. 1988;213:291–296. [Google Scholar]