Abstract
The RNF38 gene encodes a RING finger protein of unknown function. Here we demonstrate that RNF38 is a functional ubiquitin protein ligase (E3). We show that RNF38 isoform 1 is localized to the nucleus by a bipartite nuclear localization sequence (NLS). We confirm that RNF38 is a binding partner of p53 and demonstrate that RNF38 can ubiquitinate p53 in vitro and in vivo. Finally, we show that overexpression of RNF38 in HEK293T cells results in relocalization of p53 to discrete foci associated with PML nuclear bodies. These results suggest RNF38 is an E3 ubiquitin ligase that may play a role in regulating p53.
Keywords: RNF38, E3, ubiquitin protein ligase, p53, PML nuclear bodies
INTRODUCTION
The p53 tumor suppressor gene (TP53) has been termed “the guardian of the genome” [1] and it is one of the most frequently mutated genes in human cancers [2, 3, 4, 5]. Mutations in the regulators of p53, such as Arf, Mdm2 and Pirh2, are also commonly seen in cancer cells [6, 7, 8]. Considering the extensive involvement of p53 in tumorigenesis, understanding p53 regulation is important to the continued development of anti-cancer strategies.
In a search for new p53 interactors, Lunardi et al. employed an affinity purification approach with Drosophila p53 in which they identified many novel p53 binding proteins, including the little studied protein, RING finger protein 38 (RNF38) [9]. Although human RNF38 has been previously cloned [10], its function is currently unknown.
RNF38 mRNA is widely expressed in a variety of human tissues [10], and evolutionary conservation suggests an important cellular function. Interestingly, RNF38 is located on an area of chromosome 9 (9p13) that is frequently deleted in multiple cancers such as ganglioglioma [11], lung cancer [12, 13], hepatocellular carcinoma [14], and in the 9p13-24 deletion seen in the lymphoid blast transformation of chronic myeloid leukemia [15].
Many proteins containing a RING finger motif act as ubiquitin protein ligases [16, 17]. We wanted to explore the possibility that RNF38 might have E3 activity and whether or not p53 might be a substrate. p53 is known to be regulated by several ubiquitin protein ligases, the best studied of which is Mdm2/Hdm2. [18, 19, 20]. Other E3 ligases that can modify p53 include WWP1, Pirh2 and Cop1 [21, 22, 23]. Here we find that RNF38 does possess E3 ubiquitin ligase activity and can recognize p53 as a substrate for ubiquitination. In addition, overexpression of RNF38 alters the nuclear localization of p53.
MATERIALS AND METHODS
Cell Culture and transfections
All cell lines were obtained from the Tissue Culture Shared Resource at the University of Colorado Cancer Center. Human embryonic kidney 293 (HEK293) and HEK293T cells were maintained in DMEM (Hyclone) supplemented with 10% fetal bovine serum and antibiotics. H1299 cells were grown in RPMI (Hyclone) supplemented with 10% fetal bovine serum and antibiotics. Transfections were performed with Lipofectamine (Invitrogen).
Constructs
cDNA for RNF38 isoform 1 was obtained by RT-PCR from HEK293 mRNA and cloned into pcDNA 3.1(+) (Invitrogen). The RNF38 cDNA insert and all other constructs derived by PCR were fully sequenced. GFP fusions were made in pEGFP-C1 (Clontech). PCR primers and cloning details are provided in the supplementary materials. The mammalian expression vector pEBG-SrfI was obtained from Yusen Liu. Vector pMT 123 expressing HA-ubiquitin was obtained from Mathias Treier.
Expression and purification of RNF38 from Baculovirus
A six histidine tag was added to the amino terminus of RNF38 by PCR and cloned into the baculovirus recombination plasmid pVL1393. The Baculovirus Core Facility at the University of Colorado Cancer Center performed the recombination into baculovirus and growth in SF9 cells. RNF38 was purified from extracted nuclei using immobilized Co++ (“Talon”, Clontech) affinity chromatography. Full details are provided in the supplementary data.
Antibodies
Mouse monoclonal anti-p53 (1C12) was purchased from Cell Signaling. Rabbit anti-p53 (sc-6243), mouse monoclonal anti-p53 (sc-126), rabbit anti-RNF38 (sc-102096), rabbit anti-GFP (sc-8334) and mouse monoclonal anti-PML (sc-966) were purchased from Santa Cruz Biotechnology. A second rabbit anti-RNF38 antibody (AP12816a) was acquired from Abgent. Mouse anti-NR1 (#05-432) was obtained from Upstate Biotechnology. Horseradish peroxidase (HRP) conjugated rat anti-HA antibody (3F10) was from Roche Applied Science.
Binding studies
GST and GST-p53 fusion proteins were purified from E. coli BL21 transformed with pGEX-4T-2-(p53). Either 2 μg of purified RNF38 in GST binding buffer (50 mM NaCl, 20 mM Tris pH 7.5, 1 mM MgCl2, 1 mM DTT, 0.1% Thesit and 1.0 mg/mL BSA) or 850 μg of 293T whole cell extract in lysis buffer (50 mM NaCl, 20 mM Tris pH 7.5, 1 mM MgCl2, 1.0% Trition X-100, 1 mM Na3VO4, 1 mM DTT and 1 mM PMSF plus protease inhibitors) were incubated at 4° C for 30 minutes with equal amounts (2 μg) of GST or GST-p53. Preblocked (5% BSA) glutathione-Sepharose beads (G.E. Healthcare Lifesciences) were then added to the mixtures and incubated for 2 hours at 4° C with mixing. The beads were then washed with GST wash buffer (50 mM NaCl, 20 mM Tris pH7.5, 1 mM MgCl2, 1 mM DTT and 0.1% Thesit) and subjected to western blot analysis.
Ubiquitination assay – in vitro
Purified RNF38 was mixed with 50 μg/ml E1, E2 and 0.5 mg/ml HA-ubiquitin in ubiquitin buffer (50 mM KCl, 20 mM HEPES pH 7.4, 5 mM MgCl2, 1 mM DTT and 1 mM ATP) and incubated for 30 minutes at 30° C with mixing at 600 rpm. The reaction products were analyzed by western blot with anti-RNF38 (sc-102096) and anti-HA antibodies.
For in vitro p53 ubiquitination reactions, p53 was immunoprecipitated from HEK293T cells lysed in RIPA+ buffer (150 mM NaCl, 20 mM Tris pH 7.4, 1 mM EDTA, 1 mM EGTA, 1.0% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 50 mM NaF, 1 mM Na3VO4, 1 mM DTT, 1 mM PMSF plus protease inhibitors). Mouse anti-p53 (1C12) or mouse anti-NR1 control (0.75 μg) was added to 500 μg of precleared whole cell lysate and incubated overnight. Antibody complexes were then bound to preblocked (5% BSA) protein G-agarose (Sigma-Aldrich P-4691) for two hours followed by extensive washing: twice with RIPA+, twice with SNNTE (0.5 M NaCl, 50 mM Tris pH 7.5, 5 mM EDTA, 5% sucrose, 1% NP-40), twice with 200 mM glycine pH 2.5, and twice with ubiquitin buffer. The washed beads were then used in in vitro ubiquitination reactions with or without purified RNF38 as described above. Following the ubiquitination reactions, the beads were washed twice with RIPA+ and then analyzed by western blot with anti-p53 and anti-HA antibodies.
Ubiquitination assay – in vivo
HEK293T cells were transfected with pMT 123 (HA-ubiquitin) plus pEBG or pEBG-RNF38. 24 hours post-transfection cells were passaged and allowed to grow for 48 hours. At 72 hours post-transfection the cells were incubated for 4 hours in 25 μM MG132 (Sigma-Aldrich) then collected in lysis buffer (100 mM NaCl, 20 mM Tris pH 7.5, 5 mM EDTA, 10% glycerol, 1% NP-40, 1 mM DTT, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 10 mM NEM plus protease inhibitors). Immunoprecipitation of p53 was performed using 1 μg of mouse anti-p53 (sc-126) antibody to 1.5 mg of precleared whole cell extract followed by binding to preblocked protein G-agarose. Bound complexes were washed with lysis buffer then analyzed by western blot with rat anti-HA antibody.
Immunofluorescence
Cultured cells were seeded on glass coverslips, transfected and 24 hours later fixed with 4% para-formaldehyde in PBS. Cells were permeabilized with cold methanol, rinsed with PBS, and blocked with 2% nonfat milk, 0.1% Tween-20 in PBS. Primary antibody rabbit anti-p53 (sc-6243) was used at 2 μg/ml in 1% BSA, 0.5% Triton X-100 in PBS and incubated for 1 hour at room temperature. Primary antibody rabbit anti-RNF38 (AP12816a) was used at 5 μg/ml and anti-PML (sc-966) was used at 4 μg/ml incubated overnight at 4° C. Cells were washed with 0.05% Tween-20 in PBS and incubated with the appropriate fluorescent secondary (Alexa Fluor 568 goat anti-rabbit or Alexa Fluor 488 goat anti-mouse, Invitrogen) at 2 μg/ml for 1 hour. Nuclei were counterstained with 0.2 μg/ml Hoechst 33258 (Kodak). Coverslips were mounted in gelvatol and examined with a Nikon Eclipse Ti-S microscope fitted with the appropriate fluorescence filters, DS-Qi1 camera and NIS-Elements imaging software.
RESULTS
RNF38 is a nuclear protein
In their study identifying p53-interacting proteins in Drosophila, Lunardi et al. also examined the intracellular localization of 41 mammalian orthologs, including RNF38 [9]. Immunofluorescence of epitope-tagged RNF38 overexpressed in U2OS cells revealed diffuse nuclear staining. However, the data presented was a supplementary figure containing an image of a single cell with no mention of which RNF38 isoform was used or if any other staining patterns were observed. As a first step toward understanding RNF38 function, we sought to confirm and expand this result.
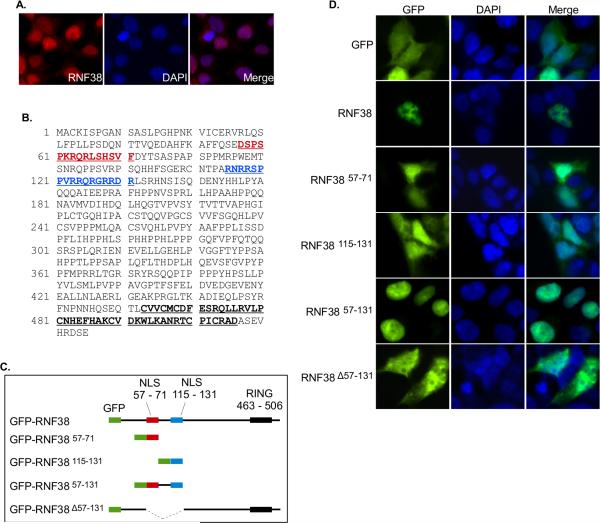
We first examined RNF38 in HEK293T cells. Endogenous RNF38 was detectable by immunofluorescence with only one of two commercially available anti-RNF38 antibodies (AP12816a) under the conditions employed. Similar to the results of Lunardi et al., we find endogenous RNF38 is also localized to the nucleus in HEK293T cells (Fig. 1A). RNF38 is predicted to have three isoforms. Isoform 1 is the largest, containing 515 amino acids, whereas isoform 2 lacks amino acids 5-54, and isoform 3 lacks amino acids 1-84. According to the manufacturer, the AP12816a antibody was raised against amino acids 47-77 of RNF38, a region present in isoform 1 but partially or entirely lacking in the other isoforms. We chose to focus on isoform 1, henceforth referred to as simply “RNF38”.
Figure 1. RNF38 is a nuclear protein with a bipartite nuclear localization signal.
(A) Endogenous RNF38 is located in the nucleus. HEK293T cells were stained with anti-RNF38 antibody, nuclei were counterstained with Hoechst 33258 and cells examined by fluorescence microscopy. (B) RNF38 contains two potential nuclear localization signals. In silico searches identified amino acids 57-71 (red) and 115-131 (blue) as possible nuclear localization signals. The RING finger domain is also indicated (463-506). (C) GFP-RNF38 fusion constructs used to test candidate nuclear localization signals. (D) Amino acid sequence 57-131 is sufficient for nuclear localization. At 24 hours post-transfection with the constructs shown in (C), HEK293T cells were fixed and treated as in (A).
The endogenous RNF38 signal was fairly weak, so to further investigate RNF38 nuclear localization, we constructed a GFP-tagged RNF38 expression vector. As shown in Figure 1, transient expression (Fig. 1D, second row) resulted in nuclear localization of GFP-RNF38. Note also that the stronger signal from overexpressed GFP-RNF38 allows more detail to be observed, and the protein appears to be excluded from nucleoli.
Identification of nuclear localization signals
To identify the motif(s) responsible for RNF38 nuclear localization we analyzed its amino acid sequence with several nuclear localization signal (NLS) prediction programs. cNLS Mapper [24] identified amino acids 57-71 as a possible NLS, and amino acids 115-131 were identified as a possible NLS by NLStradamus [25]. We then created a series of GFP fusions to examine the role these sequences might play in RNF38 localization (Fig. 1C).
Transient expression of GFP in HEK293T cells results in a uniform staining pattern throughout the nucleus and cytoplasm (Fig. 1D). In contrast, the GFP-RNF38 fusion protein appears restricted to the nucleus. GFP fusion to either amino acids 57-71 or 115-131 is insufficient to confer complete nuclear localization, though a larger GFP fusion to amino acids 57-131 is completely localized to the nucleus . This region contains both of the basic residue-rich sequences identified as putative NLSs and thus appears to function as a bipartite NLS. Deletion of this region results in loss of exclusive nuclear localization with the GFP-RNF38 57-131 fusion protein localized in both cytoplasm and nucleus.
RNF38 is an E3 ubiquitin ligase
In addition to ubiquitination of their target proteins, many E3 ubiquitin ligases undergo autoubiquitination in vitro [16, 17]. Figure 2 shows in vitro ubiquitination assays using His-tagged RNF38 purified from baculovirus infected SF9 cells. RNF38 clearly undergoes robust autoubiquitination and shows specificity in interacting with E2s. Of the three E2 proteins tested (UbcH2, UbcH5b and UbcH7), only UbcH5b supported RNF38 autoubiquitination. These findings demonstrate RNF38 is an active ubiquitin protein ligase (E3).
Figure 2. RNF38 is capable of autoubiquitination in vitro.
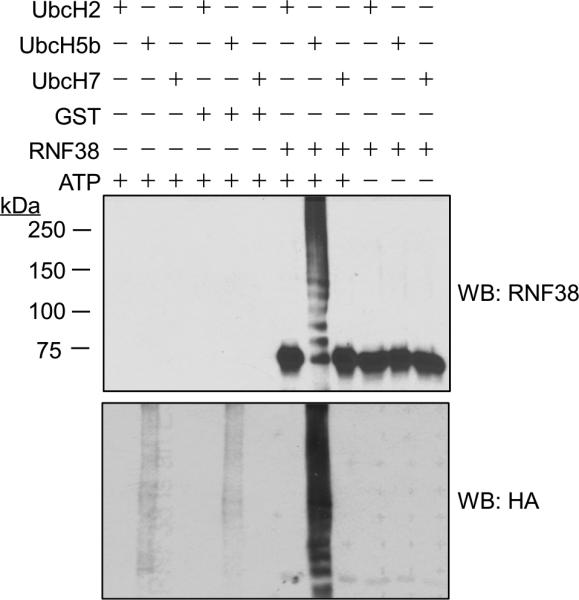
Reactions contained recombinant RNF38, E1, HA-ubiquitin, and either UbcH2, UbcH5b or UbcH7 with or without 1mM ATP. Purified GST was used as a control. Reactions were analyzed by immunoblot with anti-RNF38 (sc-102096) and anti-HA antibodies.
The RING domain limits RNF38 overexpression
We found RNF38 to be resistant to overexpression by transfection despite using a variety of expression plasmids and cell lines (including MDA-MB-231, H1299 and HEK293, data not shown). Since an active RING domain might limit expression through autoubiquitination or down-regulation of some other cellular component, we removed the C-terminal RING domain from RNF38 (RNF38 ΔRING) to explore the issue.
Truncation of the RING domain greatly increased RNF38 expression (Fig. S1A). In two cell types (HEK293T and H1299), transfection with GFP alone or GFP-RNF38ΔRING yielded many GFP-positive cells, whereas with full-length GFP-RNF38, only rare GFP-positive cells were observed. Western blot analysis of these cells confirmed the results (Fig. S1B).
Successful overexpression of full-length RNF38 constructs was detectable on western blots using derivatives of the vector pEBG in HEK293T cells (Fig. S1C). The pEBG plasmid contains an SV40 origin of replication, allowing amplification of the plasmid in HEK293T cells which express SV40 large T antigen. Treatment of cells with the proteasome inhibitor MG132 did not increase RNF38 expression, nor was a prominent ladder of ubiquitinated RNF38 species observed, suggesting that autoubiquitination may not be the mechanism limiting overexpression of full-length RNF38 (Fig. S1C). Additional experiments will be required to identify the mechanism(s) regulating RNF38 expression.
RNF38 is a p53 binding protein
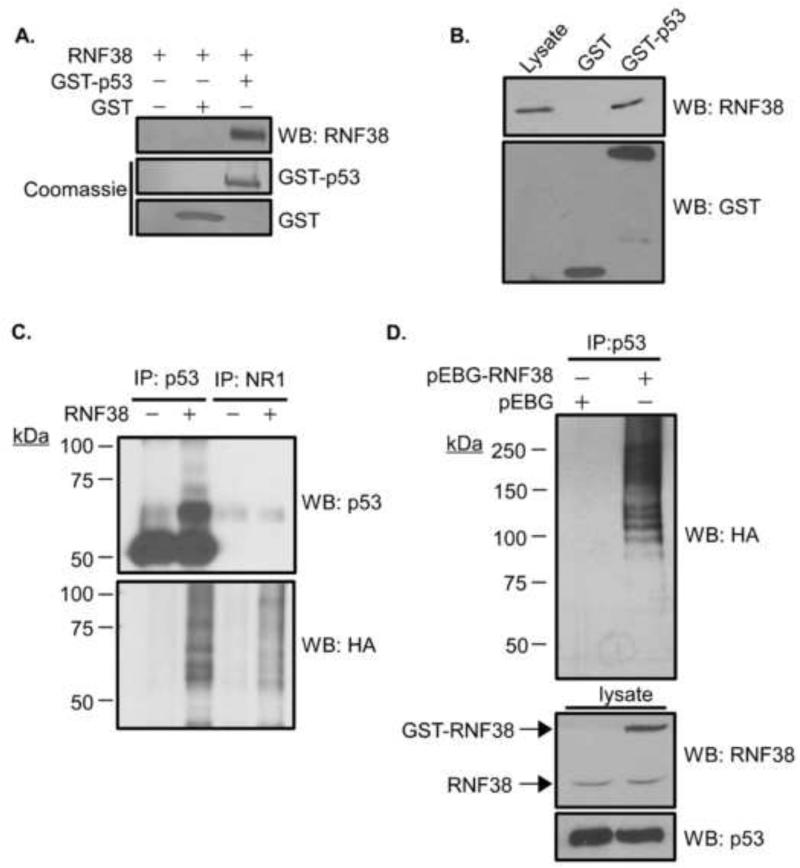
We have examined the p53-RNF38 binding interaction first reported by Lunardi et al. using a different affinity purification technique employing a fusion protein between human p53 and glutathione S-transferase (GST). Purified RNF38 binds specifically to GST-p53 but not GST alone (Fig. 3A, upper panel). Similarly, endogenous RNF38 in lysates of HEK293T cells specifically binds GST-p53 but not GST alone (Fig. 3B).
Figure 3. RNF38 binds and ubiquitinates p53.
(A) RNF38 binds to p53 in vitro. Binding interactions were tested using recombinant RNF38 incubated either alone, with GST, or GST-p53 followed by binding to glutathione-Sepharose. Bound proteins were analyzed by immunoblot with anti-RNF38 antibody (sc-102096). (B) Endogenous RNF38 binds p53. 293T cell lysate was incubated with purified GST or GST-p53. Complexes were isolated and analyzed as in (A). Lane one (Lysate) contains 3.5% of the cell lysate used for comparison. (C) p53 is ubiquitinated by purified RNF38 in vitro. p53 was immunoprecipitated from 293T cell lysate using mouse anti-p53 or a control antibody (anti-NR1). Isoloated immune complexes were added to in vitro ubiquitination reactions with recombinant HA-ubiquitin, E1, UbcH5b and with or without RNF38, as indicated. Reaction products were analyzed by immunoblot with antibodies to p53 and HA (ubiquitin). (D) Overexpression of RNF38 increases p53 ubiquitination. HEK293T cells were transfected with pMT 123 (HA-ubiquitin) plus either the pEBG expression vector or pEBG-RNF38. 72 hours post-transfection, cells were treated with 25 μM MG132 for 4 hours, lysed, and p53 recovered by immunoprecipitation. Precipitated proteins were analyzed by western blot with anti-HA antibody.
RNF38 ubiquitinates p53 in vitro and in vivo
To test whether p53 is a substrate for ubiquitination by RNF38, endogenous p53 was immunoprecipitated from HEK293T lysate and added to in vitro ubiquitination reactions. Control immunoprecipitates using an unrelated antibody (anti-NR1) were treated similarly. We found that p53 was ubiquitinated in an RNF38-dependent manner under these conditions (Fig. 3C).
Next, we asked whether we could detect RNF38-dependent ubiquitination of p53 in cells. HEK293T cells were transfected with an expression vector for HA-tagged ubiquitin and either GST or GST-RNF38. p53 was recovered by immunoprecipitation and analyzed by immunoblotting for HA-ubiquitin (Fig. 3D). GST-RNF38 clearly increased p53 ubiquitination in vivo, as evidenced by the ladder of higher molecular weight species reacting with HA (ubiquitin) antibody. We note that the p53 complexes isolated from cells appear more extensively modified than those produced in vitro, suggesting there may be additional factors or conditions involved in p53 ubiquitination in cells.
Investigation of possible effects of RNF38 on p53 turnover has been hindered by the resistance of RNF38 to overexpression. As mentioned earlier, detection of biochemical levels of exogenous RNF38 has only been achieved in HEK293T cells, in which SV40 large T antigen prevents normal p53 turnover [26]. Experiments in other systems, including H1299 cells (which lack p53 expression), have been inconclusive due to insufficient/undetectable RNF38 expression.
Overexpression of RNF38 alters p53 localization
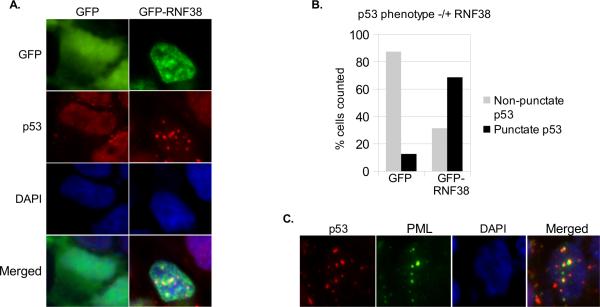
Since biochemical analysis of possible RNF38 effects on p53 turnover has been uninformative due to the technical issues described above, we turned to fluorescence microscopy to allow visualization of any effects on intracellular localization of p53. HEK293T cells were transfected with GFP-RNF38 or GFP alone, then fixed and stained for p53. In untransfected and GFP-transfected HEK293T cells, p53 is distributed diffusely throughout the nucleus with a small minority of cells (13 %) showing punctate staining (Figure 4). In cells transfected with GFPRNF38, p53 redistributed to large punctate structures in the nucleus in the majority of GFP-RNF38 positive cells (69 %).
Figure 4. RNF38 overexpression alters p53 localization.
(A) HEK293T cells were transfected with pEBG-GFP or pEBG-GFP-RNF38. At 24 hours post-transfection, cells were fixed and stained for p53. A representative image of p53 punctate structure is shown. (B) The p53 phenotype was quantified as punctate or non-punctate in GFP(RNF38) expressing cells. Approximately 200 cells each were counted. (C) RNF38 overexpression induces p53 association with PML nuclear bodies. HEK293T cells were transfected with pEBG-RNF38. 24 hours post-transfection cells were fixed and stained for p53 (sc-6243) and PML.
Since p53 has been previously identified in PML (promyelocytic leukemia-associated protein) nuclear bodies [27, 28], we analyzed the localization of PML and p53 in RNF38 transfected cells. A typical cell with a punctate p53 phenotype is shown in Figure 4C. In these cells many, though not all, p53 foci associate directly adjacent to PML nuclear bodies. The number of p53 and PML foci vary from cell to cell, and while many p53 foci colocalize with PML foci, a few of each type do not. These data suggest that overexpression of RNF38 can change the distribution of p53 within the nucleus from diffuse to punctate, and many of these foci are associated with PML nuclear bodies.
DISCUSSION
This study has begun the functional characterization of the evolutionarily conserved protein, RNF38. We have shown that RNF38 (isoform 1) is localized to the nucleus in several cell types and have identified a bipartite NLS capable of mediating this localization. We have expressed and purified RNF38 from baculovirus-infected SF9 cells and found that purified RNF38 protein is an active ubiquitin protein ligase that exhibits specificity with the E2 protein with which it interacts. We have confirmed Lunardi et al.'s findings that RNF38 is a p53-binding protein, shown that RNF38 can ubiquitinate p53 in vitro, and demonstrated that overexpression of RNF38 increases p53 ubiquitination in vivo.
Although our efforts to elucidate the dynamics of the p53-RNF38 interaction biochemically were hindered by limited RNF38 overexpression, fluorescence microscopy revealed that overexpression of RNF38 in HEK293T cells alters the intracellular distribution of p53 from a diffuse nuclear pattern to punctate foci, many of which are associated with PML nuclear bodies. Association of p53 with PML nuclear bodies has been previously described in a number of instances [27-30], and it has been suggested that PML nuclear bodies may be sites of functional regulation of p53 [31]. In this regard, our data indicating altered p53 localization upon RNF38 overexpression are intriguing. While insufficient to illuminate the exact functional interplay between these two proteins, these results do provide additional evidence for a regulatory relationship between RNF38 and p53. Taken together with our other findings, a case can be made that RNF38 may be a biologically significant regulator of p53. It remains to future studies to elaborate the full functional details of such regulation.
Supplementary Material
HIGHLIGHTS.
RNF38 is shown to be a nuclear protein with a bipartite nuclear localization signal
RNF38 protein is purified and shown to have ubiquitin protein ligase (E3) activity
We show that RNF38 binds p53 and can ubiquitinate p53 in vitro
Overexpression of RNF38 increases p53 ubiquitination in HEK293T cells
Overexpression of RNF38 in HEK293T cells alters p53 localization
ACKNOWLEDGEMENTS
We would like to thank Lori Sherman and the University of Colorado Cancer Center Protein Production-MoAB-Tissue Culture Shared Resource for their help with Baculovirus production. This study was supported by NIH grant R01 NS052770 to CKK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lane DP. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2010;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walerych D, Napoli M, Collavin L, Del Sal G. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012;33:2007–2017. doi: 10.1093/carcin/bgs232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Xiong Y. Mutations in Human ARF Exon 2 Disrupt Its Nucleolar Localization and Impair Its Ability to Block Nuclear Export of MDM2 and p53. Mol. Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]
- 7.Coutts AS, Adams CJ, La Thangue NB. p53 ubiquitination by Mdm2: A never ending tail? DNA Repair. 2009;8:483–490. doi: 10.1016/j.dnarep.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Jung Y-S, Qian Y, Chen X. Pirh2 RING-finger E3 ubiquitin ligase: Its role in tumorigenesis and cancer therapy. FEBS Lett. 2012;586:1397–1402. doi: 10.1016/j.febslet.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lunardi A, Di Minin G, Provero P, Dal Ferro M, Carotti M, Del Sal G, Collavin L. A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6322–6327. doi: 10.1073/pnas.1002447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenberg I, Hochner H, Levi T, Yelin R, Kahan T, Mitrani-Rosenbaum S. Cloning and characterization of a novel human gene RNF38 encoding a conserved putative protein with a RING finger domain. Biochem. Biophys. Res. Commun. 2002;294:1169–1176. doi: 10.1016/S0006-291X(02)00584-3. [DOI] [PubMed] [Google Scholar]
- 11.Yin X-L, Hui AB-Y, Pang JC-S, Poon WS, Ng H-K. Genome-wide survey for chromosomal imbalances in ganglioglioma using comparative genomic hybridization. Cancer Genet. Cytogenet. 2002;134:71–76. doi: 10.1016/s0165-4608(01)00611-2. [DOI] [PubMed] [Google Scholar]
- 12.Kim SK, Ro JY, Kemp BL, Lee JS, Kwon TJ, Fong KM, Sekido Y, Minna JD, Hong WK, Mao L. Identification of Three Distinct Tumor Suppressor Loci on the Short Arm of Chromosome 9 in Small Cell Lung Cancer. Cancer Res. 1997;57:400–403. [PubMed] [Google Scholar]
- 13.Girard L, Zöchbauer-Müller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide Allelotyping of Lung Cancer Identifies New Regions of Allelic Loss, Differences between Small Cell Lung Cancer and Non-Small Cell Lung Cancer, and Loci Clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- 14.Liew CT, Li HM, Lo KW, Leow CK, Lau WY, Hin LY, Lim BK, Lai PBS, Chan JYH, Wang X, Wu S, Lee JCK. Frequent allelic loss on chromosome 9 in hepatocellular carcinoma. Int. J. Cancer. 1999;81:319–324. doi: 10.1002/(sici)1097-0215(19990505)81:3<319::aid-ijc1>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 15.Nacheva EP, Brazma D, Virgili A, Howard-Reeves J, Chanalaris A, Gancheva K, Apostolova M, Valgañon M, Mazzullo H, Grace C. Deletions of Immunoglobulin heavy chain and T cell receptor gene regions are uniquely associated with lymphoid blast transformation of chronic myeloid leukemia. BMC Genomics. 2010;11:41. doi: 10.1186/1471-2164-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joazeiro CA, Weissman AM. RING Finger Proteins: Mediators of Ubiquitin Ligase Activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 18.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 19.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 20.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 Is a RING Finger-dependent Ubiquitin Protein Ligase for Itself and p53. J. Biol. Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 21.Laine A, Ronai Z. Regulation of p53 lo calization and transcription by the HECT domain E3 ligase WWP1. Oncogene. 2007;26:1477–1483. doi: 10.1038/sj.onc.1209924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-Induced Ubiquitin-Protein Ligase, Promotes p53 Degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 23.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 24.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinf. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Reilly DR. p53 and transformation by SV40. Biol. Cell. 1986;57:187–196. doi: 10.1111/j.1768-322x.1986.tb00475.x. [DOI] [PubMed] [Google Scholar]
- 27.Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000;19:6185–6195. doi: 10.1093/emboj/19.22.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen H, Maki CG. p53 and p21(Waf1) are Recruited to Distinct PML-Containing Nuclear Foci in Irradiated and Nutlin-3a-Treated U2OS Cells. J. Cell. Biochem. 2010;111:1280–1290. doi: 10.1002/jcb.22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laín S, Midgley C, Sparks A, Lane EB, Lane DP. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp. Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- 30.Jiang W-Q, Szekely L, Klein G, Ringertz N. Intranuclear Redistribution of SV40T, p53, and PML in a Conditionally SV40T-Immortalized Cell Line. Exp. Cell Res. 1996;229:289–300. doi: 10.1006/excr.1996.0374. [DOI] [PubMed] [Google Scholar]
- 31.Gostissa M, Hofmann TG, Will H, Del Sal G. Regulation of p53 functions: let's meet at the nuclear bodies. Curr. Opin. Cell Biol. 2003;15:351–357. doi: 10.1016/s0955-0674(03)00038-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.