Abstract
Mice lacking TGF-β3 exhibit an incompletely penetrant failure of the palatal shelves to fuse leading to cleft palate. The defect appears to result from impaired adhesion of the apposing medial edge epithelia of the palatal shelves and subsequent elimination of the mid-line epithelial seam. No craniofacial abnormalities were observed. This result demonstrates that TGF-β3 affects palatal shelf fusion by an intrinsic, primary mechanism rather than by effects secondary to craniofacial defects.
Members of the transforming growth factor-β (TGF-β) gene family have biological activities that control cell proliferation, migration and differentiation, regulation of extracellular matrix deposition and epithelial-mesenchymal transformation1–3. Mammals contain three highly conserved isoforms of TGF-β, termed TGF-β1, TGF-β2 and TGF-β3, which display distinctive, although at times overlapping, spatial and temporal expression patterns4–6. Previous studies suggested that TGF-β3 may play a crucial role during palatogenesis7–9, Meckel’s cartilage formation10, cardiac morphogenesis11, mammary gland development12 and wound healing13. Other tissues expressing TGF-β3 in significant levels are cartilage, bone, brain and lung4–6,14.
In mammalian palatogenesis apposition of the palatal shelves, adhesion of the medial edge epithelia (MEE) and subsequent elimination of the epithelial seam lead to a seamless mesenchymal shelf separating the oral and nasal cavities15. In vitro organ culture studies indicate that TGF-β1 and TGF-β2 accelerate palatal shelf fusion16,17 and that antisense oligodeoxynucleotides or neutralizing antibodies to TGF-β3, but not to TGF-β1 or TGF-β2, block the fusion process9. We have now created mice deficient in TGF-β3, and show that this factor has a role in palatal shelf fusion by means of an intrinsic, primary mechanism and not by effects secondary to craniofacial morphometrics. A comparison of this defect to the inflammatory disorder of TGF-β1-deficient mice18–21 indicates no overlap in their essential functions.
Mutation of TGF-β3 in ES cells
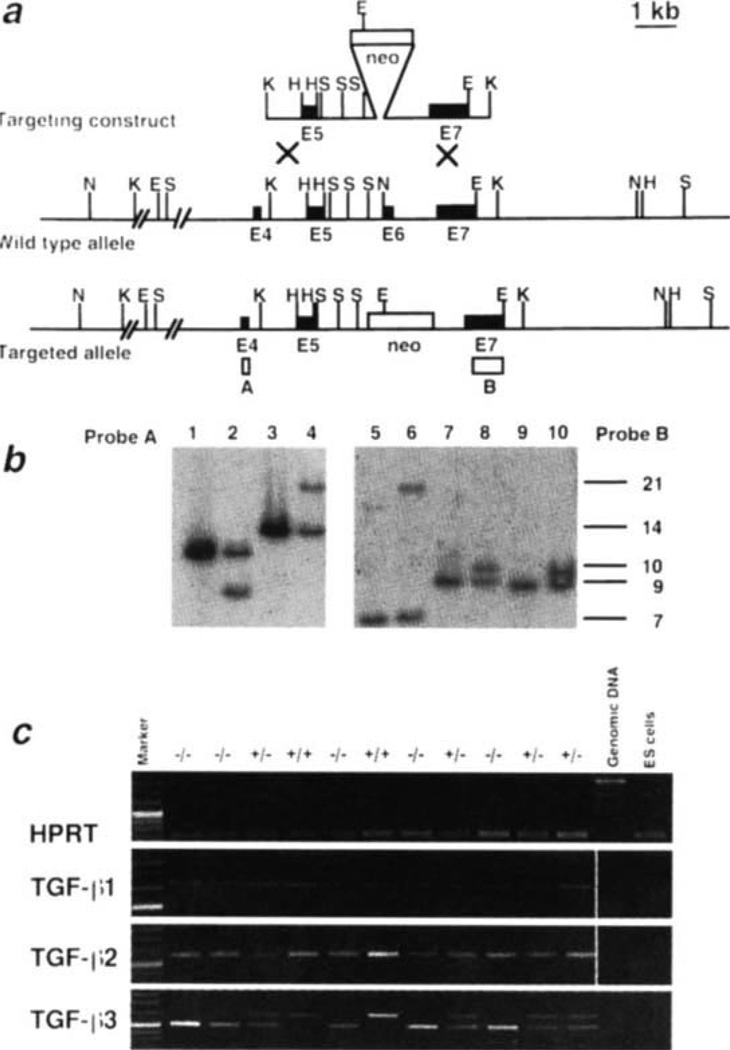
The TGF-β3 gene was mutated in ES cells (Fig. 1a) by replacing exon 6, the first full exon encoding sequences of the active domain of the protein, with the neomycin-resistance gene from pMC1neo22. Diagnostic Southern blots of the clone I98 indicated that the locus was successfully targeted; the proper genomic regions flanking both sides of the target site remained intact (Fig. 1b). Probing with a neo-gene probe indicated that there was only one integration site (not shown). Consequently, only the TGF-β3 locus has been disrupted. RT-PCR analysis of whole 11.5-(Fig. 1c) and 15.5-day embryos (not shown) indicated no TGF-β3 expression in homozygous mutant embryos, and revealed no significant change in the expression of TGF-β1 or TGF-β2 in the absence of TGF-β3.
Fig. 1.
Generation of TGF-β3-deficient mice, a, Gene targeting strategy. TGF-β3 exons are indicated by filled boxes, the neomycin gene by a shaded box. Restriction sites are EcoRI (E), HincII (H), KpnI (K), NsiI (N) and SacI (S). Probe A (open box) is a PCR-derived fragment containing exon 4, and lies outside the targeting vector. Probe B is a 1.1-kb BstXI-EcoRI fragment containing exon 7 sequences, which are included in the targeting vector. Both probes were used to test for targeted ES cells and to genotype mice, b, Southern blot analysis of genomic DNA from ES cells. Lanes 1, 3, 5, 7, 9 contain DNA from untransfected E14-1 ES cells, lanes 2, 4,6, 8,10 contain DNA prepared from ES ceil clone I98 showing homologous recombination. Enzymes used: EcoRI in lanes 1,2; NsiI in lanes 3, 4,5,6; HincII in lanes 7, 8 and SacI in lanes 9 and 10. Probe A: EcoRI, 10.5-kb and 8.6-kb band with the wild-type and mutated allele, respectively. Similarly, NsiI, a 14-kb and a 21-kb band, respectively. Probe B: NsiI gave a 7-kb and a 21-kb band with wild-type and null allele DNA, respectively; HincII, a 9-kb and a 10-kb band, respectively; SacI, 8.8-kb and 9.8-kb band, respectively, c, RT-PCR analysis of whole day 11.5 embryo RNA derived from wild-type (+/+), heterozygous (+/−) and homozygous (−/−) litter mate embryos.
Cleft palate in TGF-β3 null mutants
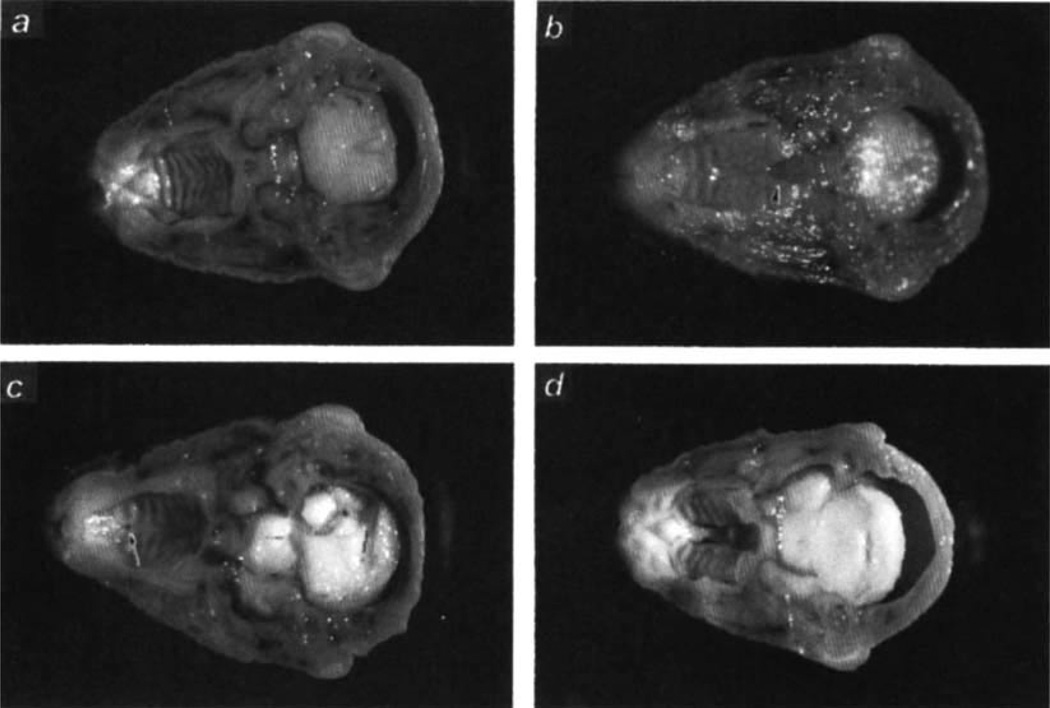
The targeted ES cell clone I98 was used to produce chi-maeric mice, which were mated with CF-1, C57BL/6 or 129/Sv mice. Heterozygous offspring showed no apparent phenotype. Intercrossing heterozygotes did not yield viable TGF-β3-null pups (Table 1). Analysis of embryos at various developmental stages (days E10.5–19.5) revealed that homozygous mutant embryos were present in the expected mendelian ratio of roughly 25% (Table 1). Close observation at parturition revealed a subset of pups that began gasping after approximately four hrs, and subsequently became cyanotic and dehydrated. All of these pups died within 24 hrs of birth, and almost all (41/47) were genotyped as homozygous mutants. The measured rate of spontaneous cleft palate on this genetic background (129/Ola × CF-1) was 3% and fully accounted for the wild-type animals with cleft palate. The TGF-β3 mutants could not successfully suckle, and the stomachs of those examined were empty. Gross morphological analyses of late gestation embryos and neonates, revealed cleft palate in all three genetic backgrounds — with variable expressivity. On the 129/Ola × CF-1 background 103 homozygous mutants were analysed for cleft palate: 92 had a posterior cleft palate (Fig. 2b), 9 had anterior clefts, that is, failure of fusion of the primary and secondary palates (Fig. 2c), and only 2 had complete clefts. The cleft palate phenotype was most severe on the C57BL/6 background with 7/15 mutants exhibiting a complete cleft palate (Fig. 2d) and 8 exhibiting only a posterior cleft. Two 129/Sv mutants were obtained; both had a posterior cleft palate. No animals had associated cleft lip.
Table 1.
Viability of offspring from TGF-β3 heterozygous intercrosses.
| Strain* | Stage | +/+ | +/− | −/− | Total |
|---|---|---|---|---|---|
| 129/Ola × CF-1 | E10.5–11.5 | 21 | 40 | 15 | 76 |
| 129/Ola × CF-1 | E12.5–13.5 | 34 | 55 | 41 | 130 |
| 129/Ola × CF-1 | E14.5–15.5 | 49 | 87 | 44 | 180 |
| 129/Ola × CF-1 | E16.5–17.5 | 13 | 14 | 15 | 42 |
| 129/Ola × CF-1 | E18.5–19.5 | 12 | 21 | 15 | 47 |
| 129/Ola × CF-1 | day 1 neonates | 15 | 31 | 19 | 65 |
| 129/Ola × CF-1 | 3 weeks of age | 78 | 155 | 0 | 233 |
| 129/Ola × C57BL/6 | 3 weeks of age | 31 | 30 | 0 | 61 |
| 129/Ola × 129/Sv | 3 weeks of age | 22 | 21 | 0 | 43 |
Mouse backgrounds: 129/Ola × CF-1, F2, one to four backcross generations on CF-1; 129/Ola × C57BL/6, F2, one to two backcross generations on C57BL/6; 129/Ola × 129/Sv, F2, one to two backcross generations on 129/Sv. Morning of appearance of copulation plug is defined as day 0.5.
Fig. 2.
Cross-sections of heads showing secondary palate, a–c, 129/Ola × CF-1 background; embryonic d 18.5. a, Wild-type (+/+) embryonic head with fused palate, b, TGF-β3-null (−/−) embryo with a cleft in the posterior palate, c, TGF-β3-null (−/−) embryo with a cleft in the anterior palate region, d, Day 1 −/− neonate with complete cleft palate; 129/Ola × C57BL/6 background. Heads were Bouin´s-fixed.
Palate development
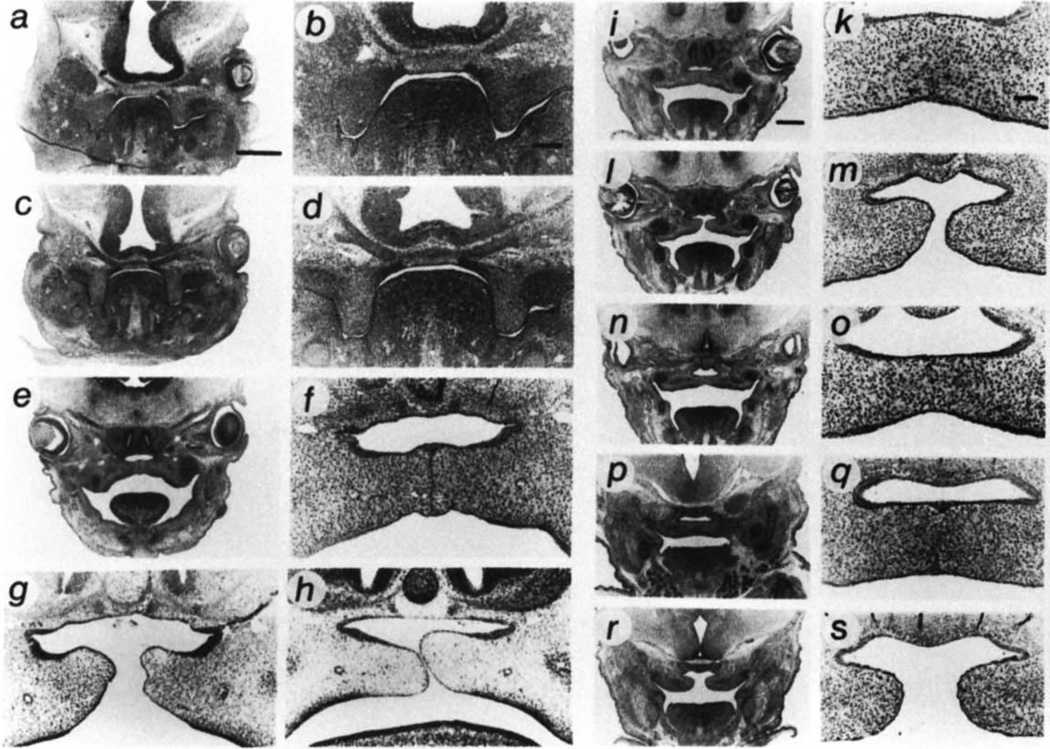
Because cleft palate can result from a number of mechanisms15,23, we analysed palate development in homozygous mutant embryos and wild-type litter mate controls to further specify the cause of the lesion. No mutants analysed had malformations of cartilage, bone or other craniofacial structures — for example, mandible — that could lead to cleft palate by means of mechanical obstruction. Data are presented for the most common lesions observed in the 129/Ola × CF-1 background. During early palatogenesis, days E11.5–E13.5 (the morning of the day of plug appearance was counted as day 0.5), mutants and controls were histologically indistinguishable (Fig. 3a–d). At this time the palatal shelves form from the maxillary processes, proliferate and undergo elevation to reach the horizontal position. The first differences between mutant and wild-type embryos became evident at day 14.5 when the palatal shelves begin to fuse, but only in the control embryos (Fig. 3e–h). In day 15.5 wild-type embryos fusion progressed in the anterior (not shown), middle (Fig. 3i, k) and in most cases in the posterior region (Fig. 3p, q). TGF-β3-null embryos showed either no fusion or fusion only in the middle (Fig 3n, o), and not in the anterior (not shown) or posterior palatal regions (Fig. 3r, s).
Fig. 3.
Developmental study of palate formation in TGF-β3-null mice (−/−) compared with wild-type (+/+) litter mates, a–o, Mid-palate region, a, b, +/+, d 12.5. c, d, −/−, d 13.5. e, f, +/+, d 14.5. g, h, −/−, d 14.5, two different embryos. i, k, +/+, d 15.5. l–o, −/−, d 15.5, two different embryos, shown to indicate variable expressivity of phenotype. p–s, Posterior palate region, p, q, +/+, d 15.5. r, s, −/−, d 15.5. The embryos were fixed in 4% paraformaldehyde, sectioned and stained with hematoxylineosin-alcian blue. Bar = 0.5 mm for a, c, e, i, l, n, p, r. Bar = 0.1 mm for b, d, f–h, k, m, o, q, s.
Analysis of the fused region in the TGF-β3-null embryos indicated that the lesion primarily affects the MEE rather than the underlying mesenchyme. After contact, adhesion and fusion of the shelves progresses rapidly in wild-type embryos and is followed by elimination of the MEE in the midline seam, a process in which epithelial triangles transiently appear at the junctions of the midline seam and the nasal and oral epithelia24. In contrast, mutant palates made contact (Fig. 3h), but the apposing MEE did not adhere (Fig. 3g). Subsequent rapid growth of the cranium during development causes unfused palatal shelves to separate, leaving a cleft. In cases where fusion did occur, the secondary palate was characterized by a lack of epithelial triangles and by an abnormal persistence of the midline seam (compare Fig. 3o–k, q). Nasal and oral epithelial differentiation were normal, that is, pseudostratified ciliated columnar and stratified squamous, respectively. In most cases (94/103), the palatal shelves did not fuse in the posterior region (Fig. 3r, s). Where fusion did occur, the pathological MEE seam persisted in the mutants, at least until day 18.5, whereas in wild-type animals the midline seam was absent by this time, leaving a continuous mesenchyme. Interestingly, in the TGF-β3-null embryos the midline seam is elongated and approximately 2 cells thick at all stages from days E15.5–E18.5. Only in rare cases was mesenchymal penetration of the seam observed, and in such cases, epithelial remnants were extensive and elongated, unlike normal round epithelial pearls. The initiating cause of the defect may lie in aberrant palatal shelf fusion rather than in a deficiency in proliferation or migration of the palatal shelf mesenchyme.
Immunocytochemical staining
This hypothesis was further substantiated by results of immunocytochemical staining for a variety of molecules. From days E11.5–E14.5 there were no differences in the immunocytochemical staining patterns or intensities between the TGF-β3-null mice and their wild-type litter mates for the following molecules: decorin, laminin, collagens IV, XII and XIV, tenascin, desmocollin, desmoplakin, E-cadherin and cytokeratins 1, 5, 8, 7 and 14. Likewise, there were no significant differences between the TGF-β3-null embryos and their homozygous wild-type litter mate controls from days E14.5–E18.5, except for differences which could be explained by either non-fusion of the palate or the persistent midline epithelial seam. Thus, for example, in the TGF-β3-null embryos, the basement membrane remained intact on either side of the seam as evidenced by positive staining for laminin and collagen IV. Equally, desmosomal components were present in the persistent midline epithelial seam. However, no differences were detected which would suggest a causative correlation with the clefting phenotype. Interestingly at E18.5, the palatal processes of the maxillary and palatine bones did not extend to the midline in the fused regions of the TGF-β3-null palates: their development was perhaps inhibited by the persistent midline epithelial seam. This caused a defect to develop similar to that seen in submucous cleft palate.
Involvement of the homeobox gene goosecoid
We have preliminary data that the homeobox gene goosecoid is expressed in the mesenchyme adjacent to the midline palatal epithelial seam and may be regulated by, or regulate, TGF-β expression (Ding and Ferguson, unpublished). We therefore performed whole mount in situ hybridization and section in situ hybridization using a mouse goosecoid probe. There were no differences in either the temporal or spatial expression of goosecoid in TGF-β3-null embryos compared to their wild-type litter mate controls. There also appeared to be little difference in the intensity of silver grains in the in situ hybridizations between the TGF-β3-null embryos and their wild-type controls.
Immuno-staining using a specific antibody to TGF-β3 protein showed the normal previously described pattern of craniofacial staining in the wild-type litter mate controls, but the complete absence of any TGF-β3 protein staining in the TGF-β3-null animals. Immunostaining for the other TGF-β isoforms (TGF-β1 and TGF-β2) in the developing embryonic heads, showed no obvious difference between the mutant animals and their litter mate controls.
Discussion
Palate fusion
Our results suggest that TGF-β3, which is expressed abundantly by the MEE7,8, is critical for the adhesion and disappearance of the MEE requisite for normal fusion of the palatal shelves. These results are consistent with previous in vitro experiments in which antisense oligonucleotides or isoform-specific antibodies to TGF-β3, but not TGF-βl or TGF-β2, inhibited fusion; this inhibition could be rescued by exogenous TGF-β39. The fusion defect in TGF-β3-deficient mice leads us to postulate that weak adhesive interactions between the MEE initiates the defect and subsequently leads to a persistence of the mid-line epithelial seam resulting in incomplete fusion. However, as qualitative analysis of adhesion molecules known to be present in fusing palates has not identified any mechanistic proof for an adhesion defect, other interpretations can not be ruled out.
Adhesion of the apposing MEE is mediated by glycoproteins and desmosomes15,25,26, and TGF-βs can regulate expression of cell adhesion and extracellular matrix molecules27,28. Our data suggest that in the TGF-β3-null mice the first adhesive interaction following palatal shelf contact is abnormal: apposing MEE appear to slide off each other rather than adhere following contact. Following contact and adhesion of the MEE, nasal and oral epithelial triangles normally form from MEE cells migrating from the mid-palate to maximize their adhesive interactions in these triangles. Significantly, in the TGF-β3-null embryos the oral and nasal epithelial triangles are either markedly reduced or absent, and the mid-line epithelial seam is abnormally persistent. In those rare instances where the mid-line epithelial seam showed mesenchymal penetration, the remnants of the seam did not form spherical epithelial pearls as normally occurs (indicating maximal adhesive interaction), but remained as elongated seam islands. In the normal fusion process, the disappearance of the seam is characterized by loss of complex cytokeratins and basement membrane components such as laminin, and by an increase of vimentin, tenascin, proteoglycan and collagen expression15. These changes were not as evident in the mutant mice. This is consistent with persistence of the mid-line epithelial seam. Although these data do not mechanistically prove an adhesion defect, they are consistent with the hypothesis that a decrease in adhesive properties results in the persistence of the elongated strands of epithelial cells in the mid-line seam, a reduction in epithelial pearl structures and a reduction in nasal and oral epithelial triangles. The absence of qualitative differences in the immuno-staining patterns for desmosome components or E-cadherin would then suggest that small quantitative differences in adhesion molecules could result in weakened interactions, or that other adhesive mechanisms, possibly new adhesion molecules, may be involved.
Three mechanisms have been postulated to mediate breakdown of the midline epithelial seam: i) epithelial-mesenchymal transformation29,30, ii) cell migration of MEE into the nasal and oral epithelia15 and iii) apoptosis25. Our results do not rule out any of these possibilities because a defect in any one could result in a persistent epithelial seam. However, we do not believe that they constitute the underlying problem in the TGF-β3-null mouse because they can only occur after the initial contact and adhesion required to form a mid-line epithelial seam. TGF-β3-deficient palatal shelves do not adhere properly. Consequently, a failure in epithelial–mesenchymal transformation is not the initiating problem in the TGF-β3-null mice. The same argument can be made for elimination of the mid-line seam by apoptosis. Preliminary studies using an ‘apoptosis tag kit’ indicate no obvious differences in apoptosis between mutant and control palates: in both cases, apoptotic cells are clearly present, though small quantitative differences could not be ruled out. Consequently, we do not believe that a defect in apoptosis is the underlying cause of the fusion defect. Together, our analysis of the developmental abnormality leading to cleft palate would indicate two possible causes: one in MEE adhesion following initial contact of the palatal shelves, and the other in the disappearance of the post-fusion epithelial cells. A defect in the disappearance of the post-fusion seam epithelial cells could also involve adhesive mechanisms. We postulate that this apparent defect in cell adhesion results either from quantitative differences in known adhesion molecules thought to play a role in palate fusion, or that it involves unknown adhesive molecules or mechanisms.
Cleft palate unaccompanied by craniofacial defects
The cleft palate phenotype of the TGF-β3 knockout mouse is quite distinct from the cleft palate phenotypes displayed in other mutant mouse strains. There are approximately 28 mutant mouse strains with cleft palate generated either by spontaneous, transgene induced or transgene insertional mechanisms. None of these have a cleft palate without accompanying craniofacial, growth or heart defects suggesting either a structural or lineage relationship23. Likewise, all knockout mice previously reported with cleft palate have disorders affecting head elements: activin-βA31,32, activin receptor type II32, follistatin33, Hoxa-2 (refs 34, 35), Msx1 (ref. 36), endothelin-1 (ref. 37), MHox38 and combined gamma retinoic acid receptor isoform knockouts39. In contrast, the defect in TGF-β3 knockout mice is independent of such defects — consistent with the idea that TGF-β3 acts specifically on the MEE and may be a downstream effector of many of these other genes. This comparison makes it clear that the TGF-β3 cleft palate phenotype results from an intrinsic primary defect in palatal shelf fusion rather than by effects secondary to craniofacial morphometric abnormalities.
Other phenotypes
Analysis of embryos and d 1 neonates did not reveal other obvious gross malformations in any other organ including cartilage, bone, brain, skeleton or heart, with the possible exception of the lung. The terminal airway system of the newborn animals appeared abnormal (not shown), but it was not clear whether this was a developmental defect in the lung or whether the lungs were altered by the respiratory distress known to accompany cleft palate in newborn mice40. Clarification of this awaits a developmental analysis of the lungs. Mice are obligate nose breathers and a cleft in the palate results in exhaustion due to lack of nourishment. Although TGF-β3 has been shown to be involved in the epithelial-mesenchymal transformation in avian cardiac morphogenesis11, preliminary results suggest that some other TGF-β may play this role in the mouse. Whether TGF-β3 plays a role in mammary gland development12 or wound healing13 will require the development of protocols to rescue the TGF-β3 knockout mice from the perinatal lethality. One explanation for the lack of obvious phenotype in most other developing tissues expressing TGF-β3 could be compensation by TGF-β1 and -β2. RT-PCR performed on cDNAs derived from RNA of whole day 11.5 and 15.5 embryos showed no evidence for upregulation of TGF-β1 or -β2 in TGF-β3 homozygous mutant embryos, although significant local changes in expression would go undetected using this approach. Maternal transfer of the TGF-β3, as has been demonstrated for TGF-βl (ref. 41), is unlikely to explain lack of other phenotypes because unlike TGF-β1 it is present in very low levels either cell-associated or free in blood42. Furthermore, no positive immunostaining for TGF-β3 protein was detected in E11.5–E18.5 heads.
Non-overlapping phenotypes of TGF-β1 and TGF-β3 knockout mice
The TGF-β3 knockout mouse displays a strikingly different phenotype from the TGF-β1 knockout mouse in which the inflammatory system is hyperactive and poorly regulated18–21. However, in both TGF-β1- and TGF-β3-null mice, the respective phenotypes are consistent with abundant and relatively exclusive expression of the growth factors7,8,43. A low level of expression at any developmental stage or in any tissue has not correlated with a mutant phenotype.
Strain dependency of expressivity and relevance to the human disease
The most severe cleft palate phenotypes were observed on the C57BL/6 background. C57BL/6 mice have increased susceptibility to teratogen-induced cleft palate15. Increased incidence of cleft palate on this background suggests that TGF-β3 interacts with other cleft palate susceptibility genes in this strain, which is also the situation in human cleft palate. This variation has been interpreted as having different genetic or environmental aetiology, although this may not be the case. The TGF-β3-null mouse shows that differing severities and types (posterior vs anterior) of palatal clefting can result from the same genetic defect. The adhesion process can be seen as a cascade involving several molecules. There is initial adhesion with some adhesion molecule(s) which is substantiated by desmosomes. Threshold levels of the adhesion molecules in combination with the duration of contact could determine the degree to which subsequent fusion steps occur. Therefore, slight localized alterations in adhesion molecule expression could determine what portion of the shelf fuses.
It is not known whether TGF-β3 plays a role in human cleft palate. Interestingly in man, as in the TGF-β3-null embryos, the preponderance of palatal clefts affect the posterior palate (soft palate and uvula). An estimated 20% of all cases of cleft lip and/or palate are thought to be familial44. Application of the candidate gene approach to human cleft palate families will reveal whether TGF-β3 is involved. So far, associations of TGF-α45,46 and the retinoic acid receptor-A44 have been shown with human nonsyndromic cleft lip with or without cleft palate. Analysis of the cleft palate phenotype in TGF-β3-mutant mice will enable us to delineate the upstream and downstream targets of TGF-β3 in the process of palate fusion. Results from such analyses will identify new candidate genes which may shed significant light on the mechanisms underlying this disfiguring human congenital disorder.
Methods
Preparation and genotying of TGF-β3 knockout mice
A replacement type targeting vector was constructed in which exon 6 of the TGF-β3 gene was replaced with a neomycin-resistance gene derived from the pMC1neo vector22. E14-1 ES cells47 were transfected with targeting vector (minus plasmid sequences). Correct targeting was found in 5/250 neomycin-resistant clones analysed by Southern blot analysis. Genomic DNA was prepared as described48. One of these clones was injected into C57BL/6 blastocysts. Chimaeric animals were mated to CF-1, C57BL/6 and 129/Sv mice. Heterozygous offspring were detected by Southern blot analysis, then intercrossed to produce homozygous TGF-β3-null mice. Embryos were genotyped using a PCR assay as described49.
RT-PCR
Total RNA was prepared by the guanidinium isothiocyanate method50 and subjected to reverse transcription using random hexamer primers. The resulting cDNAs were balanced by RT-PCR to give roughly equivalent signals for hypoxanthine phosphoribosyl transferase (HPRT) as previously described51. RT-PCR analysis using primers for TGF-β1 gave a product of 750 bp18; primers for TGF-β2 (5'-CTCCTGCATC TGGTCCCGGT-3' and 5'-GCACAGCGTC TGTCACGTCG 3') gave a 556 bp product; primers for TGF-β3 (5'–GTCACTGACA CTGTGCGCGA-3' and 5'–CTGGCCTCAG CTGCACTTAC 3') gave a 677-bp product with wild-type derived cDNA. The primer sets recognize sequences in exon 4 and exon 7, respectively. In samples from homozygous mutant animals a product of 524 bp was observed; in samples from heterozygous animals two products, 524 and 677 bp, were observed. Under these conditions no TGF-β3/neo fusion transcript could be observed (expected size 1600 bp). The 524-bp product represents the mutated TGF-β3 allele and consists of exon 4, 5 and 7 sequences with the pMC1neo gene spliced out. This splicing event results in a frameshift mutation. Therefore, all but the first 9 amino acids (encoded at the end of exon 5) of the mature portion of the TGF-β3 protein are absent, as well as cysteins involved in mature protein structural integrity. The RT-PCR analysis confirms that the mice carry the mutation created in ES cells and are unable to produce active TGF-β3 protein.
Histological preparations
Bouin’s fixation: the whole embryo was submerged into Bouin’s solution (75 ml saturated picric acid, 5 ml glacial acetic acid, 25 ml 40% formaldehyde), then used for Wilson sections52. For histology, heads were embedded in wax, sectioned and the sections deparaffinized and hydrated; the 70% and 50% ethanol steps were extended 30–60 min to remove the yellow color. Paraformaldehyde fixation: the embryos were fixed in 4% paraformaldehyde, embedded in wax and sectioned at 5 µm. Slides were stained in Alcian Blue solution (1 g Alcian blue 8GX, Sigma #3157, 100 ml distilled water and 3 ml glacial acetic acid) for 1 h. After rinsing in distilled water the slides were stained for 2 min in Harris Hematoxylin (Sigma). After thorough washing the sections were stained with Eosin Y solution for 30 s. Finally, the slides were washed, dehydrated and mounted with Permount (Fisher, SP15-100).
Immunocytochemistry
Mouse heads for immunocytochemistry were embedded in OCT and frozen in liquid nitrogen. Heads were cryo-sectioned at 7 µm using a Leitz cryostat, followed by incubation with primary antibodies to decorin, laminin, tenascin, desmoplakin, desmocollin, E-cadherin, collagens IV, XII and XIV and cytokeratins 1, 5, 8, 7 and 14. The primary antibody to TGF-β3 was a kind gift from Dr. A. B. Roberts, National Cancer Institute. These primary antibodies and the immunocytochemical protocols for their use are described in detail15,53. Specific binding was detected by an FITC-conjugated secondary antibody with or without streptavidin/biotin amplification. Sections were examined and photographed in a fluorescence microscope.
Acknowledgements
We thank K. Rajewski (Cologne) for providing the E14-1 ES cell done; K. Unskker (Heidelberg) for observations on brain structure; T. Pexieder (Lausanne) for observations on heart structure; I. Ormsby, B. O’Toole (Cincinnati) and B. Johansson (Basel) for expert technical assistance; and B. Pfeiffer and H. Spalinger (Basel) for photography. The Basel Institute for Immunology was founded and is supported by Hoffmann-La Roche Ltd., Basel, Switzerland. This research was supported in part by NIH grants HD26471, HL41496, HL46826 and ES05652 to T.D. and a Birth Defects Foundation grant to M.W.J.F. J.D. is a PhD student supported by an ORS award and Frederick Cravan Moore Scholarship of the University of Manchester.
References
- 1.Roberts AB, Spom MB. The transforming growth factor betas. In: Spom MB, Roberts AB, editors. Peptide Growth Factors and Their Receptors—Handbook of Experimental Pathology. New York: Springer–Verlag; 1990. pp. 419–472. [Google Scholar]
- 2.Kingsley DM. The TGF–beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 3.Massague J, Attisano L, Wrana J. The complex interactions of TGF–β receptors. Trends Cell Biol. 1994;4:172–178. doi: 10.1016/0962-8924(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 4.Pelton RW, Saxena B, Jones M, Moses HL, Gold LI. Immunohistochemical localization of TGF beta 1, TGF beta 2, and TGF beta 3 in the mouse embryo: expression patterns suggest multiple roles during embryonic development. J. Cell Biol. 1991;115:1091–1105. doi: 10.1083/jcb.115.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmid P, Cox D, Bilbe G, Maier R, McMaster GK. Differential expression of TGF beta 1, beta 2 and beta 3 genes during mouse embryogenesis. Development. 1991;111:117–130. doi: 10.1242/dev.111.1.117. [DOI] [PubMed] [Google Scholar]
- 6.Millan FA, Denhez R, Kondaiah P, Akhurst RJ. Embryonic gene expression patterns of TGF beta 1, beta 2 and beta 3 suggest different developmental functions in vivo. Development. 1991;111:131–143. doi: 10.1242/dev.111.1.131. [DOI] [PubMed] [Google Scholar]
- 7.Fitzpatrick DR, Denhez F, Kondaiah P, Akhurst RJ. Differential expression of TGF beta isoforms in murine palatogenesis. Development. 1990;109:585–595. doi: 10.1242/dev.109.3.585. [DOI] [PubMed] [Google Scholar]
- 8.Pelton RW, Hogan BL, Miller DA, Moses HL. Differential expression of genes encoding TGFs beta 1, beta 2, and beta 3 during murine palate formation. Devl. Biol. 1990;141:456–460. doi: 10.1016/0012-1606(90)90401-4. [DOI] [PubMed] [Google Scholar]
- 9.Brunet CL, Sharpe PM, Ferguson MWJ. Inhibition of TGF–β3 (but not TGF–β1 or TGF–β2 activity prevents normal mouse embryonic palate fusion. Int. J. dev. Biol. 1995;39:345–355. [PubMed] [Google Scholar]
- 10.Chai Y, et al. Specific transforming growth factor–beta subtypes regulate embryonic mouse Meckel’s cartilage and tooth development. Devl. Biol. 1994;162:85–103. doi: 10.1006/dbio.1994.1069. [DOI] [PubMed] [Google Scholar]
- 11.Runyan RB, Potts JD, Weeks DL. TGF–beta 3–mediated tissue interaction during embryonic heart development. Molec. Reprod. & Dev. 1992;32:152–159. doi: 10.1002/mrd.1080320211. [DOI] [PubMed] [Google Scholar]
- 12.Robinson SD, Silberstein GB, Roberts AB, Flanders KC, Daniel CW. Regulated expression and growth inhibitory effects of transforming growth factor–beta isoforms in mouse mammary gland development. Development. 1991;113:867–878. doi: 10.1242/dev.113.3.867. [DOI] [PubMed] [Google Scholar]
- 13.Shah M, Foreman DM, Ferguson MWJ. Neutralisation of TGFβ1 and TGFβ2 or exogenous addition of TGFβ3 to cutaneous rat wounds reduces scarring. J. Cell Sci. 1995;108:985–1002. doi: 10.1242/jcs.108.3.985. [DOI] [PubMed] [Google Scholar]
- 14.Pelton RW, Dickinson ME, Moses HL, Hogan BL. In situ hybridization analysis of TGF beta 3 RNA expression during mouse development: comparative studies with TGF beta 1 and beta 2. Development. 1990;110:609–620. doi: 10.1242/dev.110.2.609. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson MWJ. Palate development. Development. 1988;103(Suppl):41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- 16.Dixon MJ, Ferguson MWJ. The effects of epidermal growth factor, transforming growth factors alpha and beta and platelet–derived growth factor on murine palatal shelves in organ culture. Arch. Oral Biol. 1992;37:395–410. doi: 10.1016/0003-9969(92)90024-3. [DOI] [PubMed] [Google Scholar]
- 17.Gehris AL, Greene RM. Regulation of murine embryonic epithelial cell differentiation by transforming growth factors beta. Differentiation. 1992;49:167–173. doi: 10.1111/j.1432-0436.1992.tb00664.x. [DOI] [PubMed] [Google Scholar]
- 18.Shull MM, et al. Targeted disruption of the mouse transforming growth factor–beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni AB, et al. Transforming growth factor beta1 null mutation in mice causes excessive inflammatory response and early death. Proc. natn. Acad. Sci. U.S.A. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boivin GP, et al. Onset and progression of pathological lesions in transforming growth factor–beta 1–deficient mice. Am. J. Pathol. 1995;146:276–288. [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkarni AB, et al. Transforming growth factor–beta 1 null mice. An animal model for inflammatory disorders. Am. J. Pathol. 1995;146:264–275. [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas KR, Capecchi MR. Site–directed mutagenesis by gene targeting in mouse embryo–derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 23.Ferguson MWJ. Craniofacial malformations: towards a molecular understanding. Nature Genet. 1994;6:329–330. doi: 10.1038/ng0494-329. [DOI] [PubMed] [Google Scholar]
- 24.Carette MJ, Ferguson MWJ. The fate of medial edge epithelial cells during palatal fusion in vitro: an analysis by Dil labelling and confocal microscopy. Development. 1992;114:379–388. doi: 10.1242/dev.114.2.379. [DOI] [PubMed] [Google Scholar]
- 25.Greene RM, Pratt RM. Developmental aspects of secondary palate formation. J. Embryol. exp. Morph. 1976;36:225–245. [PubMed] [Google Scholar]
- 26.Morris–Wiman J, Brinkley L. An extracellular matrix infrastructure provides support for murine secondary palatal shelf remodelling. Anat. Rec. 1992;234:575–586. doi: 10.1002/ar.1092340413. [DOI] [PubMed] [Google Scholar]
- 27.Ferguson MWJ. Palate development: mechanisms and malformations. Ir. J. Medical Sci. 1987;156:309–315. doi: 10.1007/BF02951261. [DOI] [PubMed] [Google Scholar]
- 28.D’Angelo M, Chen JM, Ugen K, Greene RM. TGF beta 1 regulation of collagen metabolism by embryonic palate mesenchymal cells. J. exp. Zool. 1994;270:189–201. doi: 10.1002/jez.1402700208. [DOI] [PubMed] [Google Scholar]
- 29.Shuler CF, Halpern DE, Guo Y, Sank AC. Medial edge epithelium fate traced by cell lineage analysis during epithelial–mesenchymal transformation in vivo. Devl. Biol. 1992;154:318–330. doi: 10.1016/0012-1606(92)90071-n. [DOI] [PubMed] [Google Scholar]
- 30.Griffith CM, Hay ED. Epithelial–mesenchymal transformation during palatal fusion: carboxyfluorescein traces cells at light and electron microscopic levels. Development. 1992;116:1087–1099. doi: 10.1242/dev.116.4.1087. [DOI] [PubMed] [Google Scholar]
- 31.Matzuk MM, et al. Functional analysis of activins during mammalian development. Nature. 1995;374:354–356. doi: 10.1038/374354a0. [DOI] [PubMed] [Google Scholar]
- 32.Matzuk MM, Kumar TR, Bradley A. Different phenotypes for mice deficient in either activins or activin receptor type II. Nature. 1995;374:356–360. doi: 10.1038/374356a0. [DOI] [PubMed] [Google Scholar]
- 33.Matzuk MM, et al. Multiple defects and perinatal death in mice deficient in follistatin. Nature. 1995;374:360–363. doi: 10.1038/374360a0. [DOI] [PubMed] [Google Scholar]
- 34.Gendron–Maguire M, Mallo M, Zhang M, Gridley T. Hoxa–2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- 35.Rijli FM. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa–2, which acts as a selector gene. Cell. 1993;75:1333–1349. doi: 10.1016/0092-8674(93)90620-6. [DOI] [PubMed] [Google Scholar]
- 36.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nature Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 37.Kurihara Y, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin–1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- 38.Martin JF, Bradley A, Olson EN. The paired–like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- 39.Lohnes D, et al. Function of retinoic acid receptor gamma in the mouse. Cell. 1993;73:643–658. doi: 10.1016/0092-8674(93)90246-m. [DOI] [PubMed] [Google Scholar]
- 40.Brown KS. Genetics of clefting in the mouse: a critique. Prog. clin. biol. Res. 1980;46:77–89. [PubMed] [Google Scholar]
- 41.Letterio JJ, et al. Maternal rescue of transforming growth factor–beta 1 null mice. Science. 1994;264:1936–1938. doi: 10.1126/science.8009224. [DOI] [PubMed] [Google Scholar]
- 42.Anscher MS, et al. Changes in plasma TGF beta levels during pulmonary radiotherapy as a predictor of the risk of developing radiation pneumonitis. Int. J. Radial Oncol. Biol. Phys. 1994;30:671–676. doi: 10.1016/0360-3016(92)90954-g. [DOI] [PubMed] [Google Scholar]
- 43.Wahl SM. Transforming growth factor beta (TGF–beta) in inflammation: a cause and a cure. J. clin. Immunol. 1992;12:61–74. doi: 10.1007/BF00918135. [DOI] [PubMed] [Google Scholar]
- 44.Chenevix–Trench G, Jones K, Green AC, Duffy DL, Martin NG. Cleft lip with or without cleft palate: associations with transforming growth factor alpha and retinoic acid receptor loci. Am. J. hum. Genet. 1992;51:1377–1385. [PMC free article] [PubMed] [Google Scholar]
- 45.Ardinger HH. Association of genetic variation of the transforming growth factor–alpha gene with cleft lip and palate. Am. J. hum. Genet. 1989;45:348–353. [PMC free article] [PubMed] [Google Scholar]
- 46.Shiang R, et al. Association of transforming growth–factor alpha gene polymorphisms with nonsyndromic cleft palate only (CPO) Am. J. hum. Genet. 1993;53:836–843. [PMC free article] [PubMed] [Google Scholar]
- 47.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT–deficient (Lesch–Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 48.Laird PW, et al. Simplified mammalian DNA isolation procedure. Nucl. Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proetzel G. Doctoral thesis. University of Cincinnati College of Medicine; 1994. Functional analysis of transforming growth factor–beta–3 using gene targeting; pp. 1–156. [Google Scholar]
- 50.Chomczynski P, Sacchi N. Single–step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Analyt. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 51.Keller G, Kennedy M, Papayannopoulou T, Wiles MV. Hematopoietic commitment during embryonic stem cell differentiation in culture. Molec. cell. Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson JG. Methods for administering agents and detecting malformation in experimental animals. In: Wilson JG, Warkany J, editors. Teratology: principles and techniques. Chicago: University of Chicago Press; 1965. pp. 262–277. [Google Scholar]
- 53.Carette MJ, Lane EB, Ferguson MW. Differentiation of mouse embryonic palatal epithelium in culture: selective cytokeratin expression distinguishes between oral, medial edge and nasal epithelial cells. Differentiation. 1991;47:149–161. doi: 10.1111/j.1432-0436.1991.tb00233.x. [DOI] [PubMed] [Google Scholar]