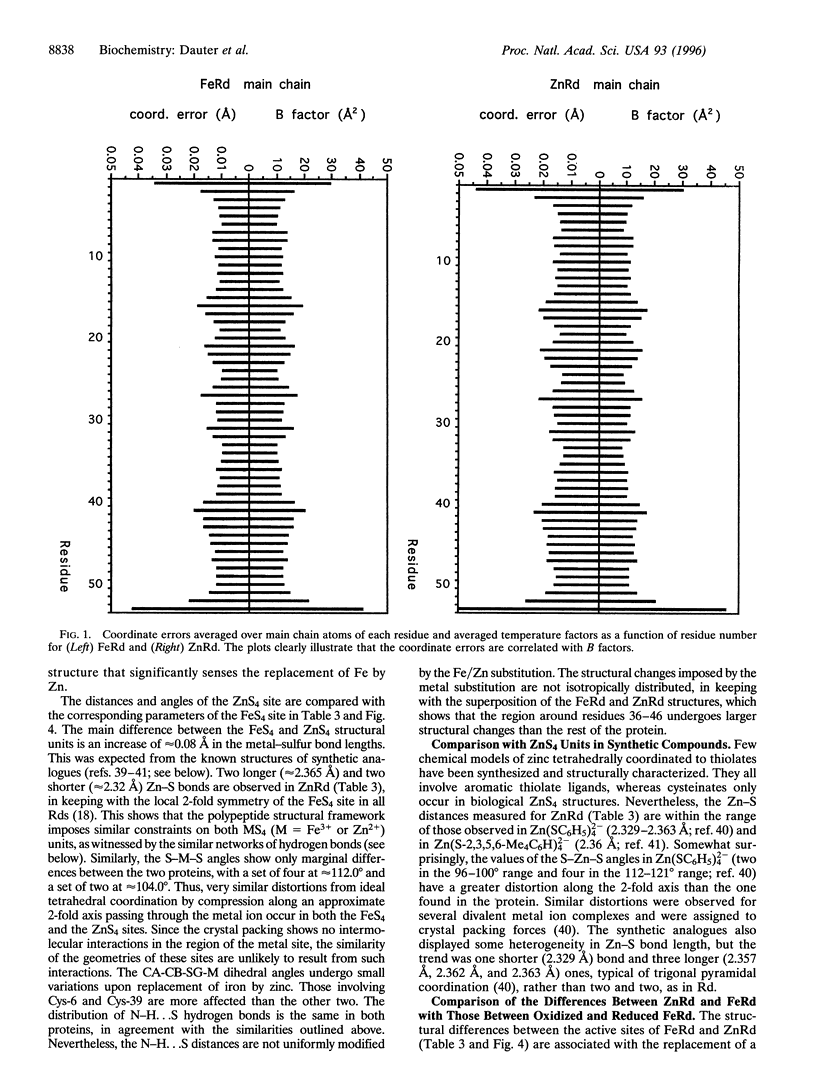
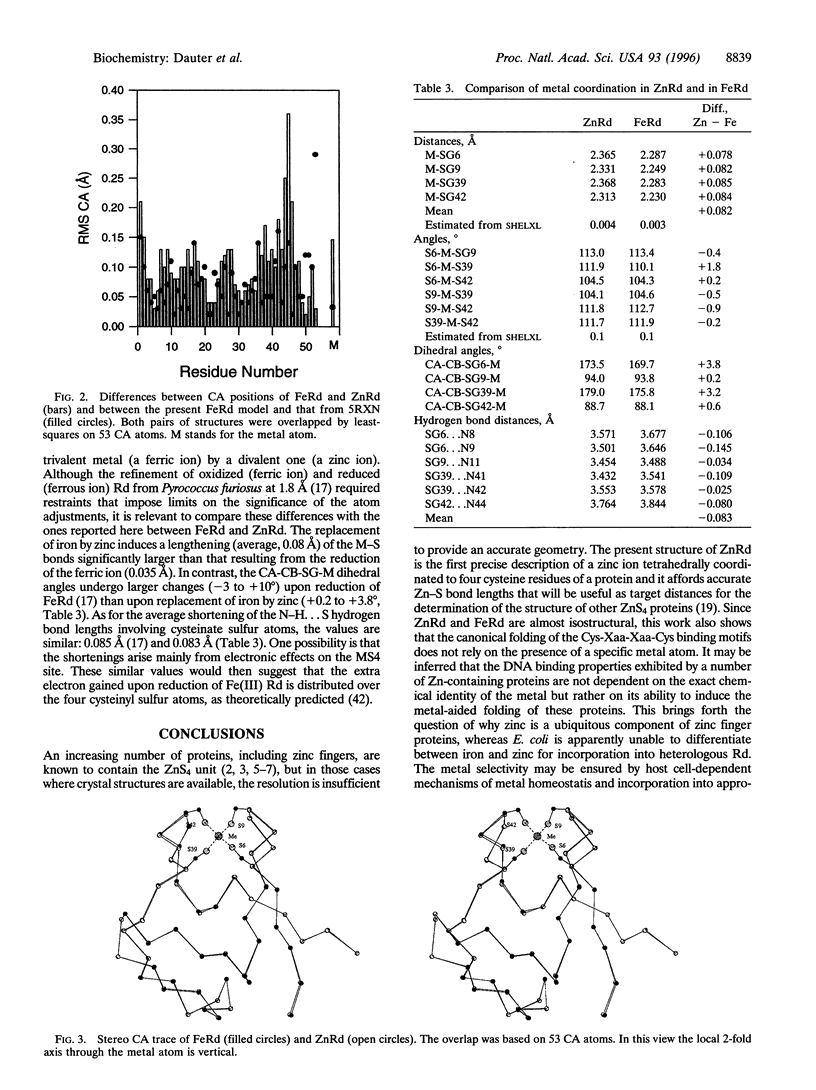
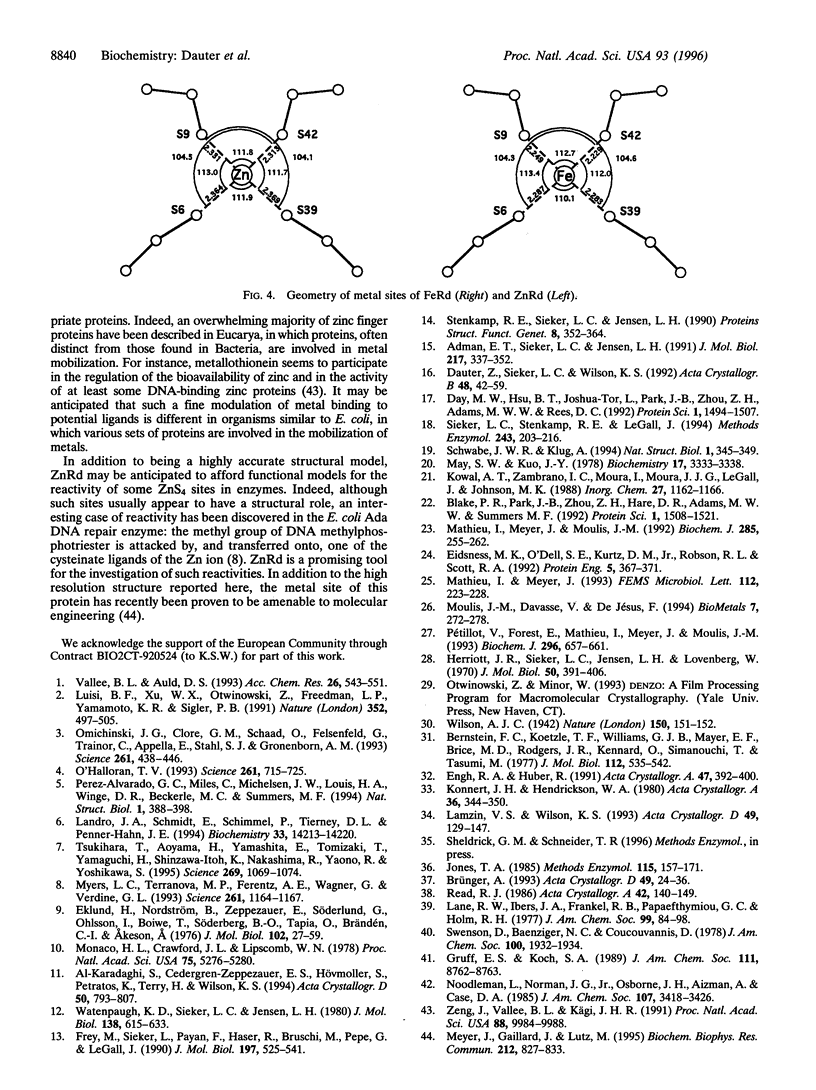
Abstract
The Zn(Scys)4 unit is present in numerous proteins, where it assumes structural, regulatory, or catalytic roles. The same coordination is found naturally around iron in rubredoxins, several structures of which have been refined at resolutions of, or near to, 1 A. The fold of the small protein rubredoxin around its metal ion is an excellent model for many zinc finger proteins. Zn-substituted rubredoxin and its Fe-containing counterpart were both obtained as the products of the expression in Escherichia coli of the rubredoxin-encoding gene from Clostridium pasteurianum. The structures of both proteins have been refined with an anisotropic model at atomic resolution (1.1 A, R = 8.3% for Fe-rubredoxin, and 1.2 A, R = 9.6% for Zn-rubredoxin) and are very similar. The most significant differences are increased lengths of the M-S bonds in Zn-rubredoxin (average length, 2.345 A) as compared with Fe-rubredoxin (average length, 2.262 A). An increase of the CA-CB-SG-M dihedral angles involving Cys-6 and Cys-39, the first cysteines of each of the Cys-Xaa-Xaa-Cys metal binding motifs, has been observed. Another consequence of the replacement of iron by zinc is that the region around residues 36-46 undergoes larger displacements than the remainder of the polypeptide chain. Despite these changes, the main features of the FeS4 site, namely a local 2-fold symmetry and the characteristic network of N-H...S hydrogen bonds, are conserved in the ZnS4 site. The Zn-substituted rubredoxin provides the first precise structure of a Zn(Scys)4 unit in a protein. The nearly identical fold of rubredoxin around iron or zinc suggests that at least in some of the sites where the metal has mainly a structural role-e.g., zinc fingers-the choice of the relevant metal may be directed by its cellular availability and mobilization processes rather than by its chemical nature.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman E. T., Sieker L. C., Jensen L. H. Structure of rubredoxin from Desulfovibrio vulgaris at 1.5 A resolution. J Mol Biol. 1991 Jan 20;217(2):337–352. doi: 10.1016/0022-2836(91)90547-j. [DOI] [PubMed] [Google Scholar]
- Al-Karadaghi S., Cedergren-Zeppezauer E. S., Hövmoller S. Refined crystal structure of liver alcohol dehydrogenase-NADH complex at 1.8 A resolution. Acta Crystallogr D Biol Crystallogr. 1994 Nov 1;50(Pt 6):793–807. doi: 10.1107/S0907444994005263. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blake P. R., Park J. B., Zhou Z. H., Hare D. R., Adams M. W., Summers M. F. Solution-state structure by NMR of zinc-substituted rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus. Protein Sci. 1992 Nov;1(11):1508–1521. doi: 10.1002/pro.5560011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T. Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr D Biol Crystallogr. 1993 Jan 1;49(Pt 1):24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- Dauter Z., Sieker L. C., Wilson K. S. Refinement of rubredoxin from Desulfovibrio vulgaris at 1.0 A with and without restraints. Acta Crystallogr B. 1992 Feb 1;48(Pt 1):42–59. doi: 10.1107/s0108768191010613. [DOI] [PubMed] [Google Scholar]
- Day M. W., Hsu B. T., Joshua-Tor L., Park J. B., Zhou Z. H., Adams M. W., Rees D. C. X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus. Protein Sci. 1992 Nov;1(11):1494–1507. doi: 10.1002/pro.5560011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidsness M. K., O'Dell S. E., Kurtz D. M., Jr, Robson R. L., Scott R. A. Expression of a synthetic gene coding for the amino acid sequence of Clostridium pasteurianum rubredoxin. Protein Eng. 1992 Jun;5(4):367–371. doi: 10.1093/protein/5.4.367. [DOI] [PubMed] [Google Scholar]
- Eklund H., Nordström B., Zeppezauer E., Söderlund G., Ohlsson I., Boiwe T., Söderberg B. O., Tapia O., Brändén C. I., Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2-4 A resolution. J Mol Biol. 1976 Mar 25;102(1):27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Frey M., Sieker L., Payan F., Haser R., Bruschi M., Pepe G., LeGall J. Rubredoxin from Desulfovibrio gigas. A molecular model of the oxidized form at 1.4 A resolution. J Mol Biol. 1987 Oct 5;197(3):525–541. doi: 10.1016/0022-2836(87)90562-6. [DOI] [PubMed] [Google Scholar]
- Herriott J. R., Sieker L. C., Jensen L. H., Lovenberg W. Structure of rubredoxin: an x-ray study to 2.5 A resolution. J Mol Biol. 1970 Jun 14;50(2):391–406. doi: 10.1016/0022-2836(70)90200-7. [DOI] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Lamzin V. S., Wilson K. S. Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr. 1993 Jan 1;49(Pt 1):129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- Landro J. A., Schmidt E., Schimmel P., Tierney D. L., Penner-Hahn J. E. Thiol ligation of two zinc atoms to a class I tRNA synthetase: evidence for unshared thiols and role in amino acid binding and utilization. Biochemistry. 1994 Nov 29;33(47):14213–14220. doi: 10.1021/bi00251a033. [DOI] [PubMed] [Google Scholar]
- Lane R. W., Ibers J. A., Frankel R. B., Papaefthymiou G. C., Holm R. H. Synthetic analogues of the active sites of iron-sulfur proteins. 14. Synthesis, properties, and structures of bis(o-xylyl-alpha,alpha'-dithiolato)ferrate(II, III) anions, analogues of oxidized and reduced rubredoxin sites. J Am Chem Soc. 1977 Jan 5;99(1):84–98. doi: 10.1021/ja00443a017. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Mathieu I., Meyer J., Moulis J. M. Cloning, sequencing and expression in Escherichia coli of the rubredoxin gene from Clostridium pasteurianum. Biochem J. 1992 Jul 1;285(Pt 1):255–262. doi: 10.1042/bj2850255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu I., Meyer J. Transcript mapping of the rubredoxin gene from Clostridium pasteurianum. FEMS Microbiol Lett. 1993 Sep 1;112(2):223–227. doi: 10.1111/j.1574-6968.1993.tb06452.x. [DOI] [PubMed] [Google Scholar]
- May S. W., Kuo J. Y. Preparation and properties of cobalt(II) rubredoxin. Biochemistry. 1978 Aug 8;17(16):3333–3338. doi: 10.1021/bi00609a025. [DOI] [PubMed] [Google Scholar]
- Meyer J., Gaillard J., Lutz M. Characterization of a mutated rubredoxin with a cysteine ligand of the iron replaced by serine. Biochem Biophys Res Commun. 1995 Jul 26;212(3):827–833. doi: 10.1006/bbrc.1995.2043. [DOI] [PubMed] [Google Scholar]
- Monaco H. L., Crawford J. L., Lipscomb W. N. Three-dimensional structures of aspartate carbamoyltransferase from Escherichia coli and of its complex with cytidine triphosphate. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5276–5280. doi: 10.1073/pnas.75.11.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulis J. M., Davasse V., De Jésus F. Characteristic features of the heterologous functional synthesis in Escherichia coli of a 2[4Fe-4S] ferredoxin. Biometals. 1994 Oct;7(4):272–278. doi: 10.1007/BF00144121. [DOI] [PubMed] [Google Scholar]
- Myers L. C., Terranova M. P., Ferentz A. E., Wagner G., Verdine G. L. Repair of DNA methylphosphotriesters through a metalloactivated cysteine nucleophile. Science. 1993 Aug 27;261(5125):1164–1167. doi: 10.1126/science.8395079. [DOI] [PubMed] [Google Scholar]
- O'Halloran T. V. Transition metals in control of gene expression. Science. 1993 Aug 6;261(5122):715–725. doi: 10.1126/science.8342038. [DOI] [PubMed] [Google Scholar]
- Omichinski J. G., Clore G. M., Schaad O., Felsenfeld G., Trainor C., Appella E., Stahl S. J., Gronenborn A. M. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993 Jul 23;261(5120):438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- Petillot Y., Forest E., Mathieu I., Meyer J., Moulis J. M. Analysis, by electrospray ionization mass spectrometry, of several forms of Clostridium pasteurianum rubredoxin. Biochem J. 1993 Dec 15;296(Pt 3):657–661. doi: 10.1042/bj2960657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Alvarado G. C., Miles C., Michelsen J. W., Louis H. A., Winge D. R., Beckerle M. C., Summers M. F. Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat Struct Biol. 1994 Jun;1(6):388–398. doi: 10.1038/nsb0694-388. [DOI] [PubMed] [Google Scholar]
- Schwabe J. W., Klug A. Zinc mining for protein domains. Nat Struct Biol. 1994 Jun;1(6):345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- Sieker L. C., Stenkamp R. E., LeGall J. Rubredoxin in crystalline state. Methods Enzymol. 1994;243:203–216. doi: 10.1016/0076-6879(94)43016-0. [DOI] [PubMed] [Google Scholar]
- Stenkamp R. E., Sieker L. C., Jensen L. H. The structure of rubredoxin from Desulfovibrio desulfuricans strain 27774 at 1.5 A resolution. Proteins. 1990;8(4):352–364. doi: 10.1002/prot.340080409. [DOI] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995 Aug 25;269(5227):1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- Watenpaugh K. D., Sieker L. C., Jensen L. H. Crystallographic refinement of rubredoxin at 1 x 2 A degrees resolution. J Mol Biol. 1980 Apr 15;138(3):615–633. doi: 10.1016/s0022-2836(80)80020-9. [DOI] [PubMed] [Google Scholar]
- Zeng J., Vallee B. L., Kägi J. H. Zinc transfer from transcription factor IIIA fingers to thionein clusters. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):9984–9988. doi: 10.1073/pnas.88.22.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]



