Abstract
Sarcospan is a component of the dystrophin-glycoprotein complex that forms a tight subcomplex with the sarcoglycans. The sarcoglycan-sarcospan subcomplex functions to stabilize α-dystroglycan at the plasma membrane and perturbations of this subcomplex are associated with autosomal recessive limb-girdle muscular dystrophy. In order to characterize protein interactions within this subcomplex, we first demonstrate that sarcospan forms homo-oligomers within the membrane. Experiments with a panel of site-directed mutants reveal that proper structure of the large extracellular loop is an important determinant of oligo formation. Furthermore, the intracellular N- and C-termini contribute to stability of sarcospan-mediated webs. Point mutation of each cysteine residue reveals that Cys 162 and Cys 164 within the large extracellular loop form disulfide bridges, which are critical for proper sarcospan structure. The extracellular domain of sarcospan also forms the main binding site for the sarcoglycans. We propose a model whereby sarcospan forms homo-oligomers that cluster the components of the dystrophin-glycoprotein complex within the membrane.
Keywords: sarcospan, sarcoglycan, dystrophin-glycoprotein complex, tetraspanin, oligomerization
INTRODUCTION
In skeletal muscle, the dystrophin-glycoprotein complex (DGC) 1 is located at the sarcolemma and is composed of peripheral and integral membrane proteins [1-5]. As a whole, the DGC links the extracellular matrix (ECM) to the intracellular actin cytoskeleton [6] and provides structural stability to the sarcolemma during muscle contraction [7, 8]. In addition to dystrophin, the core components of the DGC include the dystroglycans (α- and β-DG), the sarcoglycans (α-, β-, δ-, and γ-SG), syntrophin, and sarcospan (SSPN) (for review, see [9]). The SGs are single-pass transmembrane proteins that form a tight subcomplex with SSPN [10, 11]. Dystrophin binds β-DG and F-actin, thereby creating a structural axis across the membrane bilayer [12-14]. α-DG is a receptor for extracellular matrix proteins such as laminin, agrin, perlecan, and neurexin. Disruption of structural connections (either laterally within the bilayer or across the bilayer) results in muscle disease and underscores the importance of protein-protein interactions within the DGC.
Several forms of muscular dystrophy are caused by primary mutations in the genes encoding components of the DGC [15-19]. Primary mutations in the dystrophin gene cause Duchenne muscular dystrophy (DMD), which is characterized by the loss of dystrophin protein and concomitant loss of the entire DGC. Autosomal recessive limb-girdle muscular dystrophies (AR-LGMD) 2C-2F result from mutations in the SG genes that lead to either partial or complete loss of the SGs (for review, [20]). The SGs are tightly associated with SSPN to form a SG-SSPN subcomplex within the DGC [10, 11, 21]. In either partial or complete SG-deficiency, SSPN is completely absent from the sarcolemma, indicating that the entire tetrameric SG-subcomplex is required for proper membrane localization and stabilization of SSPN [10, 11]. The SG-SSPN subcomplex contributes to DGC complex stability by anchoring α-DG at the membrane [22-24]. Without the SG-SSPN subcomplex, the sarcolemma is very fragile and more susceptible to contraction-induced injury (for review, [20]). As a tetraspanin-like protein, SSPN is poised to serve as an important stabilizing molecule within the DGC [25]. Although several classes of proteins contain four transmembrane domains, dendrogram analysis suggests that SSPN is most closely related to the tetraspanin superfamily [25]. Tetraspanins possess four transmembrane domains (TM), a small extracellular loop (SEL), and four conserved cysteine residues within a large extracellular loop (LEL) [26-29]. Tetraspanins interact with a diverse array of proteins including integrins, immunoreceptors, and signaling molecules (for review [30, 31]). The primary role of tetraspanins is to organize proteins at the cell surface into unique membrane microdomains referred to as ‘tetraspanin webs’ (for review [31]). Tetraspanin webs arise from the interaction of several tetraspanins with each other through either homo- or hetero-oligomerization. By clustering proteins at the cell surface, tetraspanins control a wide range of cellular functions including adhesion, motility, and signaling (for review, [26]). Defects in tetraspanin function are associated with many diseases including X-linked mental retardation [32], lymphoid B-cell abnormalities [33], and tumor metastasis [34]. In addition, tetraspanin CD81 serves as a co-receptor for the hepatitis C virus [35].
We hypothesize that SSPN functions similar to tetraspanins within the sarcolemma. As a first test of this model, we sought to determine whether SSPN forms protein webs within the plasma membrane. Using independent methods, we found that SSPN homo-oligomerizes through critical domains located in the intracellular regions and the LEL. SSPN mutants were employed in order to delineate the key regions of SSPN that are important for mediating interactions with the SGs. A model is proposed in which SSPN functions as a facilitator of protein interactions within the DGC.
MATERIALS AND METHODS
SSPN Transgenic Mice
Transgenic constructs were engineered with the human skeletal α-actin promoter and the VP1 intron upstream of human SSPN, using previously described methods [36, 37]. Skeletal muscle from 4-week old SSPN transgenic mice (SSPN-TG, line 31.6) was homogenized in 10 volumes of lysis buffer (3% SDS, 0.113 M sucrose, and 0.066 M Tris-HCl, pH 6.8). Prior to homogenization, protease inhibitors (0.6 μg/mL pepstatin A, 0.5 μg/mL aprotinin, 0.5 μg/mL leupeptin, 0.75 mM benzamidine, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF)) were added to the lysis buffer. Samples were rotated at 4°C for 1 h and centrifuged at 15,000 × g for 15 min to clear lysates. 60 μg of protein lysates were loaded onto 5-20% SDS-polyacrylamide gradient gels under reducing (loading sample buffer containing 0.25% (7.13 mM) β-mercaptoethanol) or non-reducing (loading sample buffer without β-mercaptoethanol) conditions. Samples were boiled for 5 min. prior to electrophoresis. Gels were transferred to nitrocellulose (Millipore) and immunoblotted with affinity-purified, polyclonal antibodies to human SSPN (Rabbit 15; 1:250).
Immunoprecipitation from C2C12 cells
Before beginning the immuonoprecipitation protocol, MBP (New England Biolabs) and SSPN (Rabbit 18) antibodies were covalently cross-linked to Protein-G separose (Amersham Pharmacia) beads using dimethyl pimelimidate (DMP; Sigma). Clarified 1% digitonin solubilized C2C12 myoblast cell lysates (0.9 mg) were incubated with SSPN or MBP cross-linked beads for 18 h, at 4°C. Immobilized immune complexes were then washed with 0.1% digitonin. Immunoprecipitated proteins were eluted with 3% SDS loading sample buffer. Precipitated proteins were then analyzed by SDS-Page and immunoblotted as described below.
SSPN Recombinant Fusion Proteins
GST Fusions
GST-NT was engineered by subcloning a cDNA fragment encoding amino acids 1-26 of murine SSPN into BamHI and EcoRI sites of the pGEX4T1 vector (Amersham-Pharmacia Biotech Inc, Piscataway, NJ). Proteins expressed from the pGEX4T1 without an insert (GST) or with SSPN N-terminus (GST-NT) have molecular masses of 27.9- and 29.3- kDa, respectively. GST fusion proteins were purified from BL21 E. coli (Stratagene, La Jolla, CA) using protocols supplied by the manufacturer (Amersham-Pharmacia). In brief, recombinant protein expression was induced in bacteria during exponential growth by treatment with 1 mM isopropyl-1-thio-β-D-galactopyranoside at 37 °C for 3 h. The cells were harvested and lysed by sonication in PBS plus 1% Triton X-100. Clarified lysates were affinity purified using glutathione-sepharose column chromatography.
MBP Fusions
cDNA fragments encoding amino acids 1-26 (N-terminus), 115-157 (LEL), or 186-216 (C-terminus) of murine SSPN were subcloned into BamHI and PstI sites of the pMALC4 vector (New England Biolabs, Beverly, MA) in order to create MBP-NT, MBP-LEL, and MBP-CT recombinant fusion proteins, respectively. The predicted molecular mass of each fusion protein is as follows: MBP (50.8-kDa), MBP-NT (45.9-kDa), MBP-LEL (47.9-kDa), and MBP-CT (46.7-kDa). MBP fusion proteins were purified as described above except that cells were lysed by sonication in cold MBP buffer (20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, 1% Triton X-100). Clarified lysates were applied to amylose sepharose. After washing, purified fusion proteins were eluted with 10 mM maltose in MBP buffer, according to the manufacturer's instructions (New England Biolabs).
Overlay Assay
Approximately 40 μg of purified fusion protein was subjected to 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were transferred to PVDF, blocked in blotto (5% non-fat dry milk in TBST), and incubated with approximately 50 μg of fusion protein in 1% BSA plus TBST overnight. Subsequently, membranes were washed in TBST and incubated with either a polyclonal antibody to the C-terminus of mouse SSPN (Rabbit 18; 1:100) or anti-GST monoclonal antibodies for 1h at RT. Immunoblots were washed and incubated for 1 h with the appropriate horseradish peroxidase-conjugated secondary antibodies at a dilution of 1:500 (Amersham-Pharmacia, Piscataway, NJ). After extensive washing in TBST, immunoreactive bands were detected using the West Pico Enhanced Chemiluminescence Protocol (Pierce, Rockford, IL).
SSPN Mutant Construction
Deletion and point mutants were engineered using the Quikchange Protocol (Stratagene, La Jolla, CA). Reverse complimentary PCR primers were designed to span the 5’ and 3’ regions flanking the desired 18 nt (6 codons) region for deletion. Mutant plasmids were generated by PCR amplification of a pcDNA3 expression construct containing human SSPN cDNA (pcDNA3.SSPN) using primers with engineered mutations. PfuTurbo DNA polymerase (Stratagene) was used for PCR. The methylated, parental pcDNA3.SSPN was removed by digestion with DpnI. The remaining, mutant plasmid was transformed into competent XL1-blue E. coli cells. To facilitate protein detection, all SSPN and SG constructs were designed to express proteins with a myc-tag at the C-terminus with the exception of ΔTM4-CT. Mutant ΔTM4-CT was cloned into the pCMV-Tag2A vector (Stratagene), which was engineered to express a flag epitope tag followed by the N-terminus of SSPN (amino acids 1-157). The sequences of all constructs were verified by direct DNA sequence analysis performed by the UCLA Sequencing Core Facility (Los Angeles, CA) in order to confirm that the reading frame was preserved across the deletion and that the plasmids were void of unintended mutations.
Transient Transfections of CHO cells
CHO cells were electroporated with wild-type (WT), mutant SSPN, or SG pcDNA3 expression constructs (~15 μg of each plasmid DNA) at 340 V and 950 μF using a BioRad electroporator (Biorad, Hercules, CA), as previously described [10, 22]. 30 h after transfection, proteins were extracted in cold lysis buffer containing 50 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 1% digitonin (Biosynthag AG, Staad, Switzerland), plus the following protease inhibitors: 0.6 μg/ml pepstatin A, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.1 mM PMSF, 0.75 mM benzamidine. Lysates were clarified by centrifugation at 14,000 × g for 20 mins.
Sucrose Gradient Fractionation
Transfected CHO cell lysates (300 μg) were layered on top of 10-45% sucrose gradients prepared by adding 8 mls of a 10% sucrose solution into an open top Polyclear™ centrifuge tubes (Seton, Los Gatos, CA) using a 14 gauge Hamilton syringe and underlaying with an equal volume of a 45% sucrose solution. Gradients were mixed using the Biocomp Gradient ip station (Biocomp, Fredericton, Canada). Sucrose gradients were centrifuged at 150,000 × g in a Beckman Optima L-90K ultracentrifuge (Beckman, Palo Alto, CA) for 18 h at 4°C using an SW41 rotor. Fourteen 1-ml fractions were collected and 80 μl of each fraction was analyzed by SDS-PAGE and immunoblotting. Migration distances of native, high molecular mass standards (Amersham Bioscience, Piscataway, NJ) were determined by applying markers to sucrose gradients.
Protein Gel Electrophoresis and Immunoblotting
Protein samples were treated with either reducing (3% SDS, 0.115 M sucrose, 50 mM Tris-HCl, pH 7.4, 1% β-mercaptoethanol) or non-reducing (3% SDS, 0.115 M sucrose, 50 mM Tris-HCl, pH 7.4, 2% N-ethyl-maleimide) sample buffer. Samples were electrophoresed through isocratic (12%) or gradient (5-15%) polyacrylamide gels. To visualize proteins, gels were stained with Coomassie blue (50% methanol, 10% glacial acetic acid, 2% Coomassie brilliant blue R250). For immunoblotting, gels were transferred to either PVDF or nitrocellulose membranes (Millipore, Billerica, MA). Membranes were blocked for 1 h in 3% blotto buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.2% Tween-20, 3% non-fat dry milk) and incubated overnight with primary antibodies. Immunoblots were washed in blotto and incubated for 1 h with the appropriate horseradish peroxidase-conjugated secondary antibodies at a dilution of 1:500 (Amersham-Pharmacia). Immunoblots were washed in TBST and immunoreactive bands were detected as described above.
Immunoprecipitation from CHO cells
Protein extracts were prepared from CHO cells transfected with SG and SSPN constructs, as described above. Lysates were first precleared by incubating with Protein-G sepharose (Amersham-Pharmacia) for 1 h at 4°C. Sepharose beads and any non-specifically bound proteins were removed by brief centrifugation at 9,000 × g. Precleared supernatants (100 μl) were used for immunoprecipitation with δ-SG antibody (NCL-d-SG, Novocastra laboratories Ltd, Newcastle-upon-Tyne, United Kingdom), as previously documented [10, 38]. Protein samples were separated by reducing SDS-PAGE and transferred to PVDF membranes for immunoblotting with anti-(c-myc)-peroxidase antibody (1:500, Roche, Indianapolis, IN), which recognizes the myc tag on SSPN and all four SGs.
RESULTS AND DISCUSSION
The important role of tetraspanins in cell adhesion, proliferation, migration and fusion, as well as host-parasite interactions is a newly emergent area of biology. By associating laterally with one another in the plasma membrane, tetraspanins function to cluster membrane proteins and promote protein-protein interactions that control a variety of cell activities (for review [30]). We sought to determine whether SSPN, like nearly all tetraspanins, is able to form oligomers within the membrane bilayer.
Oligomerization of SSPN in Skeletal Muscle
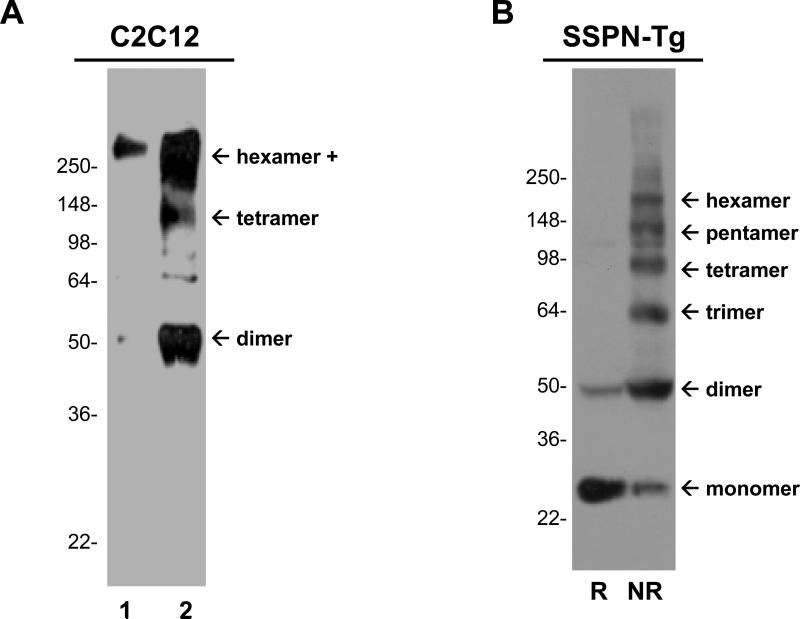
As a first test for the ability of SSPN to form oligomers, we analyzed SSPN isolated from skeletal muscle myoblasts under non-reducing conditions to maintain potential disulfide conditions. C2C12 myoblasts are proliferating mononuclear cells that express SSPN and DG, but do not express the tetrasmeric SGs. Cell lysates were subjected to immunoprecipitation with SSPN antibodies to facilitate SSPN detection. Immunoblots demonstrate that SSPN forms higher molecular mass structures that migrate at sizes consistent with dimer (50-kDa) and tetramer (100-kDa) homo-oligomers (Fig. 1A). These structures were not visualized with a negative control polyclonal antibody (Fig. 1A). Antibodies were covalently cross-linked to the Protein-G matrix to assure that the antibody used for immunoprecipitation was not present in the eluate fractions. As a second test for SSPN-SSPN interaction, we analyzed SSPN protein extracted from skeletal muscle under non-reducing conditions to maintain potential disulfide interactions. For these experiments, a transgenic mouse model that was engineered to overexpress low levels of human SSPN (SSPN-TG) was employed. SSPN-Tg mice (line 31.6) that express approximately 2 to 3-fold more SSPN compared to non-transgenic (non-Tg) controls do not display any muscle pathology. Protein lysates from SSPN-Tg muscle were subjected to SDS-PAGE and immunoblotting with antibodies specific to exogenous, human SSPN. In the absence of reducing agents, SSPN displays an oligomerization pattern consisting of dimers (50-kDa), trimers (75-kDa), tetramers (100-kDa), pentamers (125-kDa), and hexamers (150-kDa) (Fig. 1B). Treatment of muscle protein lysates with β-mercaptoethanol restores SSPN to its reduced, 25-kDa monomeric state (Fig. 1B). We were unable to perform these experiments with non-Tg muscle due to the low-titer of our antibodies to endogenous mouse SSPN.
Figure 1. Thiol-mediated oligomerization of SSPN.
A, Skeletal muscle C2C12 myoblasts were subjected to immunoprecipitation under non-reducing conditions. Myoblasts express SSPN, dystrophin, and the DGs, but do not express any of the SG subunits. Myoblasts were incubated with protein-G sepharose that was covalently cross-linked with non-specific polyclonal antibodies as a negative control (lane 1) or with SSPN antibodies (lane 2). Protein eluates were subjected to SDS-PAGE and immunoblotting with SSPN antibodies. B, Examination of SSPN-SSPN oligomers in skeletal muscle from transgenic mice that were engineered to over-express SSPN at very low levels (2-3-fold). The SSPN-transgenic (SSPN-Tg) mice did not display any obvious muscle phenotype. 60 μg of skeletal muscle lysates were electrophoretically separated on 5-20% SDS-polyacrylamide gradient gels in either reducing (R) or non-reducing (NR) conditions. Reducing conditions were created by the addition of 7 mM β-mercaptoethanol during electrophoresis. Immunoblot transfers were probed with anti-human SSPN antibodies. Under reducing conditions, SSPN migrates at its predicted molecular mass of 25-kDa. In the absence of reducing agent, SSPN forms monomers (25-kDa), dimers (50-kDa), trimers (75-kDa), tetramers (100-kDa), pentamers (125-kDa), and hexamers (150-kDa). Molecular size markers (x103 Da) are indicated on the left.
Use of recombinant proteins to analyze SSPN-SSPN interactions
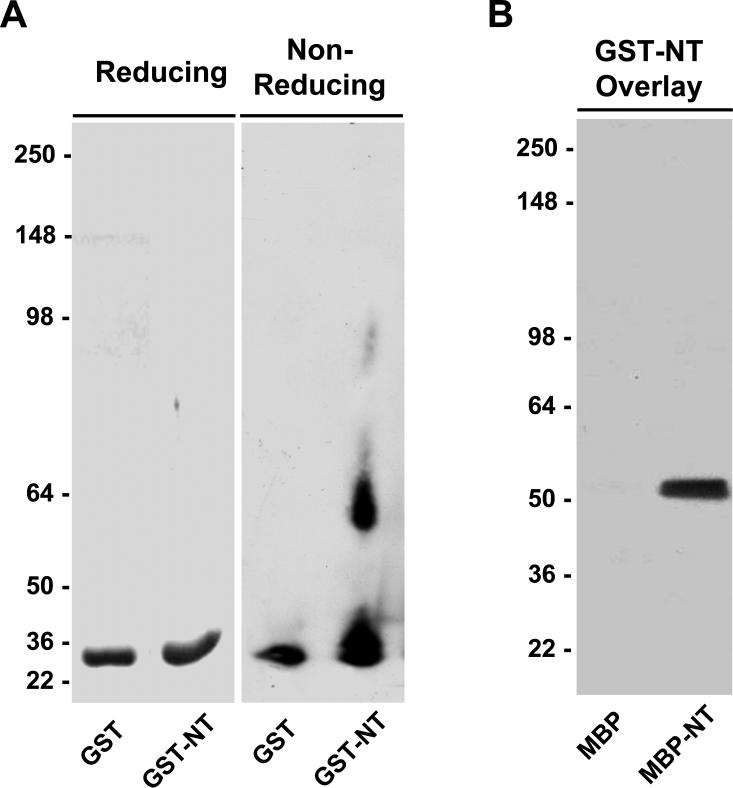
Studies with site-directed mutants demonstrate that tetraspanin-tetraspanin interactions are mediated by multiple intermolecular binding sites (reviewed by [39]). In order to first identify regions of SSPN that are critical for mediating oligomerization, we analyzed recombinant fusion proteins designed to represent the N-terminal (NT), large extracellular loop (LEL), and C-terminal (CT) regions of SSPN. Fusions were created to maltose binding protein (MBP) or glutathione-S-transferase (GST). Recombinant proteins were expressed in bacteria at robust levels without forming insoluble inclusion bodies. Purified GST fusion proteins were subjected to analysis under reducing and non-reducing polyacrylamide gel electrophoresis. In both of these conditions, GST migrated as a monomer (Fig. 2A). However, dimerization occurred under non-reducing conditions when the NT region of SSPN was fused to GST (Fig. 2A). Protein overlay assays were employed to further analyze interactions between the N-terminal regions of SSPN. Purified GST-NT fusion proteins were incubated with nitrocellulose transfers containing reduced MBP and MBP-NT. Binding of GST-NT was detected by probing overlays with GST antibodies. GST-NT interacted with MBP-NT, but not MBP alone (Fig. 2B). This data supports a role for intermolecular N-terminal interactions between SSPN molecules.
Figure 2. SSPN N-terminus constitutes dimerization domain.
A, Recombinant proteins representing glutathione-S-transferase (GST) fused to the N-terminus of SSPN (GST-NT) were analyzed for oligomerization. Under reducing conditions both GST and GST-NT migrated at the predicted molecular masses of 27.9- and 29.3-kDa, respectively. Under non-reducing conditions, purified GST-NT exhibited robust dimerization. B, Overlay assays were performed by incubating 50 μg/ml of purified GST-NT fusion protein with nitrocellulose transfers containing MBP (50.8 kDa) alone and MBP-NT (45.9 kDa). Binding of GST-NT was detected using antibodies that cross-react with the GST tag. GST-NT specifically binds MBP-NT and not MBP alone. Molecular size markers (x 103 Da) are indicated on the left.
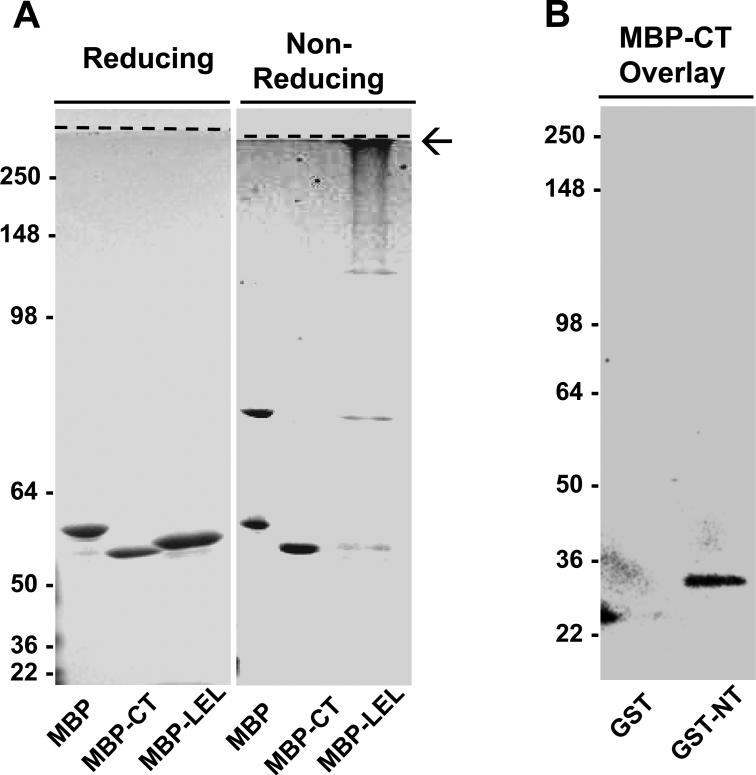
Analysis of SSPN C-terminal (CT) fusion proteins was also performed by reducing and non-reducing polyacrylamide gel electrophoresis. Under non-reducing conditions, MBP alone formed monomers and dimers (Fig. 3A). MBP dimer formation was inhibited when MBP was fused to SSPN C-terminus (MBP-CT, Fig. 3A). In addition, GST-CT did not bind to MBP-CT in overlay assays (data not shown), suggesting that the C-terminal regions of SSPN do not interact with each other. However, the C-terminus of SSPN was found to interact with the SSPN N-terminal region in protein overlay assays. MBP-CT was overlayed onto nitrocellulose transfers of GST and GST-NT. Specific binding of MBP-CT to GST-NT was detected using SSPN antibodies that cross-react with the C-terminal region of SSPN (Fig. 3B). These data suggest that the intracellular C-terminus alone is incapable of oligomerization, but that the N-terminus can associate with both N-and C-terminal regions.
Figure 3. Identification of two interaction sites within SSPN: N-terminus to C-terminus and LEL to LEL.
A, Recombinant proteins engineered by the fusion of maltose binding protein (MBP) to either the N-terminus (MBP-NT), the large extracellular loop (MBP-LEL), and the C-terminus (MBP-CT) of SSPN were analyzed for oligomerization. Purified fusion proteins were separated by 15% SDS-PAGE under reducing or non-reducing conditions, as indicated above each panel. Polyacrylamide gels were stained with Coomassie-blue for protein detection. Under non-reducing conditions MBP, MBP-CT and MBP-LEL fusion proteins migrated at the predicted molecular masses of 50.8-kDa, 46.7-kDa and 47.9-kDa, respectively. However, under non-reducing conditions, the MBP-LEL showed dramatic aggregation (arrow) and accumulated at the interface between the resolving and stacking gels (dashed line). MBP-CT did not exhibit thiol-mediated oligo formation. B, Overlay assays were performed by incubating 50 μg/ml of purified MBP-CT fusion protein with nitrocellulose transfers containing GST and GST-NT. Anti-SSPN C-terminal antibodies were employed for the detection of MBP-CT binding. MBP-CT displayed interaction with GST-NT. The predicted molecular mass of GST and GST-NT migrated is 27.9- and 29.3-kDa, respectively. Molecular weight markers are indicated on each blot (x 103 Da).
The LEL region of SSPN exhibited dramatic aggregation under non-reducing conditions, as illustrated by accumulation of MBP-LEL fusion protein at the interface between the resolving and stacking gels (Fig. 2A). Protein overlay assays between individual LEL domains were not possible since GST-LEL fusion proteins could not be purified without severe protein degradation. Taken together, our data suggest that SSPN-SSPN clustering is mediated by several points of contact, including NT to NT, NT to CT, and LEL to LEL.
Site-directed mutagenesis of SSPN reveals complexity in SSPN web formation
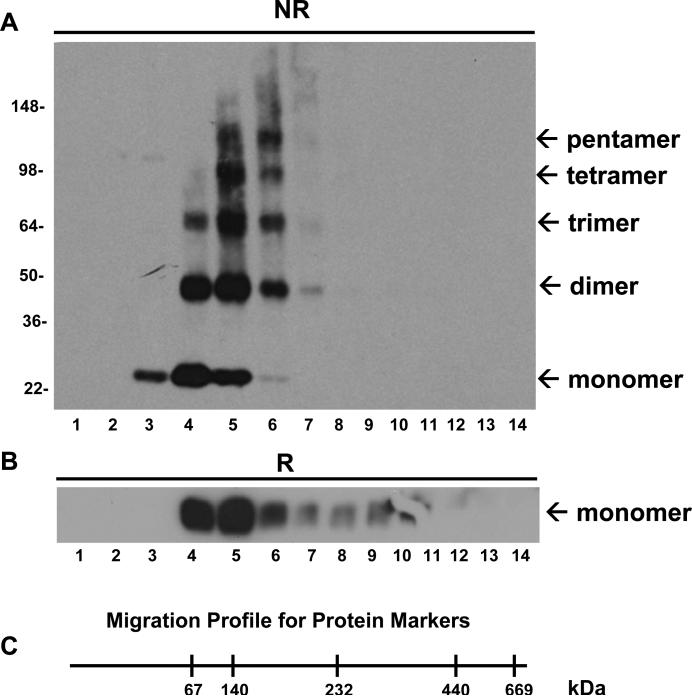
In order to further define regions of SSPN that are critical for oligomer formation, we utilized an in vitro CHO cell expression system so that we could express mutant polypeptides of SSPN. Although artificial introduction and expression of proteins into cultured cells does not always replicate the in vivo state, such approaches can serve as a ‘next-best’ model and provide important insights into protein interactions. We demonstrate that wild-type SSPN forms thiol-mediated oligomers in cultured CHO cells. Expression vectors encoding human SSPN with a myc-tag engineered at the C-terminus were introduced into CHO cells, which do not express endogenous SSPN or SGs [10, 38]. Proteins were extracted with digitonin and subjected to centrifugation through a 10-45% linear sucrose gradient under non-reducing conditions. Analysis of fractions by immunoblotting with SSPN antibodies reveals the presence of several distinct species of SSPN (Fig. 4A). SSPN oligomers exhibit a characteristic tetraspanin migration profile during sucrose gradient centrifugation. SSPN migrates to fractions 3 to 6, which represent the low-density sucrose fractions where tetraspanin webs accumulate [40]. Analysis of molecular mass markers on non-denaturing sucrose gradients reveals that proteins of 60-200 kDa migrate within fractions 3 to 6, which is consistent with the predicted mass of SSPN oligomers (Fig. 4C). Under non-reducing conditions, SSPN exists as a monomer (25-kDa), dimer (50-kDa), trimer (75-kDa), and tetramer (100-kDa). Pentamer (125-kDa) structures were also evident, although at very low levels. Upon treatment with β-mercaptoethanol, these higher order species were completely reduced to a single band at the expected mass of the SSPN monomer (Fig. 4B). Identical results were obtained when immunoblots were probed with antibodies to the myc-tag, which was engineered onto the C-terminus of SSPN (data not shown).
Figure 4. Reconstitution of SSPN oligomerization.
A, SSPN oligomerization was reconstituted using a heterologous cell expression system. CHO cells were transfected with expression vectors encoding myc-tagged human SSPN. Whole cell protein lysates (100 μg) were applied to 10-45% linear sucrose gradients without the addition of reducing agents. Protein fractions were immunoblotted under non-reducing (NR) conditions with anti-SSPN antibodies. SSPN protein migrates at monomer, dimer, trimer, and tetramer masses. Minor pentamer (125-kDa) formation was observed. B, Protein lysates were also subjected to reducing SDS-PAGE and immunoblotting with SSPN antibodies (R) to demonstrate that SSPN migrates as a monomer (25-kDa) under reducing conditions. Molecular weight markers are indicated (x 103 Da). C, Migration distances of native, high molecular mass standards on sucrose gradients under non-reducing conditions are shown. Molecular masses are provided in kDa.
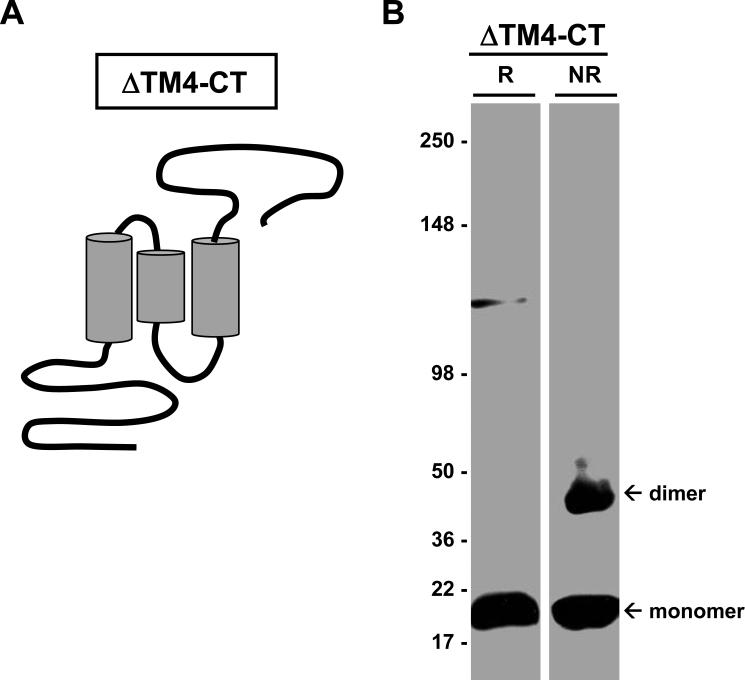
In order to further localize regions within SSPN that contribute to dimerization, we utilized the in vitro CHO cell expression system to analyze a panel of SSPN mutants created by site-directed mutagenesis. We first engineered deletion mutants to remove large regions within SSPN, including the N-terminus, each transmembrane domain, the large extracellular loop, and the C-terminus. Of the many mutants created, only one mutant (ΔTM4-CT) was stably expressed in CHO cells. Mutant ΔTM4-CT lacks the entire TM4 and the CT region, as illustrated in Figure 5A. CHO cells transfected with ΔTM4-CT were analyzed by sucrose gradient centrifugation and immunoblotting with SSPN antibodies. Under non-reducing conditions, ΔTM4-CT forms robust monomers and dimers (Fig. 5B). Trimer and tetramer formation was never observed for this mutant, even upon over-development of immunoblots. These data suggest that the C-terminal region of SSPN is not required for dimer formation.
Figure 5. Removal of the C-terminus does not alter dimer formation.
A, Mutant ΔTM4-CT lacks the 4th transmembrane domain (TM4) and the C-terminal region (CT) of SSPN, as illustrated. B, CHO cells transfected with ΔTM4-CT displayed dimer formation under non-reducing conditions (NR). Under reducing conditions (R), ΔTM4-CT migrates at its predicted molecular mass (19-kDa). Immunoblots were probed with anti-flag antibodies, which detect the flag epitope tag engineered onto the N-terminus of SSPN. Molecular weight markers are indicated (x 103 Da).
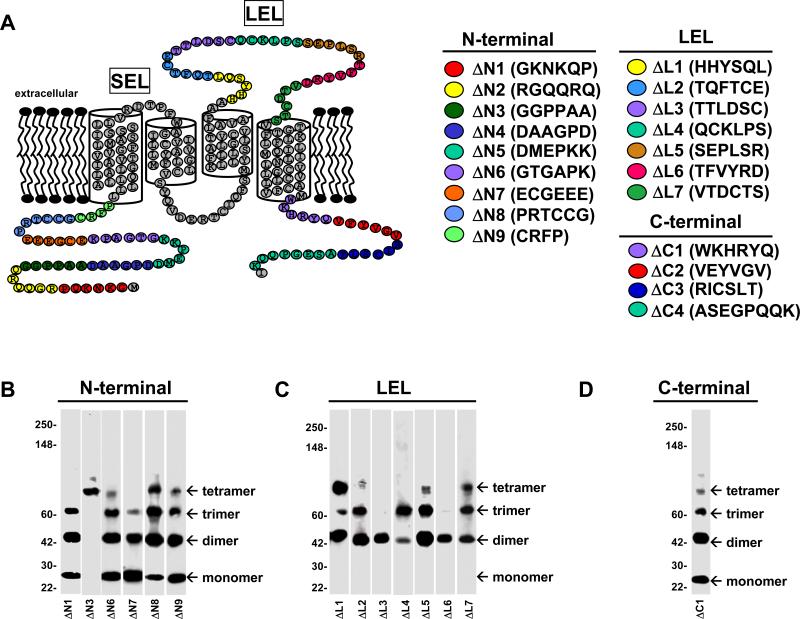
Since large deletions within SSPN caused protein instability, a panel of mutants was created by removing only six consecutive residues at a time. Our efforts were focused on the intracellular N- and C-terminus as well as the LEL, based on the ability of these isolated regions to form oligomers when expressed as recombinant fusion proteins. We first engineered deletion mutants in the N-terminal portion of SSPN (Fig. 6A). Plasmids encoding the N-terminal deletion mutants (ΔΝ1-ΔΝ9) were introduced into CHO cells and protein lysates were separated by centrifugation through linear sucrose gradients. Western blot analysis was performed on immunoblots of all 14-fractions collected from the sucrose gradients for each mutant. Immunoblots were probed with SSPN antibodies and fraction 5, which represents the peak fraction, is shown for each mutant. Table 1 summarizes the oligomerization behaviors exhibited by each mutant. Mutants ΔΝ2, ΔΝ4, and ΔΝ5 were not stably expressed at detectable levels in CHO cells and could not be analyzed. However, the remaining N-terminal deletion mutants expressed at robust levels. ΔΝ1 and ΔΝ6-ΔΝ9 formed robust monomer, dimer, and trimer structures (Fig. 6B). However, tetramer formation was only weakly evident in several of these mutants, suggesting that the N-terminus is important for stabilizing SSPN tetramers (Fig. 6B). Mutant ΔΝ3 exhibited robust tetramers, but completely lacked lower-order structures (Fig. 6B). These data support the notion that elements within the N-terminus are important for stabilizing SSPN web structures.
Figure 6. Site-directed mutagenesis reveals regions required for SSPN-SSPN interactions.
A, Schematic diagram representing SSPN mutants. Site-directed mutagenesis was used to remove six consecutive amino acids from wild-type SSPN in order to create each deletion mutant. Nine mutants were created in the intracellular N-terminus of SSPN (ΔΝ1-ΔΝ9), seven mutants were generated in the LEL (ΔL1-ΔL7) and four mutants were created in the C-terminus (ΔC1-ΔC4). Deleted residues are color-coded for each mutant, as shown. B-D, CHO cells were transfected with plasmids encoding each mutant and protein lysates were subjected to sucrose gradient centrifugation. Protein fractions were analyzed by immunoblotting with SSPN antibodies under non-reducing conditions to test for oligomerization. The peak fraction (No. 5) from each gradient is shown. Mutants ΔN2, ΔΝ4, and ΔΝ5 as well as mutants ΔC2-ΔC4 were not stably expressed in CHO cells and could not be analyzed. Table 1 summarizes the oligomer behavior of each SSPN mutant. Molecular size markers (x 103 Da) are indicated on the left.
Table 1.
Summary of Oligomerization for SSPN Mutants
| DELETION MUTANTS | CYS→ALA POINT MUTANTS | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mutant | Mo. | Di. | Tri. | Tet. | Mutant | Mo. | Di. | Tri. | Tet. |
| ΔN1 | + | + | + | - | C39A | + | + | + | + |
| ΔN2 | N/S | C47A | + | + | + | + | |||
| ΔN3 | - | - | - | + | C48A | + | + | + | + |
| ΔN4 | N/S | C50A | + | + | + | + | |||
| ΔN5 | N/S | C95A | + | + | + | + | |||
| ΔN6 | + | + | + | Weak | C106A | + | + | + | + |
| ΔN7 | + | + | + | - | C116A | + | + | + | + |
| ΔN8 | + | + | + | + | C136A | + | + | + | - |
| ΔN9 | + | + | + | Weak | C155A | - | + | + | + |
| ΔL1 | - | + | - | - | C162A | - | + | + | + |
| ΔL2 | - | + | + | - | C164A | - | + | + | + |
| ΔL3 | - | + | - | - | C184A | - | + | + | + |
| ΔL4 | - | + | + | - | C205A | + | + | + | + |
| ΔL5 | - | + | + | Weak | C209A | - | - | - | + |
| ΔL6 | - | + | - | - | C213A | + | + | + | + |
| ΔL7 | - | + | + | Weak | C231A | N/S | |||
| ΔC1 | + | + | + | Weak | C136A + C209A | + | + | + | + |
| ΔC2 | N/S | C155A + C162A | - | + | + | + | |||
| ΔC3 | N/S | C155A + C164A | - | + | + | + | |||
| ΔC4 | N/S | C155A + C184A | - | + | + | - | |||
| ΔTM4-CT | + | + | - | - | C162A + C184A | + | + | + | + |
Our next mutagenesis efforts were directed toward the LEL region of SSPN (Fig. 6A). Plasmids encoding seven LEL deletion mutants (ΔL1- ΔL7) were expressed in CHO cells and protein lysates were applied to sucrose gradients for isolation of SSPN clusters. Under non-reducing conditions, LEL mutants were incapable of forming stable monomers, suggesting that structural elements within the LEL stabilize the monomeric state (Fig. 6C). In addition to perturbing monomer stability, two of the LEL deletion mutants (ΔL3 & ΔL6) only formed dimers, suggesting that these portions of the LEL contribute to trimer and tetramer formation (Fig. 6C). Only very weak tetramer structures were observed in mutants ΔL2-ΔL6. Mutant ΔL1 exhibited robust dimers and tetramers, but very low levels of trimer. Taken together, data from LEL mutagenesis studies suggest that distinct regions within the LEL mediate monomer, trimer, and tetramer structures. Furthermore, perturbations within the LEL do not alter formation of dimers, suggesting that the dimerization domain lies outside of the LEL or that there are multiple dimerization domains.
Four mutants were generated in the C-terminal region (ΔC1-ΔC4) and only one of the mutants (ΔC1) was stably expressed in CHO cells. Analysis of mutant ΔC1 reveals that it forms monomers, dimers, and trimers, but lacks robust tetramer formation (Fig. 6C).
Analysis of cysteine residues in SSPN web formation
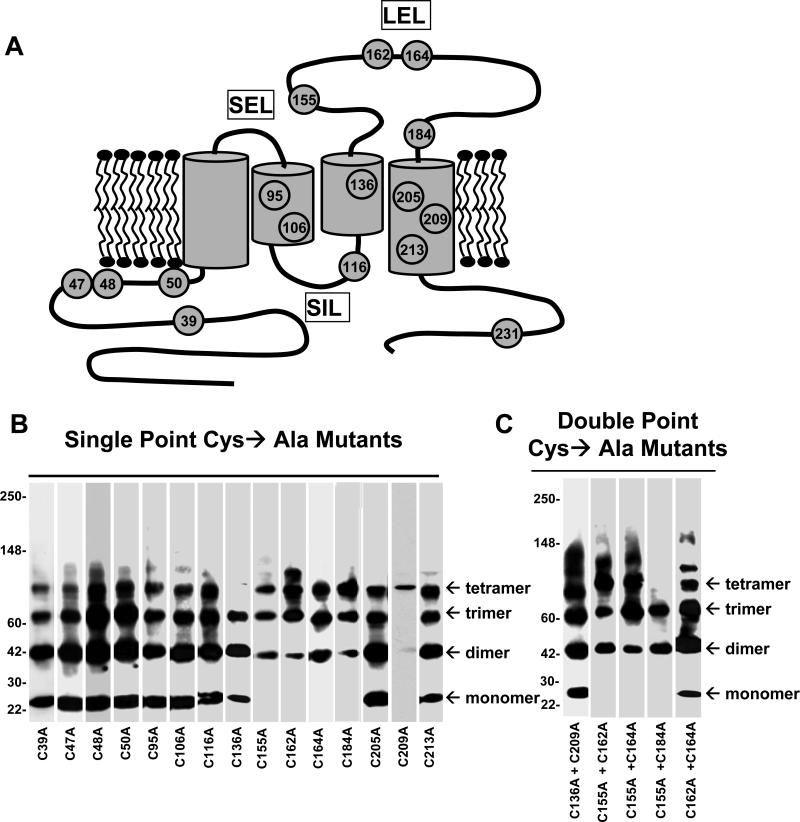
Based on the observation that SSPN forms oligomers under non-reducing conditions, we hypothesized that SSPN-SSPN interactions are mediated by intermolecular disulfide bridges between Cys groups located within the protein binding interface. In order to identify the Cys residues involved in such interactions, we performed mutagenesis to convert each Cys residue into Ala, which lacks a reactive thiol group (Fig. 7A). The Cys→Ala mutants in the N-terminus (C39A, C47A, C48A & C50A) displayed normal oligomer patterns indicating that loss of individual Cys residues in the N-terminus does not perturb SSPN-SSPN interactions (Fig. 7B). Mutants in TM2 (C95A & C106A) exhibited normal oligomer behavior (Fig. 7B). Mutant C116A, present in the small intracellular loop (SIL), also displayed normal oligomerization. However, tetramer structures were not detected in mutant C136A, which is located within TM3 in close proximity to the LEL (Fig. 7A & B). The lack of tetramer observed for this mutant suggests that Cys 136 may be directly involved in intermolecular disulfide crosslinking to form SSPN tetramers. Normal oligomer pattern formation was found in two of the three mutants in the TM4 (C205A & C213A). However, one of the TM4 mutants (C209) only exhibited tetramer structures, suggesting that Cys 209 contributes to stability of lower-order structures (Fig. 7B). The only C-terminal mutant (C231A) was not stably expressed and could not be analyzed.
Figure 7. Mutagenesis of Cys residues within SSPN.
A, Schematic representation of Cys→Ala mutants in SSPN. Site-directed mutagenesis was employed to convert each Cys residue (shown in gray) to Ala. B & C, CHO cells were transiently-transfected with expression constructs representing each mutant and analyzed by sucrose gradient centrifugation, as previously described. All fractions were analyzed by immunoblotting with SSPN antibodies and the peak fraction (No. 5) is shown for each mutant. Single Cys→Ala point mutants are shown in panel B and double Cys→Ala mutants are shown in panel C. Mutant C231A was not stably expressed in CHO cells and could not be analyzed.
The LEL of SSPN contains four conserved cysteines and data from the LEL deletion mutants suggests that these residues may form intramolecular disulfide crossbridges important for monomer stability. We postulate that mutagenesis within the LEL disrupts proper intramolecular Cys-Cys packing, since close alignment of Cys residues is critical for formation of covalent thiol bonds. This model supports the notion that the Cys groups within the LEL are highly reactive and also in close proximity to neighboring SSPN molecules. Interestingly, the conserved Cys residues within the LEL of nearly all tetraspanins participate in intramolecular disulfide crosslinking, which is critical for correct formation of protein interaction interfaces (for review [30, 31]). Crystal structures of the large extracellular loop of tetraspanin CD81 support a model whereby three α-helices, stabilized by intramolecular disulfide bonds, form a site for homo-oligomerization [27].
Mutation of any of the four Cys amino acids in the LEL (C155A, C162A, C164A, C184A) resulted in normal dimer, trimer, and tetramer formation (Fig. 7B). However, none of these mutants possessed a stable monomer under non-reducing conditions (Fig. 7B). The lack of stable monomer suggests that the Cys residues within the LEL are normally involved in intramolecular disulfide bridges. Mutagenesis of a single Cys within the LEL leaves its partner Cys available for intermolecular disulfide interactions with a neighboring SSPN. In order to identify the intramolecular Cys pairing within the LEL, we created double mutants that possessed two Cys→Ala mutations within a single SSPN molecule. The rationale was that if two Cys residues are involved in intramolecular disulfide bridge formation, then removal of both Cys would allow for reappearance of the monomer due to absence of a free, unpaired Cys residue. Double mutants C155A + C162A, C155A + C164A, C155A + C184A did not produce monomer structures (Fig. 7C). However, double mutagenesis of Cys 162 and Cys 164 caused the reappearance of SSPN monomers (Fig. 7C). This data supports a model whereby Cys 162 and Cys 164 forms either covalent intra- or inter-molecular thiol linkage. Our data also suggest that both Cys 155 and Cys 184 are involved in disulfide bonding with residues not found in the LEL, since double mutant C155A + C184A did not form monomers (Fig. 7C). Similar to many of the deletion mutants in the LEL, double mutant C155A + C184A did not exhibit tetramer formation, suggesting that these residues are critical for tetramer stability.
A double mutagenesis approach was also employed to examine the contribution of Cys 209 and Cys 136 to proper oligomer structure. C209A displayed only tetramer formation while C136A specifically lacked tetramer formation (Fig. 7B). Topology algorithms predict that these residues are in close proximity within the membrane bilayer [25]. Double mutant C136A + C209A resulted in normal oligo pattern formation, including the reappearance of stable monomers (Fig. 7C). These data suggest that Cys 136 and Cys 209 are closely aligned within the plasma membrane and may form intramolecular disulfide bonds.
Analysis of SG binding sites within SSPN
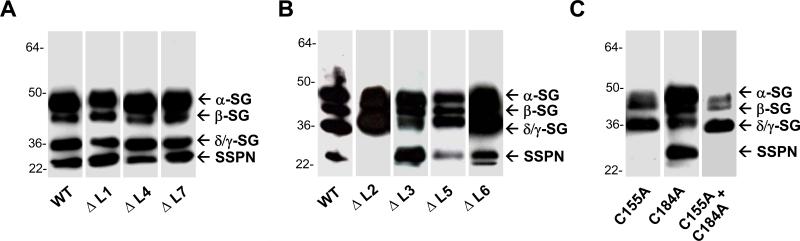
Previous work from mouse models, in vitro binding experiments, and patient studies has established that SSPN forms a tight subcomplex with the SGs [10, 11]. In order to determine which regions of SSPN are responsible for mediating interactions with the SGs, we tested the ability of each SSPN mutant to interact with the tetrameric SG-subcomplex. Myc-tagged cDNA constructs of the SGs (α-, β-, γ-, and δ-subunits) and SSPN were transiently introduced into CHO cells by electroporation. Immunoblots of cellular protein lysates with anti-myc antibodies demonstrated that the SGs and SSPN are expressed at relatively equal quantities (data not shown). CHO cells transfected with the SGs plus wild-type SSPN were immunoprecipitated using monoclonal antibodies to δ-SG (Fig. 8A), as previously documented [10, 38]. Each of the N-terminal deletion mutants immunoprecipitated normally with the SGs (data not shown), indicating that the N-terminus does not form the major site for SG interaction. LEL deletion mutants ΔL1, ΔL3, ΔL4, and ΔL7 displayed normal complex formation with the SGs (Fig. 8A). However, two LEL mutants (ΔL2 & ΔL5) failed to interact with the SGs (Fig. 8B), indicating that mutations within the LEL perturb SG-SSPN subcomplex formation. Mutant ΔL6 bound weakly to the SGs, as revealed by overexposure of immunoblots (Fig. 8B). Interestingly, mutant ΔL2, which lacks Cys 155, did not interact with the SGs.
Figure 8. Analysis of SSPN interaction with the SG subcomplex.
A, The SG-SSPN subcomplex was reconstituted in vivo by co-transfecting CHO cells with expression vectors representing myc-tagged human α-, β-, γ-, and δ-SGs plus SSPN, as previously documented [10, 38]. Immunoprecipitation of δ-SG from lysates of cells expressing all four SGs plus SSPN results in co-precipitation of the SG-SSPN subcomplex (WT, lane 1). Each LEL deletion mutant was co-transfected with the SGs and analyzed by immunoprecipitation with δ-SG antibodies. ΔL1, ΔL3, ΔL4, and ΔL7 co-precipitate normally with the SG subcomplex. B, Mutants ΔL2, ΔL5, and ΔL6 did not properly immunoprecipitate with the SGs. Mutants ΔL2 and ΔL5 did not associate with the SGs, while ΔL6 displayed weak interaction with the SGs. C, Analysis of Cys→Ala SSPN mutants by co-precipitation with the SGs. Samples were analyzed on either gradient (5-15%, panel A) or isocratic (12%, panel B) polyacrylamide gels. Proteins were detected by immunoblotting with anti-myc antibodies, which recognize the 9E10 tag on each protein construct. Mutants C155A and C155A+C184A exhibited a dominant negative effect and caused disruption of the SG subcomplex, as shown by the loss of δ- and γ-SG from the immune complex. Immunoblots of mutants ΔL2 and ΔL6 have been over-exposed for detection of SSPN protein.
We also analyzed whether mutation of SSPN's cysteine residues would affect SG interactions by co-transfecting all four SGs with the SSPN Cys→Ala mutants. We found that SSPN and the SGs expressed at wild-type levels (data not shown). Cell lysates were subjected to immunoprecipitation with δ-SG antibodies. Stability of the SSPN-SG subcomplex is reflected by the presence of the entire subcomplex in the immunoprecipitate. Two SSPN mutants caused de-stablization of the tetrameric SGs. Analysis of SSPN Cys→Ala single point mutants reveals that one SSPN mutant (C155A) was not immunoprecipitated with δ-SG antibodies (Fig. 8C). In fact, α- and β-SG were only weakly associated with δ-SG, suggesting that SSPN mutant C155A disrupted interactions between the tetrameric SG-subcomplex in a dominant-negative fashion (Fig. 8C). This disruption was particularly impressive given that β- and δ-SG form a core around which the α- and γ-SG are attached [41]. Likewise, SSPN double mutant C155A + C184A also disrupted SG-SSPN complex formation in a dominant-negative fashion. Cells were co-transfected with SSPN double mutant C155A + C184A and all four SGs, which were expressed at normal levels (data not shown). However, only δ/γ-SG was found in the δ-SG immunopreciptiate, suggesting that expression of SSPN double mutant C155A + C184A causes disruption of the tetrameric SG-subcomplex.
Model of SSPN-mediated protein interactions within DGC
Our study represents the first biochemical test of whether SSPN exhibits tetraspanin-like properties. SSPN has several, but not all, of the conserved tetraspanin domains. For this reason, it has been debated whether or not SSPN possesses tetraspanin-like functions [26]. All tetraspanins form higher order structures through either homo- or hetero-tetraspanin interactions [26,28]. These clusters of tetraspanins create distinctive domains within the plasma membrane that contribute to signaling, adhesion, membrane fusion, and proliferation [28,30]. The data presented in the current report demonstrates that SSPN does indeed form oligomers within the membrane bilayer. These studies have lead us to hypothesize that SSPN-SSPN interactions form: 1) scaffolds onto which the DGC is assembled and 2) distinct domains within the plasma membrane that function to cluster possible signaling molecules. SSPN is expressed, along with DG, in mononuclear C2C12 myoblasts. The SGs are only expressed upon differentiation of myoblasts into myotubes. We propose that the SSPN-SG subcomplex is transported to the sarcolemma where it interacts with SSPN-DG, which is already at the sarcolemma. Given our demonstration that SSPN forms homo-oligomers, it is tantalizing to speculate that the DGs and SGs are ‘snapped’ into place by SSPN-SSPN interactions.
Our mutant analysis shows that specific amino acids within SSPN have distinct functions (Fig. 9A). Despite our extensive mutagenesis, only mutants in the N-terminus of SSPN caused disruption of dimer formation. Thus, the N-terminus appears to be critical for stability of SSPN dimer structures. Mutants within the LEL exhibited abnormal trimer and tetramer oligmers, suggesting that the higher ordered structures are formed from intermolecular interactions between the LEL regions of neighboring SSPNs. Furthermore, point mutation of any one of the cysteine residues in the LEL completely abolished the presence of monomeric SSPN. This is an interesting observation that reveals the importance of thiol-mediated bonds in the stability of SSPN's structure. Taken together, these data support a model whereby SSPN forms tetraspanin-like structures via multiple points of contact between neighboring SSPN molecules (Fig. 9A & B).
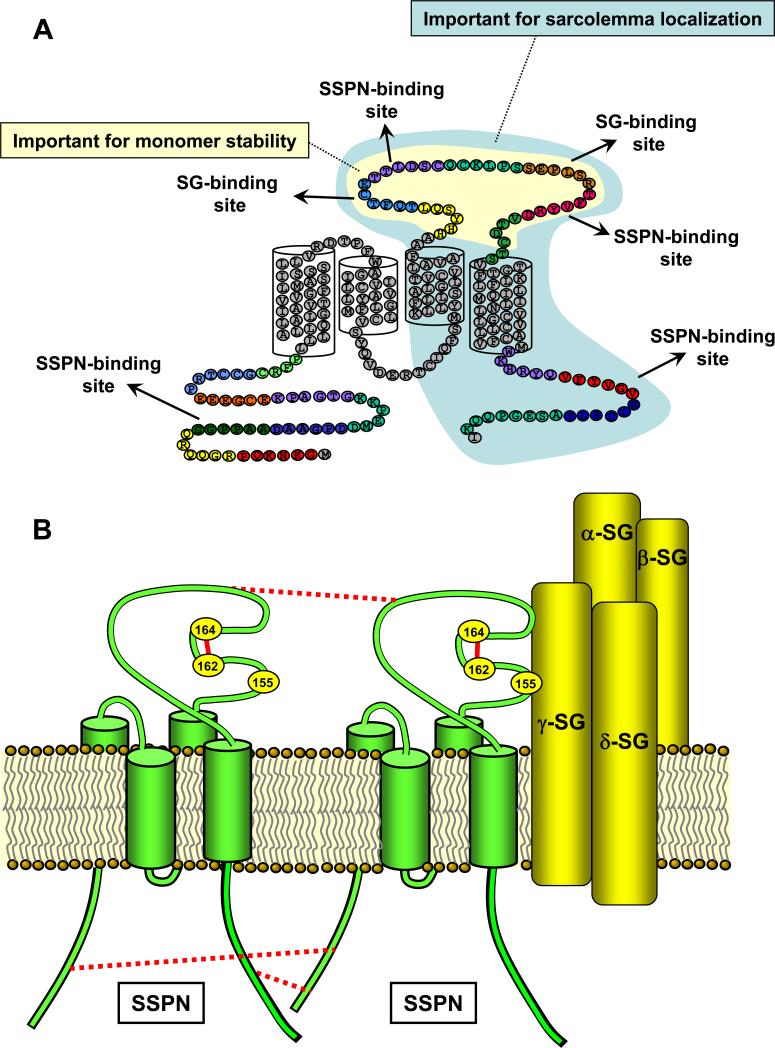
Figure 9. SSPN functions to facilitate protein interactions within the DGC.
A, Evidence presented in the current report demonstrates that SSPN possess distinct binding regions for the SGs. These regions are located within the LEL, as shown. In addition, domains that form the SSPN-SSPN interface are identified within the extracellular and intracellular regions of SSPN, suggesting that the formation of SSPN webs occurs through a complex set of protein interactions. SSPN monomers were very susceptible to mutations in nearly any region within the LEL (shaded in yellow). We speculate that mutation within the LEL disrupts the Cys packing so that intramolecular thiol linkages are disrupted. This observation suggests that the LEL structure is an important determinant in SSPN function. Perturbations in the LEL disrupt SSPN structure (as measured by monomer stability) as well as SSPN-SSPN and SSPN-SG interactions. We also speculate that the C-terminal half of SSPN (shaded in light blue) plays a role in sarcolemma localization based on the observation that microspan protein (which lacks this region) is localized to the sarcoplasmic reticulum. B, A model of SSPN-SG interactions within the DGC is proposed. Previous studies have shown that the SG-SSPN subcomplex functions to stabilize α-DG at the sarcolemma [22-24]. We propose that an intramolecular disulfide bridge is formed between Cys 162 and Cys 164 in the LEL (solid red lines). This bond is important for proper LEL structure and interaction with the SG subcomplex. Cys 209 forms a thiol linkage with Cys 136 (not shown). Cys 184 and Cys 155 are likely involved in disulfide pairing, although we were unable to identify their partner Cys residues. Furthermore, Cys 155 appears to be an important component of the SG binding interface. Mutant studies support a model whereby SSPN webs are stabilized by interactions in the NT, LEL, and CT regions (dashed red lines). SSPN forms tetraspanin-like webs within the plasma membrane, which may function to promote interactions between the SG and DG subcomplexes. SSPN is depicted in green and the SGs are depicted in yellow.
In addition to analyzing SSPN-SSPN interactions, we employed SSPN mutants to determine which regions of SSPN mediate interaction with the SGs. Our studies indicate that two distinct regions within the LEL are crucial for stable formation of the SG-SSPN subcomplex within the DGC (Fig. 9A). We were unable to analyze interaction of SSPN with individual SGs due to the instability of SSPN protein when co-transfected with an individual SG molecule. In addition to interacting with the SGs, SSPN also interacts with β-DG through distinct protein interaction sites (Miller and Crosbie, unpublished results). The extracellular Cys-rich region of the SG proteins plays critical roles in assembly and stability of the SG subcomplex [41, 43], suggesting that thiol-mediated protein interactions are important for the stability of the entire DGC. Our data suggest that the Cys residues within SSPN may also be important for stability of the DGC based on our observations that expression of two Cys→Ala SSPN mutants caused de-stablization of the tetrameric SG-subcomplex. We propose a model whereby SSPN webs form a core around which the SGs and DGs are attached. The SSPN mediated clustering of the SGs and DGs is poised to promote efficient anchorage of α-DG at the membrane and contribute to DGC stability both laterally within the bilayer and across the bilayer to the extracellular matrix.
We have recently reported the characterization of microspan, which is produced from alternate splicing of SSPN mRNA [42]. Microspan is approximately half the size of SSPN and it possesses TM 1 and 2, but lacks TM 3 and TM4 as well as the LEL. Interestingly, microspan protein is localized to the sarcoplasmic reticulum, which is an extensive network of internal membrane sacs that house Ca2+ required for skeletal muscle contraction. From this observation, in combination with our mutant studies, we can surmise that the LEL is important for targeting to the sarcolemma (Fig. 9A).
Our studies demonstrate that cysteine residues in SSPN are involved in both intra- and inter-molecular disulfide bonds. We speculate that thiol bonds between SSPN and SGs may be one mechanism by which the SSPN-SG subcomplex is held together. If so, then it is tantalizing to speculate that disease mutations that convert an extracellular Cys residue in the SGs [43] may, in turn, perturb normal interaction of the SGs with SSPN. In other words, our studies may illuminate some of the mechanistic details of sarcoglycan-deficient limb girdle muscular dystrophy pathogenesis.
ACKNOWLEDGEMENTS
We thank James Hansen for mutant construction, Steven Lee for recombinant protein purifications, Kari Morrissey for figure preparation and the members of the Duchenne Muscular Dystrophy Research Center (UCLA) for their comments and support. This research was supported in part by a Howard Hughes Medical Institute grant through the Undergraduate Biological Sciences Education Program to the University of California, Los Angeles. E.W. was a recipient of the HHMI UBSEP fellowship. A. K. Peter was supported by the Molecular, Cellular and Integrative Physiology pre-doctoral training fellowship (NIH: T32 GM65823) as well as fellowships from the Ursula-Mandel and Edith Hyde Foundations. G. Miller was supported by a Research Development grant from the Muscular Dystrophy Association USA (MDA3704). This research was also supported by a grant from the NIH to R. H. Crosbie (AR48179-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: DG, dystroglycan, DGC, dystrophin-glycoprotein complex, LEL, large extracellular loop, SEL, small extracellular loop, SIL, small intracellular loop, SSPN, sarcospan, SG, sarcoglycan, TG, transgenic, TM, transmembrane domain.
REFERENCES
- 1.Campbell KP, Kahl SD. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–62. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- 2.Ervasti JM, Kahl SD, Campbell KP. Purification of dystrophin from skeletal muscle. J Biol Chem. 1991;266:9161–5. [PubMed] [Google Scholar]
- 3.Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–9. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- 4.Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–31. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida M, Ozawa E. Glycoprotein complex anchoring dystrophin to sarcolemma. J Biochem. 1990;108:748–52. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
- 6.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–23. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weller B, Karpati G, Carpenter S. Dystrophin-deficient mdx muscle fibers are preferentially vulnerable to necrosis induced by experimental lengthening contractions. J Neurol Sci. 1990;100:9–13. doi: 10.1016/0022-510x(90)90005-8. [DOI] [PubMed] [Google Scholar]
- 8.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci U S A. 1993;90:3710–4. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michele DE, Campbell KP. Dystrophin-glycoprotein complex: post-translational processing and dystroglycan function. J Biol Chem. 2003;278:15457–60. doi: 10.1074/jbc.R200031200. [DOI] [PubMed] [Google Scholar]
- 10.Crosbie RH, Lebakken CS, Holt KH, Venzke DP, Straub V, Lee JC, Grady RM, Chamberlain JS, Sanes JR, Campbell KP. Membrane targeting and stabilization of sarcospan is mediated by the sarcoglycan subcomplex. J Cell Biol. 1999;145:153–65. doi: 10.1083/jcb.145.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosbie RH, Lim LE, Moore SA, Hirano M, Hays AP, Maybaum SW, Collin H, Dovico SA, Stolle CA, Fardeau M, Tome FM, Campbell KP. Molecular and genetic characterization of sarcospan: insights into sarcoglycan-sarcospan interactions. Hum Mol Genet. 2000;9:2019–27. doi: 10.1093/hmg/9.13.2019. [DOI] [PubMed] [Google Scholar]
- 12.Rybakova IN, Amann KJ, Ervasti JM. A new model for the interaction of dystrophin with F-actin. J Cell Biol. 1996;135:661–72. doi: 10.1083/jcb.135.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rybakova IN, Ervasti JM. Dystrophin-glycoprotein complex is monomeric and stabilizes actin filaments in vitro through a lateral association. J Biol Chem. 1997;272:28771–8. doi: 10.1074/jbc.272.45.28771. [DOI] [PubMed] [Google Scholar]
- 14.Amann KJ, Renley BA, Ervasti JM. A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. J Biol Chem. 1998;273:28419–23. doi: 10.1074/jbc.273.43.28419. [DOI] [PubMed] [Google Scholar]
- 15.Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 16.Sunada Y, Bernier SM, Kozak CA, Yamada Y, Campbell KP. Deficiency of merosin in dystrophic dy mice and genetic linkage of laminin M chain gene to dy locus. J Biol Chem. 1994;269:13729–32. [PubMed] [Google Scholar]
- 17.Peng HB, Ali AA, Daggett DF, Rauvala H, Hassell JR, Smalheiser NR. The relationship between perlecan and dystroglycan and its implication in the formation of the neuromuscular junction. Cell Adhes Commun. 1998;5:475–89. doi: 10.3109/15419069809005605. [DOI] [PubMed] [Google Scholar]
- 18.Sugita S, Saito F, Tang J, Satz J, Campbell K, Sudhof TC. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154:435–45. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowe MA, Deyst KA, Leszyk JD, Fallon JR. Identification and purification of an agrin receptor from Torpedo postsynaptic membranes: a heteromeric complex related to the dystroglycans. Neuron. 1994;12:1173–80. doi: 10.1016/0896-6273(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 20.Allamand V, Campbell KP. Animal models for muscular dystrophy: valuable tools for the development of therapies. Hum Mol Genet. 2000;9:2459–67. doi: 10.1093/hmg/9.16.2459. [DOI] [PubMed] [Google Scholar]
- 21.Duclos F, Broux O, Bourg N, Straub V, Feldman GL, Sunada Y, Lim LE, Piccolo F, Cutshall S, Gary F, Quetier F, Kaplan JC, Jackson CE, Beckmann JS, Campbell KP. β-sarcoglycan: genomic analysis and identification of a novel missense mutation in the LGMD2E Amish isolate. Neuromuscul Disord. 1998;8:30–8. doi: 10.1016/s0960-8966(97)00135-1. [DOI] [PubMed] [Google Scholar]
- 22.Holt KH, Lim LE, Straub V, Venzke DP, Duclos F, Anderson RD, Davidson BL, Campbell KP. Functional rescue of the sarcoglycan complex in the Bio 14.6 hamster using δ-sarcoglycan gene transfer. Mol Cell. 1998;1:841–848. doi: 10.1016/s1097-2765(00)80083-0. [DOI] [PubMed] [Google Scholar]
- 23.Durbeej M, Cohn RD, Hrstka RF, Moore SA, Allamand V, Davidson BL, Williamson RA, Campbell KP. Disruption of the beta-sarcoglycan gene reveals pathogenetic complexity of limb-girdle muscular dystrophy type 2E. Mol Cell. 2000;5:141–51. doi: 10.1016/s1097-2765(00)80410-4. [DOI] [PubMed] [Google Scholar]
- 24.Coral-Vazquez R, Cohn RD, Moore SA, Hill JA, Weiss RM, Davisson RL, Straub V, Barresi R, Bansal D, Hrstka RF, Williamson R, Campbell KP. Disruption of the sarcoglycan-sarcospan complex in vascular smooth muscle: A novel mechanism for cardiomyopathy and muscular dystrophy. Cell. 1999;98:465–474. doi: 10.1016/s0092-8674(00)81975-3. [DOI] [PubMed] [Google Scholar]
- 25.Crosbie RH, Heighway J, Venzke DP, Lee JC, Campbell KP. Sarcospan: the 25kDa transmembrane component of the dystrophin-glycoprotein complex. J Biol Chem. 1997;272:31221–31224. doi: 10.1074/jbc.272.50.31221. [DOI] [PubMed] [Google Scholar]
- 26.Hemler ME. Specific tetraspanin functions. J Cell Biol. 2001;155:1103–7. doi: 10.1083/jcb.200108061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitadokoro K, Bordo D, Galli G, Petracca R, Falugi F, Abrignani S, Grandi G, Bolognesi M. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 2001;20:12–8. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seigneuret M, Delaguillaumie A, Lagaudriere-Gesbert C, Conjeaud H. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J Biol Chem. 2001;276:40055–64. doi: 10.1074/jbc.M105557200. [DOI] [PubMed] [Google Scholar]
- 29.Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. 2001;58:1189–205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy S, Shoham T. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 2005;20:218–24. doi: 10.1152/physiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- 31.Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 32.Zemni R, Bienvenu T, Vinet MC, Sefiani A, Carrie A, Billuart P, McDonell N, Couvert P, Francis F, Chafey P, Fauchereau F, Friocourt G, des Portes V, Cardona A, Frints S, Meindl A, Brandau O, Ronce N, Moraine C, van Bokhoven H, Ropers HH, Sudbrak R, Kahn A, Fryns JP, Beldjord C, Chelly J. A new gene involved in X-linked mental retardation identified by analysis of an X;2 balanced translocation. Nat Genet. 2000;24:167–70. doi: 10.1038/72829. [DOI] [PubMed] [Google Scholar]
- 33.Maecker HT, Do MS, Levy S. CD81 on B cells promotes interleukin 4 secretion and antibody production during T helper type 2 immune responses. Proc Natl Acad Sci USA. 1998;95:2458–62. doi: 10.1073/pnas.95.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeyama S, Koyama M, Yamaoko M, Sasada R, Miyake M. Suppression of cell motility and metastasis by transfection with human motility-related protein (MRP-1/CD9) DNA. J Exp Med. 1993;177:1231–7. doi: 10.1084/jem.177.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–41. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 36.Crawford GE, Faulkner JA, Crosbie RH, Campbell KP, Froehner SC, Chamberlain JS. Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J Cell Biol. 2000;150:1399–410. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer MJ, Guyon JR, Sorimachi H, Potts A, Richard I, Herasse M, Chamberlain J, Dalkilic I, Kunkel LM, Beckmann JS. Stable expression of calpain 3 from a muscle transgene in vivo: immature muscle in transgenic mice suggests a role for calpain 3 in muscle maturation. Proc Natl Acad Sci USA. 2002;99:8874–8879. doi: 10.1073/pnas.132269299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holt KH, Campbell KP. Assembly of the sarcoglycan complex. Insights for muscular dystrophy. J Biol Chem. 1998;273:34667–70. doi: 10.1074/jbc.273.52.34667. [DOI] [PubMed] [Google Scholar]
- 39.Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol. 2005;5:136–48. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 40.Charrin S, Manie S, Oualid M, Billard M, Boucheix C, Rubinstein E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 2002;516:139–44. doi: 10.1016/s0014-5793(02)02522-x. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Shi W, Zhang Y, Sokol R, Cai H, Lun M, Moore B, Farber M, Stepanchick J, Bonnemann C, Chan Y. Identification of functional domains in sarcoglycans essential for their interaction and plasma membrane targeting. Exp Cell Res. 2006;312:1610–25. doi: 10.1016/j.yexcr.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 42.Miller G, Peter AK, Espinoza E, Heighway J, Crosbie RH. Over-expression of microspan, a novel component of the sarcoplasmic reticulum, causes a severe muscle pathology with triad abnormalities. J Muscle Res Cell Motil. 2006 doi: 10.1007/s10974-006-9069-2. in press. [DOI] [PubMed] [Google Scholar]
- 43.Hack AA, Groh ME, McNally EM. Sarcoglycans in muscular dystrophy. Microsc Res Tech. 2000;48:167–80. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<167::AID-JEMT5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]