Abstract
Noisy and overlapped mass spectrometry data hinders the sequence coverage that can be obtained from Hydrogen Deuterium exchange analysis, and places a limit on the complexity of the samples that can be studied by this technique. Advances in instrumentation have addressed these limits, but as the complexity of the biological samples under investigation increases, these problems are reencountered. Here we describe the use of binomial distribution fitting with asymmetric linear squares regression for calculating the accurate deuterium content for mass envelopes of low signal or that contain significant overlap. The approach is demonstrated with a test data set of HIV Env gp140 wherein inclusion of the new analysis regime resulted in obtaining exchange data for 42 additional peptides, improving the sequence coverage by 11%. At the same time, the precision of deuterium uptake measurements was improved for nearly every peptide examined. The improved processing algorithms also provide an efficient method for deconvolution of bimodal mass envelopes and EX1 kinetic signatures. All these functions and visualization tools have been implemented in the new version of the freely available software, HX-Express v2.
Introduction
In the last 20 years Hydrogen Deuterium exchange coupled to mass spectrometry (HDX-MS) has become a valuable tool for studying protein conformation, dynamics and protein-protein/protein-ligand interactions [1, 2]. The kinetics of the exchange between backbone amide hydrogens with solvent deuterium yields significant insight into the relative degree of order/disorder, hydrogen bonding, and solvent accessibility within proteins [3, 4]. As most HDX studies examine dozens to hundreds of peptic fragments, data analysis often becomes the most time consuming step. In the last decade many H/D analysis software packages have emerged, both open source and commercial; AutoHD [5], HX-Express [6], DEX [7], Hydra [8], HeXicon [9], Ex-MS [10], HD-Benchtop [11], Mass Analyzer [12], HD-examiner (Sierra Analytics), and DynamX (Waters), to name a few. Although many of these software packages and analysis algorithms offer automation that significantly reduces the analysis time, these approaches may extract data for only the resolved, cleanly identifiable peptides, leaving significant portions of data unanalyzed. To obtain maximal sequence coverage and resolution it is necessary to analyze as many fragments as possible. To this end, it may be necessary to analyze peptides with low signal to noise or spectra in which different species overlap in chromatographic and m/z space. Species that overlap in the chromatographic dimension can sometime be partially resolved with selective integration of the chromatographic peak and in most cases require close manual inspection.
Since the earliest experiments it has been realized that uncorrelated local dynamics yield a binomial exchange profile (EX2 regime) while more global correlated unfolding events result in a bimodal exchange pattern (EX1 regime) [13]. Some spectra may also exhibit mixed EX1/EX2 exchange profiles resulting in complicated mass envelopes [14]. Deconvolution of such bimodal behavior within the exchange profile is critical for accurate interpretation of HDX data [15], but this has only recently started to become incorporated into automated HDX analysis software [9, 10]. For these and other reasons, we have advocated a semi-automated approach [6] for analysis of HDX data and inspection of all mass spectra regardless of quality.
Even with semi-automated analysis, a large number of fragments are still either too weak or too contaminated by overlapping peaks to obtain a “clean” deuterium uptake profile. These difficulties can be partially alleviated by using instrumentation with very high resolving power and spectral deconvolution [10, 16] or by utilizing an additional separation stage via ion mobility [17]. Even with these advances in instrumentation, examination of larger and more complicated biological systems will invariably lead right back to problems associated with overlapping species. Recently, computational approaches utilizing machine learning with global examination of the exchange data have made some progress to address this problem and expand sequence coverage [18].
Here we implement an alternative, and in our view simpler approach for extracting deuterium levels for overlapped peptides. Our strategy combines and streamlines 1) binomial distribution fitting described by Chik et al [19]; and 2) asymmetric least squares regression, as commonly used for baseline correction algorithms [20]. This extremely efficient approach also serves as a rapid method for deconvolution of bimodal spectra (i.e., EX1 kinetic signatures) even for data that is of low signal to noise or contains overlapped species. These features are all available in the newly updated version of HX-Express [6], with additional tools to aid in visualizing bimodal profiles.
Methods
Hydrogen deuterium exchange data for HIV Env gp140 was collected on a Waters Synapt Q-TOF as described previously [21]. Spectra were manually integrated over the chromatographic elution profile based on extracted ion chromatograms, smoothed (typically 2 rounds of 4 channel, Savitsky-Golay method) and imported from MassLynx (Waters) into HX-Express [6] within Microsoft Excel. Spectra were processed using isotopic peak detection and centroid fitting before implementation of binomial fitting. The source code of HX-Express [6] was modified to include the functions described in the following sections and the new version is freely available from www.hxms.com/HXExpress. Deuterium uptake was calculated by calculating the percent exchange for each time point relative to a “zero” and a fully deuterated standard to account for both “IN” and back-exchange [22].
Binomial fitting and asymmetric linear squares regression
Modules for binomial fitting were developed and implemented with Visual Basic scripts as follows. Isotopic peaks in the spectra were converted to integer values by subtracting the monoisotopic peak m/z and multiplying by the charge state (z). The peptide natural abundance isotopic distribution was either read directly from the undeuterated spectra or calculated from the amino acid (and carbohydrate) composition [5]. The number of slow-exchanging amides was based on the peptide sequence and used as the number of events (n) in calculation of the binomial distribution function (eq. 1). n was initially estimated as the number of amino acids minus the number of prolines, minus 1 for the N-terminal residue. In some cases peptides contained fast exchanging residues, which back-exchange rapidly [23] and therefore n was set slightly lower. For glycopeptides the glycan groups need to be taken into account as some carbohydrate groups (primarily N-Acetyl hexosamines) will also retain deuterium [24]. Examination of the fit to a fully deuterated standard was used to assess whether the value of n generated the correct envelope width, and with the majority of peptides the initial estimate was accurate. For a few highly protected peptides, better fits were achieved with a slightly lower n for the early time points, presumably because some amides have yet to exchange by then.
The centroid shift in each spectrum relative to the undeuterated profile served as an initial estimate of the binomial distribution probability (“p”). Each theoretical peak (Imcalc) was reconstructed by applying the natural abundance profile to each peak in the binomial distribution with up to 3 points of zero padding on both sides of the mass envelope as described by Chik et al [19]. The peak intensities were scaled by a weighting term (A), using the intensity of the highest data point as an initial guess. Least squares regression was performed using the Gauss-Newton algorithm implemented within the Excel Solver module (Microsoft, Redmond WA) to minimize the discrepancy (χ2) between the isotopic peaks and the calculated binomial profile by varying p and A (eq. 2). For data sets showing overlap with an interfering ion, the asymmetry term (λ) was user defined (typically between 2 and 10, based on visual assessment) and applied to points where the fit exceeded the data. The resulting degree of deuteration was calculated as either the percentage relative to the undeuterated (pUN) and fully deuterated (pTD) values (pt-pUN)/(pTD-pUN) or average deuterium uptake (pt·n), and was plotted in the summary page for each time point (t) and condition.
| Eq 1: |
| Eq 2: |
Full spectral reconstruction and fitting
The initial step of binomial fitting to raw spectral data was performed as described above except the theoretical spectrum was always reconstructed by calculating the natural abundance isotopic distribution from chemical composition. The integer offset for each isotopic peak was adjusted slightly to account for the accurate mass increase from deuterium as described by Kan et al [10]. A Gaussian distribution was used to represent each isotopic peak as described by Pascal et al [25] (Eq. 3). Fitting was performed by minimizing χ2, by varying p, A, σ, and an additional baseline offset value. Initial estimates for p and A were just as described above. The initial value for the Gaussian peak width (σ) was estimated from the instrumental resolution. The peak positions (μ) were also allowed to vary within a specified tolerance, estimated from instrumental accuracy.
| Eq 3: |
Bimodal deconvolution
Spectra showing additional broadening relative to the binomial distribution were further analyzed by fitting to a combination of two binomial functions. The p value for the more highly deuterated species (p2) was bracketed as larger than the less deuterated species (p2 > p1) and less than or equal to the fully deuterated labeling probability (p2 ≤ pTD). Macros for performing the fitting, summarizing the results, and generating exchange plots were implemented within Visual Basic v6.5 (Microsoft) and integrated into the user-selectable menu options of HX-Express v2.
Results and Discussion
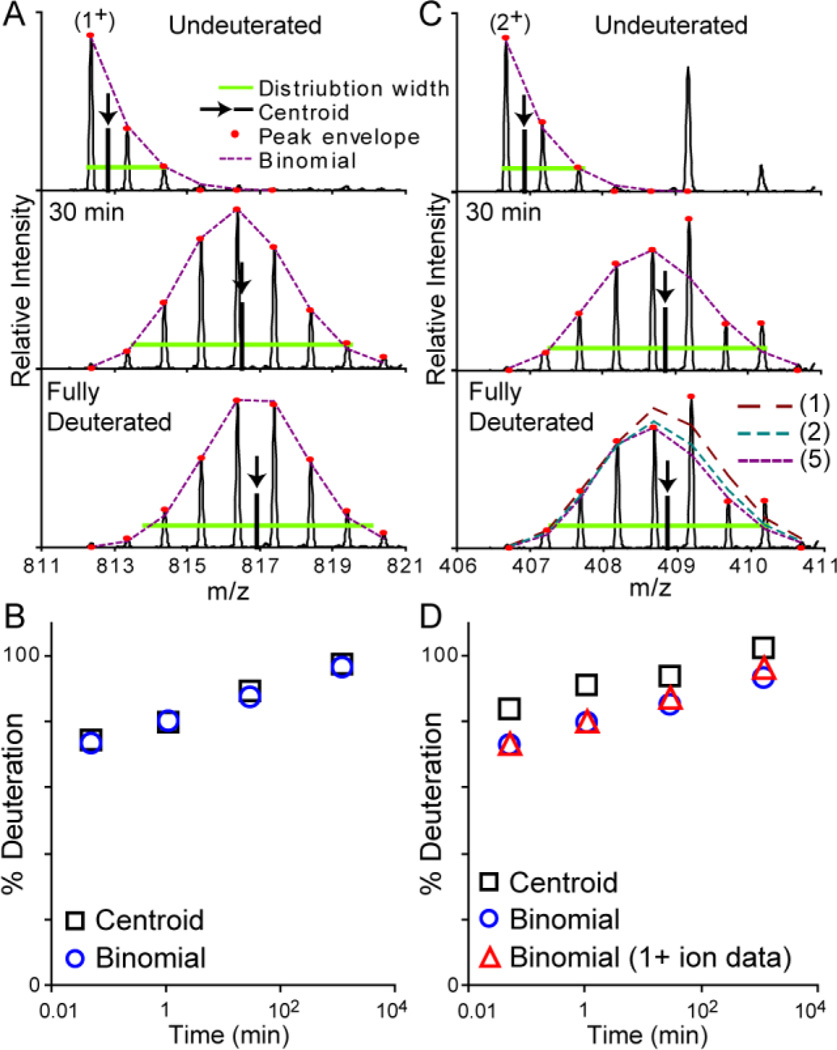
A total of 179 peptic peptides from HIV gp140 were analyzed in HX-Express v2 using deuterium calculation with both conventional centroiding and binomial distribution fitting. For most well-resolved peaks with good signal-to-noise the two methods result in identical deuterium uptake profiles. An example of this is shown in Figure 1A for the 1+ ion of a peptide. The data also contained a strong signal for the 2+ ion, but with a nearby 1+ contaminant peak that could not be resolved chromatographically (Figure 1C). The more conventional centroid processing approach results in a deuterium uptake plot that is skewed higher due to the presence of the overlapping fragment within the mass envelope at later deuterium exchange time points. The same effect persists even when fitting a binomial distribution to these time points. Including an asymmetry term (λ) into the calculation, in this case λ=5, results in a greater error contribution for peaks of lower intensity than the predicted distributions, thereby favoring the distribution to fit to the lower intensity “uncontaminated” peaks. The net benefit is evident when comparing the data for the uncontaminated 1+ ion to the 2+ ion fit using the asymmetric term (Figure 1D).
Figure 1.
Example of binomial fitting of an overlapped peptide. A) The deuterated mass envelope of the 1+ ion contains no overlapping peaks and fits well to a binomial distribution. B) The exchange profile from this data extracted from the binomial fitting or conventional centroiding is essentially identical. C) The 2+ ion of this same fragment contains an overlapping 1+ ion from another species. An asymmetry term (λ) of 5 was used for the binomial fitting of these overlapped mass envelopes. The fully deuterated spectra shows the effects of using λ=1 (no asymmetry), 2, and 5. D) The binomial fitting with λ=5 (circles) results in an accurate depiction of the 2+ mass envelope, and the measured deuterium uptake is identical to that from the 1+ ion (triangles), unlike that obtained from centroid analysis (squares).
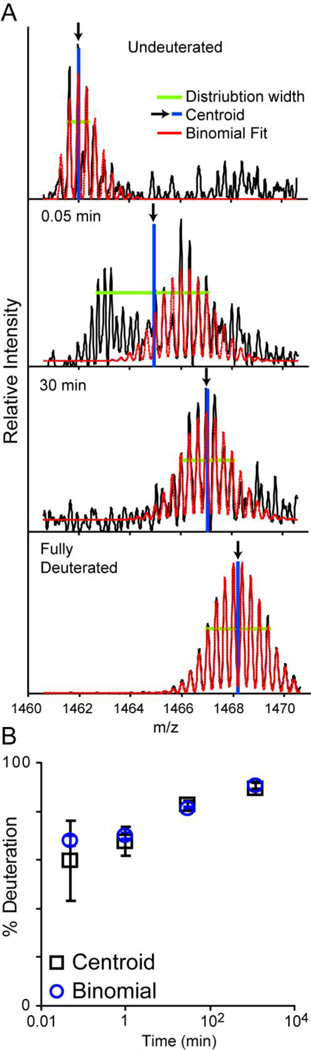
The binomial fitting routine converts the position of each isotopic peak as integer offsets from the monoisotopic peak and with noisier data the isotopic peak picking algorithms may fail to capture the full isotopic envelope. This problem occurred for nearly one-fifth of the peptides within our HIV gp140 data set. To alleviate the problems associated with noisy data and poorly picked peaks, the binomial fitting scheme was expanded to reconstruct the full spectrum and fit to the raw spectral data, instead of just the single points for each isotopic peak. An example of this is shown in Figure 2. In this case the data was not only of low signal-to-noise but the peptide isotope distributions for early deuteration time points were overlapped with signals from another peptide (Figure 2A, 3 sec time point). The binomial distribution restricts the width of the mass envelope and the charge state; and instrumental accuracy and resolution further restrain the possible solutions. While processing data of such weak signal invariably requires close visual inspection, the binomial fitting results in a more precise fit compared to conventional centroiding, which can be seen from the error bars from duplicate measurements (Figure 2B). Examination of the residuals of the modeled distribution subtracted from the raw spectra is also useful for assessing the quality of the fit, and is automatically generated in HX-Express v2 (Supplementary Figure 1).
Figure 2.
Binomial fitting to raw spectral data. A) A 3+ ion with relatively low signal is fit using the reconstructed spectra from the binomial distribution (dashed red lines). The fits for each time point along with the residuals are shown in Supplementary Figure 1. Arrows show the centroid position based on the distribution width. The lower m/z mass envelope observed in the 0.05 min time point is from a contaminating 4+ ion. B) Binomial fitting show much less variance in the final exchange profile relative to centroid analysis of the raw data. Error bars represent the standard deviation from duplicate measurements.
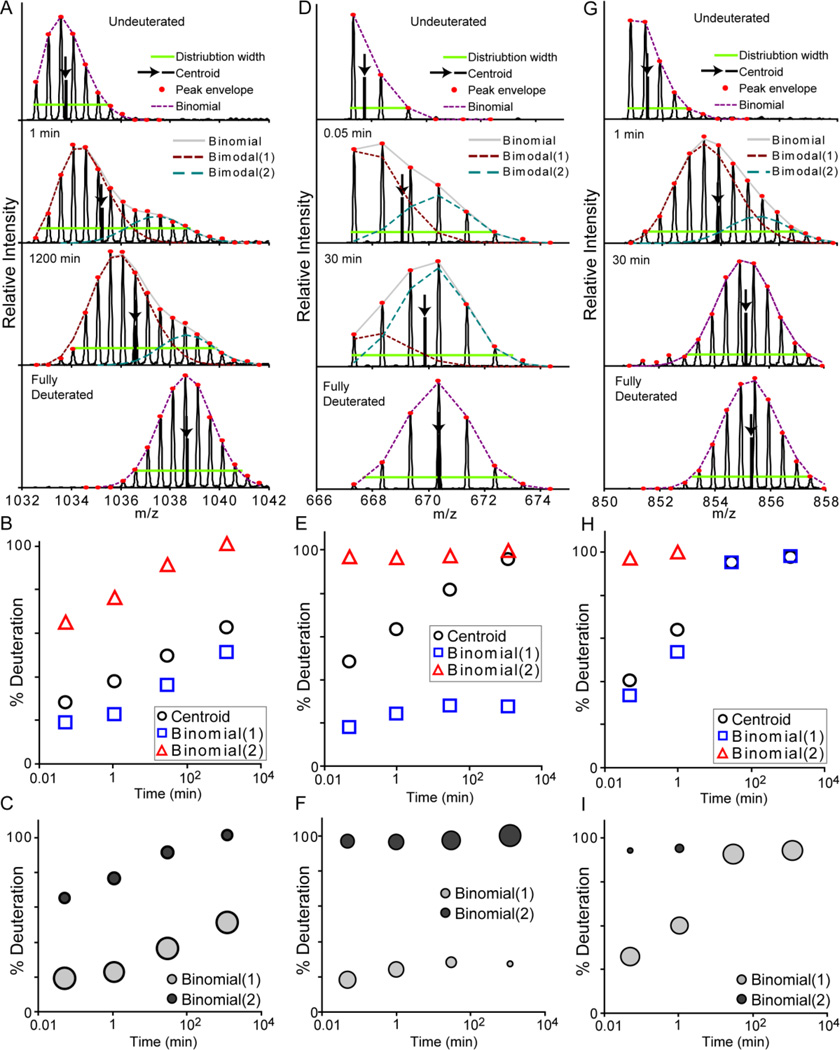
The binomial fitting approach also serves to detect the presence of bimodal exchange profiles, i.e., EX1 kinetic signatures or multiple distinct conformations. Peak width analysis can also reveal this type of behavior [14], but since a small degree of mass envelope broadening occurs even within the EX2 exchange regime, it is not always effective at detecting subtle deviations from EX2 behavior. Fitting with the binomial distribution intrinsically accounts for this broadening effect and therefore provides a robust method for detecting even subtle bimodal behavior. Figure 3 shows three examples of profiles with bimodal distributions. As described for even the earliest measurements of peptide hydrogen exchange MS [26], a combination of multiple binomial distribution functions can be fit to the data for deconvolution of the species present in the mass envelopes. In HX-Express v2, this process quickly and effectively resolves the two species and presents them for visual inspection and validation (Figure 3A, D, G). The deuterium uptake curves for each species can be determined, and provide far more insight into the system than is obtainable by simple centroid analysis (Figure 3B, E, H).
Figure 3.
Deconvolution of bimodal patterns with two binomial distributions. Three examples are shown of fragments (A–C), (D–F) and (G–I) exhibiting bimodal distributions in the deuterated mass envelopes. A) Deconvolution of the spectra from the first peptide by fitting with two binomial distributions resolves the exchange profiles of both species (B). C) The bubble plot shows the exchange profiles with the relative size of each point reflecting the relative population of each species. For this peptide, the size of the bubbles is relatively constant through the time course, indicating the presence of two distinct conformations. D) In the case of the second peptide, the exchange profile indicates predominant EX1 exchange kinetics, and the bubble plot shows the shift between the exchange profiles over time (E, F). G) The third peptide exhibits a bimodal profile only at the early time points. The increase in the population of the highly deuterated species suggests that this is attributed to EX1 kinetics rather than distinct conformations in solution (H, I).
Bubble plots are a useful way to visualize the data obtained from bimodal deconvolution. These plots show the exchange profile of both species with the bubble size correlating to the relative population of each species at each point (Figure 3C, F, I). This information can be useful for differentiating between EX1 kinetics vs. the presence of distinct conformations. For the first peptide (Figure 3C), the relative populations of the two species remain constant over the course of deuterium exchange, indicating the presence of two distinct, non-interchanging conformations. The second peptide (Figure 3F) shows a change in the relative population over time, indicating EX1 kinetics. The behavior of the third peptide (Figure 3I) is less definitive. The rise in the relative population of the highly deuterated species from 0.14 +/− 0.02 to 0.24 +/− 0.04 (Average +/− standard deviation) between the 3 sec and 1 min time points indicates the presence of mixed EX1/EX2 kinetics rather than two distinct conformations. The relative populations are also included in the final output in HX-Express v2, so they can be used to estimate kinetic parameters associated with EX1 events. For even more convoluted data, which is rare, a triple binomial fitting algorithm is available. The bimodal deconvolution routine can also be used in combination with fitting to raw spectral data, however this is roughly 10 times slower, requiring up to 10–20 seconds of processing time per spectrum on a modern desktop computer.
Lastly we note that the accuracy of bimodal deconvolution is dependent on the ability to resolve the two species. For two species with only a minor difference in exchange kinetics, the mass envelopes will be poorly resolved, and the certainty in characterizing each species becomes limited. Having a larger number of data points along the mass envelope helps constrain the binomials for more accurate fits. For this reason we use the highest possible level of deuterium content in the exchange step to distribute the mass envelope over a larger m/z range. While is detrimental to the signal to noise and raises the risk of overlap [27], in many cases it is necessary for the accurate characterization of the exchange profile.
Conclusions
The application of binomial distributions combined with asymmetric least squares fitting serves as a useful tool for obtaining accurate deuterium content from noisy or overlapped mass spectrometry data. This type of data is encountered frequently in our laboratory, especially in the analysis of large glycoproteins which are typically only available in small quantities. For such difficult systems, simply increasing the quantity of protein introduced into the mass spectrometer is generally not an option. In the case of HIV Env gp140, binomial fitting procedures, as implemented with HX-Express v2, provided data for 8 additional (unique) peptides, improving the sequence coverage from 80% to 91%. We note that a certain level of peptide-to-peptide parameter optimization is required along with close visual inspection of the fitting to ensure no erroneous fits are generated. While this approach is modestly slower than analysis software packages offering user-free automation, semi-automated analysis provides more confidence in the final results. HX-Express v2 maintains a semi-automated analysis approach, and now includes tools for binomial fitting, bimodal deconvolution, asymmetric least squares regression for handling overlapped data, and visualization tools for interpreting bimodal exchange behavior.
Supplementary Material
Acknowledgments
We wish to thank Jamie R. Williamson and Ryan S. Littlefield for insightful discussion and assistance with programming. Members of the Lee and Engen laboratories are acknowledged for their help in testing and debugging HX-Express v2. This work was supported by NIH grants F32-GM097805 (MG), R00-GM080352 and R01-GM099989 (KKL), and R01-GM086507 and R01-GM101135 (JRE).
References
- 1.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17(11):1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal Bioanal Chem. 2010;397(3):967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engen JR, Smith DL. Investigating protein structure and dynamics by hydrogen exchange MS. Anal Chem. 2001;73(9):256A–265A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 4.Mandell JG, Baerga-Ortiz A, Falick AM, Komives EA. Measurement of solvent accessibility at protein-protein interfaces. Methods Mol Biol. 2005;305:65–80. doi: 10.1385/1-59259-912-5:065. [DOI] [PubMed] [Google Scholar]
- 5.Palmblad M, Buijs J, Hakansson P. Automatic analysis of hydrogen/deuterium exchange mass spectra of peptides and proteins using calculations of isotopic distributions. J Am Soc Mass Spectrom. 2001;12(11):1153–1162. doi: 10.1016/S1044-0305(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 6.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17(12):1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 7.Hotchko M, Anand GS, Komives EA, Ten Eyck LF. Automated extraction of backbone deuteration levels from amide H/2H mass spectrometry experiments. Protein Sci. 2006;15(3):583–601. doi: 10.1110/ps.051774906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slysz GW, Baker CA, Bozsa BM, Dang A, Percy AJ, Bennett M, Schriemer DC. Hydra: software for tailored processing of H/D exchange data from MS or tandem MS analyses. BMC Bioinformatics. 2009;10:162. doi: 10.1186/1471-2105-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreshuk A, Stankiewicz M, Lou X, Kirchner M, Hamprecht FA, Mayer MP. Automated detection and analysis of bimodal isotope peak distributions in H/D exchange mass spectrometry using HeXicon. International Journal of Mass Spectrometry. 2010;302:125–131. [Google Scholar]
- 10.Kan ZY, Mayne L, Chetty PS, Englander SW. ExMS: data analysis for HXMS experiments. J Am Soc Mass Spectrom. 2011;22(11):1906–1915. doi: 10.1007/s13361-011-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, Marciano D, Novick S, Goswami D, Chalmers MJ, Griffin PR. HDX workbench: software for the analysis of H/D exchange MS data. J Am Soc Mass Spectrom. 2012;23(9):1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Zhang A, Xiao G. Improved protein hydrogen/deuterium exchange mass spectrometry platform with fully automated data processing. Anal Chem. 2012;84(11):4942–4949. doi: 10.1021/ac300535r. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2(4):522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J Am Soc Mass Spectrom. 2006;17(11):1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Ramachandran P, Kumar R, Gross ML. H/D Exchange Centroid Monitoring is Insufficient to Show Differences in the Behavior of Protein States. J Am Soc Mass Spectrom. 2013;24(3):450–453. doi: 10.1007/s13361-012-0555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazazic S, Zhang HM, Schaub TM, Emmett MR, Hendrickson CL, Blakney GT, Marshall AG. Automated data reduction for hydrogen/deuterium exchange experiments, enabled by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2010;21(4):550–558. doi: 10.1016/j.jasms.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iacob RE, Murphy JP, 3rd, Engen JR. Ion mobility adds an additional dimension to mass spectrometric analysis of solution-phase hydrogen/deuterium exchange. Rapid Commun Mass Spectrom. 2008;22(18):2898–2904. doi: 10.1002/rcm.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lou X, Kirchner M, Renard BY, Kothe U, Boppel S, Graf C, Lee CT, Steen JA, Steen H, Mayer MP, Hamprecht FA. Deuteration distribution estimation with improved sequence coverage for HX/MS experiments. Bioinformatics. 2010;26(12):1535–1541. doi: 10.1093/bioinformatics/btq165. [DOI] [PubMed] [Google Scholar]
- 19.Chik JK, Vande Graaf JL, Schriemer DC. Quantitating the statistical distribution of deuterium incorporation to extend the utility of H/D exchange MS data. Anal Chem. 2006;78(1):207–214. doi: 10.1021/ac050988l. [DOI] [PubMed] [Google Scholar]
- 20.Eilers PH. Parametric time warping. Anal Chem. 2004;76(2):404–411. doi: 10.1021/ac034800e. [DOI] [PubMed] [Google Scholar]
- 21.Guttman M, Kahn M, Garcia NK, Hu SL, Lee KK. Solution Structure, Conformational Dynamics, and CD4-Induced Activation in Full-Length, Glycosylated, Monomeric HIV gp120. J Virol. 2012;86(16):8750–8764. doi: 10.1128/JVI.07224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 23.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttman M, Scian M, Lee KK. Tracking hydrogen/deuterium exchange at glycan sites in glycoproteins by mass spectrometry. Anal Chem. 2011;83(19):7492–7499. doi: 10.1021/ac201729v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pascal BD, Chalmers MJ, Busby SA, Mader CC, Southern MR, Tsinoremas NF, Griffin PR. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinformatics. 2007;8:156. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z. Protein hydrogen exchange determined by mass spectrometry: a new tool for probing protein high-order structure and structural changes. Ph.D. Thesis. Purdue University; 1995. [Google Scholar]
- 27.Slysz GW, Percy AJ, Schriemer DC. Restraining expansion of the peak envelope in H/D exchange-MS and its application in detecting perturbations of protein structure/dynamics. Anal Chem. 2008;80(18):7004–7011. doi: 10.1021/ac800897q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.