Abstract
The Immunological Genome Consortium has generated a public resource (www.immgen.org) that provides a compendium of gene expression profiles of ~270 leukocyte subsets in the mouse. This effort established carefully standardized operating procedures that resulted in a transcriptional dataset of unprecedented comprehensiveness and quality. The findings have been detailed recently in a series of publications providing molecular insights into the development, heterogeneity, and/or function of these cellular lineages and distinct subpopulations. Here, we review the key findings of these studies, highlighting what has been gained and how the knowledge can be used to accelerate progress toward a comprehensive understanding of the immune system.
Introduction
Unraveling the vast complexity of the immune system remains an enormous challenge, but its function and dysfunction underlies protection and pathology in a myriad of diseases, making it a high-value research target. Genome-scale systems biology approaches have become increasingly accepted as a powerful complement to reductionist approaches, which excel at testing very specific relationships but fail to capture unanticipated (and typically unmeasured) effects [1,2]. In addition, these “global” approaches can more effectively address certain types of questions. For example, akin to the comparison of phylogenetics based on a single ribosomal RNA sequences [3] versus whole genomes [4], population relationships and heterogeneity can be evaluated on a whole-genome scale instead of being based on small subsets of molecules. Transcriptomics, the study of whole genome gene expression, is also a powerful approach for discovering new molecules involved in known processes, as well as involvement of known pathways in new processes. These unexpected connections, which would otherwise go undiscovered, are an important aspect of accelerating our understanding of immune complexity.
Below, we summarize the major findings of the first iteration of the Immunological Genome (ImmGen) project [5–8], which has obtained microarray-based transcriptomes for most of the well-defined leukocyte subsets of the C57BL/6 mouse. Projects within the Consortium were categorized into the major leukocyte subdivisions to leverage the expertise of individual labs, and the focus of each study reflected the interests and questions of the associated field. Here, we provide a perspective on the types of insights that have been made through large-scale transcriptome analysis, the ways in which these data can be mined by the community, and what can be anticipated from future studies.
Improved molecular definitions of leukocyte subsets and relatedness
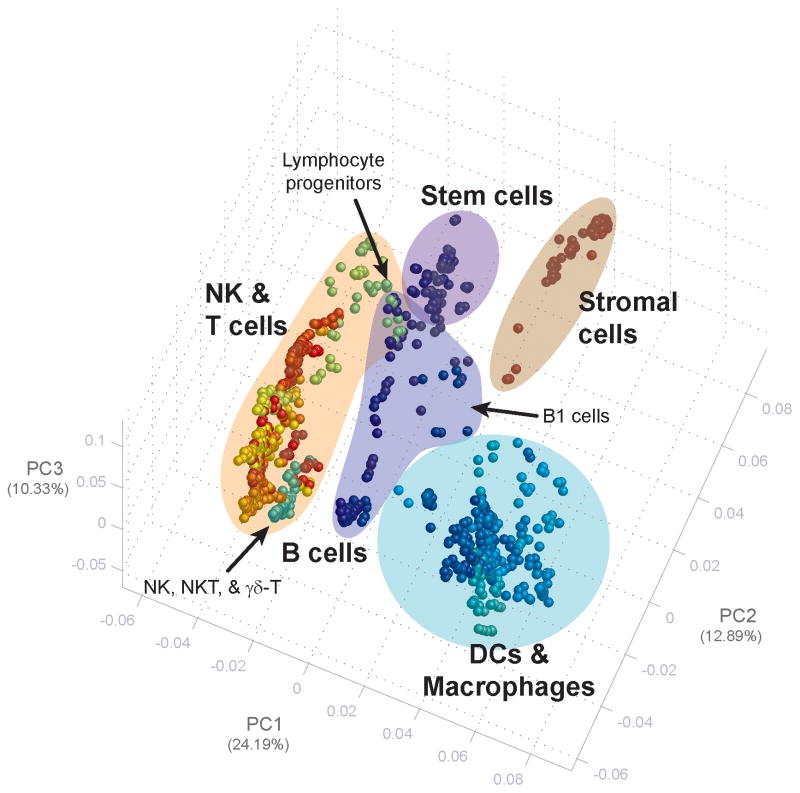
One of the most common threads running through the studies was description of population signatures and relatedness (Figure 1). In large part, this was a natural extension of one of the early challenges faced by the Consortium: how to choose leukocyte subsets for profiling in the first place, and what constitutes a subset? Similar to sequencing a genome, defining a baseline transcriptome is to some degree immutable, in that the data will serve as a resource for many years to come. Thus, faced with the desire to choose the “best” subsets, it became evident that many subsets, both in their cell surface characteristics that allowed their purification and their functions, remained poorly defined.
Figure 1. Principal components analysis for publicly available ImmGen populations.
The top three principal components (PC), explaining the predominant trends across populations, were calculated from the 15% most variable genes across all populations as previously described [15]. PC1 is enriched in genes that distinguish innate from adaptive populations; PC2 is enriched in genes that distinguish progenitors from mature leukocytes.
There is no better example of this challenge than in the myeloid compartment, where no single marker can clearly distinguish all macrophage subsets from dendritic cell (DC) lineages [9]. Previous transcriptome studies had unsuccessfully attempted to find single population-defining markers and reached the conclusion that none existed [10,11]. These studies, however, were based on data pooled from multiple sources without standardized sample preparation, raising the question of whether more stringent quality control would improve sensitivity. As a result, a significant focus of the ImmGen macrophage and dendritic cell studies was the identification of improved common and subset-specific phenotyping markers [12,13]. Several new macrophage- and DC-enriched markers were identified, and new combinatorial markers have been proposed; although similar to previous studies, no single marker could definitively distinguish these myeloid subsets. Although the widespread utility of these new markers awaits further confirmation, it is clear that these findings are not an endpoint, but are important steps toward improved phenotypic and functional definition of myeloid subsets. Furthermore, it highlights the need to better understand the vast diversity of myeloid cell biology.
Other significant advances of the ImmGen project were the first genome-wide molecular definition of poorly characterized leukocyte subtypes, their relationships to other subsets, and new markers to aid in their identification. For example, the Vγ2+ subtype of immature gamma delta (γδ) T cells was found to be unusually distinct from other subtypes, although this distinction became less prominent upon maturation [14]. New cell surface phenotyping markers were identified for memory natural killer (NK) cells [15], which could previously only be identified through allogeneic markers after adoptive transfer [16]. Extensive overlap between molecular signatures of NK and iNKT cells identified more extensive commonalities than previously appreciated, and both populations were also shown to exhibit a number of newly discovered similarities to activated CD8+ effector cells [15,17]. Conversely, closely related subsets were found to have unexpected complexity in their relationships; for example, CD4− and CD4+ iNKT subsets in the spleen were found to be more distinct from one another than the same subsets in the liver [17]. Fibroblastic reticular cells in lymph node stroma were found to express high levels of cadherin-11, providing an additional marker for their discrimination from other stromal and hematopoietic cells [18]. The same study also identified a previously undefined population - the “double-negative cells” (lacking expression of gp38 and CD31) - as contractile pericytes based on their expression profiles being enriched for functions related to smooth muscle cells and cardiomyocytes [18]. Together, these improved molecular definitions and surface phenotyping markers should greatly accelerate our understanding of the heterogeneity within these cell types and their distinct functional capacities.
New transcriptional correlates of physiological processes
In contrast to the subsets described above, many leukocytes have better defined cell surface phenotypes that have facilitated their functional characterization. However, our understanding of common processes is largely based on limited subsets of molecules identified through genetics or biochemistry. Advantages of transcriptomics include the ability to identify genes associated with a process that may also be essential for viability (a limitation of genetic screens), high sensitivity (a limitation of biochemistry), and high-throughput. These characteristics have allowed association of many new genes with otherwise well-described processes.
In an excellent example of identifying new genes associated with well described processes, the well characterized progression of CD8+ T cells through effector and memory phases was defined with great molecular precision [19]. These T cells were profiled from 12 hours through 100 days post-infection during responses to infection with Listeria monocytogenes (Lm) and vesicular stomatitis virus (VSV), providing a detailed analysis of CD8+ T cell activation through short-term effector, short-term memory, late effector-memory, and memory cell formation, with new molecular markers identified for each stage. This included transcription factors associated with each stage, presenting a framework for understanding the regulatory “decisions” made throughout T cell activation. Furthermore, the authors found that the response of CD8+ T cells occurred surprisingly quickly (by 12 hr post-infection), and that responses to Lm and VSV were stereotyped, although different in their magnitude. Reassuringly, antigen-specific responses to Lm infection using the ovalbumin-specific OT-I T cell receptor transgenic system were found to be similar to those identified by Lm antigen-specific H-2 tetramer staining, which had not previously been investigated at the whole transcriptome level. Comparisons similar to those for CD8+ T cells were made for NK cells responding to mouse cytomegalovirus infection, along with comparisons to their analogous stages of CD8+ cell responses, identifying conserved mechanisms by which these effector populations respond to infection [15].
In another example, the under-characterized lymph node stromal cells revealed rich production of cytokines, chemokines, and growth factors, with new cellular source assignments being made for some known stromal cell-expressed genes (Il7 and Vegfa), but with the majority being newly discovered genes [18]. In addition, thorough descriptions of stromal expression of important matrix-contributing genes, such as integrins, collagens, proteoglycans, and laminins, were discovered and assigned to specific stromal subsets, generating a rich resource for discovering novel stroma-leukocyte interactions.
One final notable example was the mining of dendritic cell transcriptomes for to identify genes encoding immunomodulatory receptors [13]. These molecules are of great interest for their role in the increasingly appreciated process of immune homeostasis [20], which makes them potential targets for therapeutic intervention. Although their physiological relevance remains to be proven, the knowledge of their expression itself has revolutionized a functional screening-based process that commonly requires years to decades into a focused candidate-based approach that can be executed much more rapidly.
Regulatory network modeling
Identification of differentially regulated genes can reveal candidate effectors for a process, but large gene lists often exceed our ability to perform secondary assays to evaluate functional relevance. To address this, it is useful to focus on identification of transcription factors that may regulate a set of genes (often referred to as a “module”) involved in a biological process, and to work down the regulatory hierarchy to identify specific effectors. However, even this strategy can be daunting when faced with dozens of candidate transcription factors.
A powerful approach that has demonstrated great utility in addressing this problem is regulatory network modeling [7,21–23]. Although there are differences in its precise implementation, the general approach is to identify co-variance between the expression of transcription factors with other genes across a large dataset. Various metrics of similarity can be used to quantify this co-variance, and statistical thresholding allows identification of putative regulatory interactions. This approach is effective for large datasets, where subtle co-variance can become more apparent due to the large number of samples and lead to the identification of “hub” regulators.
The ImmGen dataset, due to its precisely standardized methodology and large size, affords a unique opportunity for regulatory network modeling. New methodology specifically tailored to the data was developed and applied across all subsets [7]. The network data were used by many groups to identify regulators associated with processes of interest, such as dendritic cell and γδT cell development [13,14], CD8+ T cell activation [19], NK cell differentiation and activation [15], and macrophage identity [12]. Testing the hundreds of predictions made in these studies will be the topics of future study for many years.
Moving beyond the transcriptome
Due to the practical limits of publication, the existing analyses have barely scratched the surface of the information generated. Opportunities exist for many individuals to mine the data for new hypotheses or to support existing ones, and the availability of user-friendly tools, including some on mobile devices (www.immgen.org and freely available from app stores), brings analysis of the data within reach of researchers without bioinformatics skills. It is arguable that the only way to maximize effective utilization of the data is through a concerted effort from the entire community, rather than leaving analysis to specialists who are not necessarily versed in the details of a given biological system. It is important to consider that many genes without known contributions to leukocyte function have been glossed over in previous studies in order to minimize speculation, but that these genes are a rich source of further investigation.
During the course of ImmGen data generation, other areas were explored that have not yet been described in publications. One such subject is the important role of natural genetic variation in modulating the response of the transcriptome to external stimuli, which is broadly recognized to occur, but more often studied in humans than mice and is lacking a genome-wide, systematic analysis. Although the ImmGen baseline compendium was largely generated using a fixed genetic background (C57BL/6), analysis is underway to chart the impact of the natural genetic variation in mice on the transcriptome. In a collaborative effort with The Jackson Laboratory, expression data have been generated for two immune cells types (bone marrow granulocytes and splenic CD4+ T cells) across 40 genotyped strains of the Mouse Phenome Database. This analysis is revealing a significant impact of genetic variation by identifying hundreds of immune expression quantitative trait loci (commonly known as eQTLs), which are currently being exploited to enhance and refine the structure of gene regulatory networks (S. Mostafavi et al., in preparation).
In addition to deeper data mining, the numerous new predictions already made by the ImmGen studies will provide many testable hypotheses for years to come. In parallel, additional transcriptome studies will continue, generating an even more complete picture of leukocyte function. Unlike the largely static primary sequence of a genome, the “transcriptome” is dynamic, responding to environmental, developmental, and epigenetic cues. For immunologists, these dynamics are often of primary interest – the protective or pathological responses during disease, immune responsiveness to pharmacological intervention, or normal operation of any of a number of homeostatic processes. Thus, although baseline profiles for many resting leukocyte subsets have been obtained, we are in the early stages of exploring the wide variety of possible perturbed states, many of which can result in the emergence of new leukocyte subsets not present at steady-state. A second iteration of the ImmGen Project to examine such perturbed states is currently underway, but choosing a sliver that can serve as a representative of the enormous possible search space remains a great challenge.
Importantly, the ImmGen effort has demonstrated that careful standardization can produce high-quality data that can far exceed that which can be generated by any single lab. This is a key proof-of-principle in considering human studies, where on top of the parameters described above, the bounds of the search space are extended even further by the genetic and environmental heterogeneity that will be inherent in human studies. The inter-study comparability of data from many prior studies has been poor due to technical variation in sample preparation, so this will be an important consideration as we move toward similar undertakings with human samples. Many of these lessons can also be applied to the execution of other systems biology approaches that attempt to catalog post-transcriptional aspects of cellular diversity, which could also reveal additional layers of population heterogeneity.
Another technical consideration that has also emerged is the applicability of deep sequencing to transcriptomics studies. Technologies such as RNA-Seq have seen an explosion of development in the years since the ImmGen project was initiated [24], and the ability of de novo sequencing to discover new isoforms generated by alternative RNA splicing and non-coding RNAs makes it attractive for more completely describing transcriptomes. However, there remain some major hurdles to widespread adoption of deep sequencing for transcriptome analysis. First, the cost of preparation for deep sequencing currently exceeds that of microarrays on a per-sample basis. Although these costs can be reduced by multiplexing samples in lanes (“indexing” or “barcoding”), this procedure, as typically employed, negates the potential sensitivity advantages of deep sequencing. Second, the computational and personnel infrastructure required for analysis of deep sequencing data far exceeds that required for large-scale microarray analysis, which for large datasets can already be substantial. Nonetheless, we anticipate that future improvements will address these issues to make RNA-Seq better suited to ImmGen-scale analyses.
In the grand scale of systems immunology, it is important to remember that identifying steady-state transcriptomes is but one step in understanding the complexity of cellular diversity and function. Even with regard to transcription alone, similar efforts will be required to understand histone modifications and occupancy, methylation states, and site-specific binding of transcription factors and their associated complexes (contextualized in more detail in [2]) on a subset-specific level. Technology has also advanced in the area of single-cell analysis, with the first dedicated commercial systems for single-cell transcriptomics becoming available this year. Analyzing the genome-wide transcriptome of individual cells of populations that are observed to be homogeneous by a limited set of flow cytometric markers is arguably the best approach available for efficiently determining whether or not further levels of subset heterogeneity exist. The technology faces a number of challenges related to reproducibility, controlling for cell cycle stage, and cost, and multiplying the existing ~270 populations by the analysis of another hundred-fold (i.e., 100 individual cells per population) is a daunting endeavor. However, such a compendium would comprise a strong foundation for many future studies, lending confidence to investigators that the population under study is in fact homogeneous, with an ultimate impact on the precision of cell-targeted therapeutics.
Conclusions
The transcriptomes obtained by the ImmGen Consortium are a significant step toward understanding leukocyte population structure and heterogeneity in function. In addition to these global phenomena, the association of new genes with immunological processes of interest will accelerate research in multiple areas. The lessons learned from the undertaking demonstrate that similar approaches for human immunology, either by microarrays or deep sequencing technologies, are a feasible and worthwhile goal for consortium biologists.
Highlights.
High-quality transcriptomes have been obtained for ~270 mouse leukocyte subsets
Discoveries include new phenotyping markers, new subsets, and novel regulators
Current transcriptome studies are underway to evaluate perturbed states
Acknowledgments
We thank all colleagues and collaborators in the ImmGen project for enriching interactions and contributions, and the successive members of the ImmGen core team (J. Ericson, K. Rothamel, S. Davis, R. Paik, R. Cruse). C.C.K. is supported by NIAID R00 AI085035; LLL is an American Cancer Society Professor and supported by NIH grants AI066897, AI068129, and CA095137. The ImmGen project is supported by grant R24-AI072073 from NIH/NIAID.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1**.Benoist C, Germain RN, Mathis D. A plaidoyer for “systems immunology”. Immunol Rev. 2006;210:229–234. doi: 10.1111/j.0105-2896.2006.00374.x. An eloquent perspective on the value of systems immunology and the challenges it faces in a historically hypothesis-focused system. [DOI] [PubMed] [Google Scholar]
- 2.Germain RN, Meier-Schellersheim M, Nita-Lazar A, Fraser IDC. Systems biology in immunology: a computational modeling perspective. Annu Rev Immunol. 2011;29:527–585. doi: 10.1146/annurev-immunol-030409-101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dagan T. Phylogenomic networks. Trends Microbiol. 2011;19:483–491. doi: 10.1016/j.tim.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Benoist C, Lanier L, Merad M, Mathis D. Consortium biology in immunology: the perspective from the Immunological Genome Project. Nat Rev Immunol. 2012;12:734–740. doi: 10.1038/nri3300. [DOI] [PubMed] [Google Scholar]
- 6.Hyatt G, Melamed R, Park R, Seguritan R, Laplace C, Poirot L, Zucchelli S, Obst R, Matos M, Venanzi E, et al. Gene expression microarrays: glimpses of the immunological genome. Nat Immunol. 2006;7:686–691. doi: 10.1038/ni0706-686. [DOI] [PubMed] [Google Scholar]
- 7.Shay T, Kang J. Immunological Genome Project and systems immunology. Trends Immunol. 2013 doi: 10.1016/j.it.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heng TSP, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 9.Chow A, Brown BD, Merad M. Studying the mononuclear phagocyte system in the molecular age. Nat Rev Immunol. 2011;11:788–798. doi: 10.1038/nri3087. [DOI] [PubMed] [Google Scholar]
- 10.Mabbott NA, Kenneth Baillie J, Hume DA, Freeman TC. Meta-analysis of lineage-specific gene expression signatures in mouse leukocyte populations. Immunobiology. 2010;215:724–736. doi: 10.1016/j.imbio.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Hume DA, Summers KM, Raza S, Baillie JK, Freeman TC. Functional clustering and lineage markers: insights into cellular differentiation and gene function from large-scale microarray studies of purified primary cell populations. Genomics. 2010;95:328–338. doi: 10.1016/j.ygeno.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 12*.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. Proposes the use of MerTK and CD64 to distinguish macrophages from dendritic cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. Investigates a large number of dendritic cell subsets at the molecular level, including identification of candidate regulators of dendritic cell diversity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narayan K, Sylvia KE, Malhotra N, Yin CC, Martens G, Vallerskog T, Kornfeld H, Xiong N, Cohen NR, Brenner MB, et al. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat Immunol. 2012;13:511–518. doi: 10.1038/ni.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, Best JA, Goldrath AW, Lanier LL. Molecular definition of the identity and activation of natural killer cells. Nat Immunol. 2012;13:1000–1009. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, Kang J, Brenner MB, Monach P, Shinton SA, Hardy RR, et al. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol. 2013;14:90–99. doi: 10.1038/ni.2490. Addresses questions about subset diversity in iNKT cells and the relatedness of these subsets to one another and other lymphocyte populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malhotra D, Fletcher AL, Astarita J, Lukacs-Kornek V, Tayalia P, Gonzalez SF, Elpek KG, Chang SK, Knoblich K, Hemler ME, et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat Immunol. 2012;13:499–510. doi: 10.1038/ni.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Best JA, Blair DA, Knell J, Yang E, Mayya V, Doedens A, Dustin ML, Goldrath AW, Monach P, Shinton SA, et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013 doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germain RN. Maintaining system homeostasis: the third law of Newtonian immunology. Nat Immunol. 2012;13:902–906. doi: 10.1038/ni.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 22.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 23.Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]