Abstract
Cytosolic DNA induces type-I interferons and other cytokines that are important for antimicrobial defense but can also result in autoimmunity. This DNA signaling pathway requires the adaptor protein STING and the transcription factor IRF3, but the mechanism of DNA sensing is unclear. Here we showed that mammalian cytosolic extracts synthesized cyclic-GMP-AMP (cGAMP) in vitro from ATP and GTP in the presence of DNA but not RNA. DNA transfection or DNA virus infection of mammalian cells also triggered cGAMP production. cGAMP bound to STING, leading to the activation of IRF3 and induction of interferon-β. Thus, cGAMP represents the first cyclic di-nucleotide in metazoa and it functions as an endogenous second messenger that triggers interferon production in response to cytosolic DNA.
Host defense against foreign genetic elements is one of the most fundamental functions of a living organism. The presence of self or foreign DNA in the cytoplasm is sensed by eukaryotic cells as a danger signal or a sign of foreign invasion(1). DNA can be introduced into the cytoplasm by bacterial or viral infection, transfection, or ‘leakage’ from the nucleus or mitochondria under some pathological conditions that cause autoimmune diseases such as lupus. In mammalian cells, cytosolic DNA triggers the production of type-I interferons (IFNs) and other cytokines through the endoplasmic reticulum protein STING (also known as MITA, MPYS or ERIS) (2). STING recruits and activates the cytosolic kinases IKK and TBK1, which activate the transcription factors NF-κB and IRF3, respectively. NF-κB and IRF3 then enter the nucleus and function together to induce IFNs and other cytokines. DNA-dependent RNA polymerase III has been shown to be a sensor that detects and transcribes AT-rich DNA such as poly[dA:dT] into an RNA ligand capable of stimulating the RIG-I pathway to induce IFNs(3, 4). However, most DNA sequences do not activate the RNA polymerase III – RIG-I pathway. Instead, cytosolic DNA activates the STING-dependent pathway in a sequence-independent manner. How cytosolic DNA activates the STING pathway remains elusive.
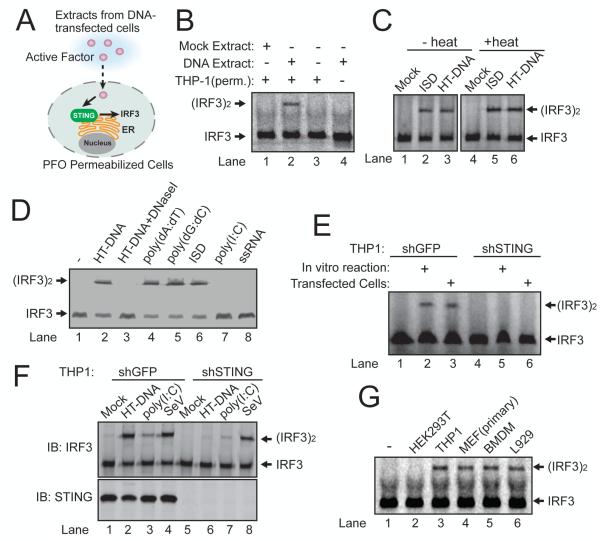
We hypothesized that DNA binds to and activates a putative cytosolic DNA sensor, which then directly or indirectly activates STING, leading to the activation of IRF3 and NF-κB (fig. S1A). To test this model, we developed an in vitro complementation assay using the murine fibrosarcoma cell line L929, which is known to induce interferon-β (IFNβ) in a STING-dependent manner(5) (Fig. 1A). We used a L929 cell line stably expressing a short hairpin (sh)RNA against STING such that DNA transfection would only activate factors upstream of STING, including the putative DNA sensor (fig. S1, A and B). The L929-shSTING cells were transfected with different types of DNA and then cytoplasmic extracts from these cells were mixed with the human monocytic cell line THP1 or murine macrophage cell line Raw264.7, which was permeabilized with perfringolysin O (PFO; Fig. 1A). PFO treatment pokes holes in the plasma membrane (6), allowing the cytoplasm to diffuse in and out of cells, while retaining organelles including endoplasmic reticulum (which contains STING) and Golgi apparatus inside the cells(7). If an upstream activator of STING is generated in the DNA transfected cells, the cytoplasm containing such an activator is expected to activate STING in the PFO-permeabilized cells, leading to the phosphorylation and dimerization of IRF3.
Figure 1. DNA-dependent generation of a heat-resistant small molecule activates the STING pathway.
(A) Illustration of an activity assay for cellular factors that activate the STING pathway. PFO: perfringolysin O. (B) Cytosolic extracts from mock or ISD-transfected L929-shSTING cells were incubated with PFO permeabilized THP1 cells together with 35S-IRF3. Dimerization of IRF3 was analyzed by native gel electrophoresis followed by autoradiography. (C) Similar to (B), except that in lanes 4-6, cytosolic extracts were heated at 95°C for 5 min to denature proteins and then the heat-resistant supernatant was incubated with PFO-permeabilized THP1 cells. HT-DNA: herring testis DNA. (D) L929-shSTING cytosolic extracts were incubated with the indicated nucleic acids in the presence of ATP and then the heat-resistant supernatant was assayed for its ability to stimulate IRF3 dimerization in permeabilized Raw264.7 cells. (E) THP1 cells stably expressing shRNA against GFP (control) or STING were permeabilzed with PFO and then incubated with the heat-resistant supernatant from the reaction mixture containing DNA-supplemented L929 cytosolic extracts (lanes 2 and 5) or from DNA-transfected L929 cells (lanes 3 and 6). IRF3 activation was analyzed by native gel electrophoresis. (F) THP1 cells described in (E) were transfected with HT-DNA, poly[I:C] or infected with Sendai virus (SeV), followed by measurement of IRF3 dimerization. (G) Cytosolic extracts from the indicated cell lines were incubated with HT-DNA and then heat-resistant supernatants were assayed for their ability to stimulate IRF3 dimerization in permeabilized Raw264.7 cells. Unless noted otherwise, all results in this paper were representative of at least two independent experiments.
Cytoplasmic extracts from L929-shSTING cells transfected with a DNA sequence known as interferon-stimulatory DNA (ISD, Fig. 1B, lane 2), poly[dA:dT], a GC-rich 50-base pair dsDNA (G:C50), poly[dI:dC] or herring testis DNA (HT-DNA; fig. S1C) activated IRF3 in permeabilized THP1 cells, indicating that this activity was independent of DNA sequence.
To determine if the STING activator is a protein, we incubated the cytoplasmic extracts at 95°C to denature most proteins and then incubated the ‘heat supernatant’ with permeabilized THP1 cells. Surprisingly, the heat supernatant from the ISD or HT DNA transfected cells caused IRF3 dimerization (Fig. 1C). This activity was resistant to treatment with Benzonase (fig. S1D), which degrades both DNA and RNA, or proteinase K (fig. S1D). Thus, the STING activator is probably not a protein, DNA or RNA.
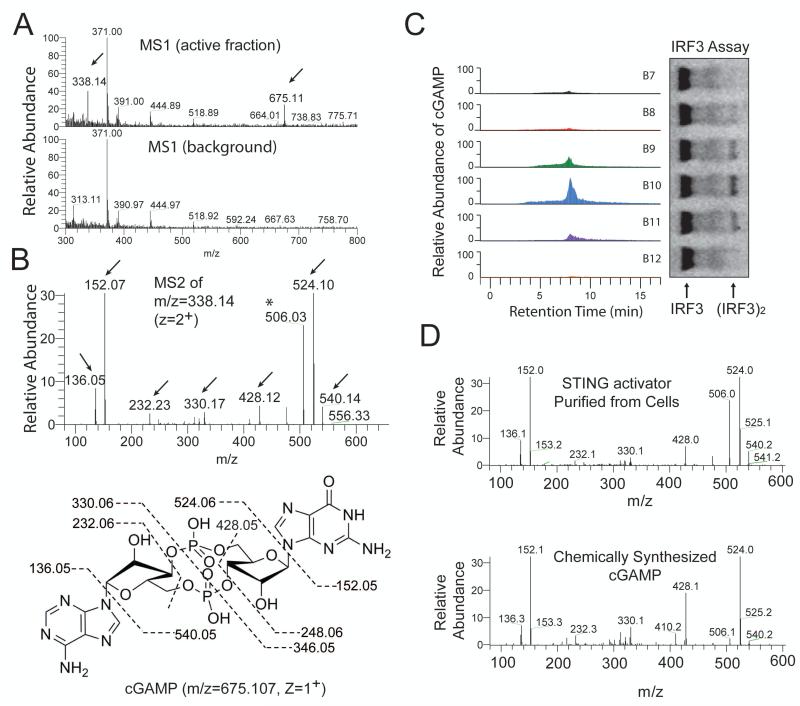
To test if DNA could stimulate the generation of the heat-resistant STING activator in vitro, we incubated HT DNA with L929-shSTING cytoplasmic extracts (S100) in the presence of ATP (fig. S1E). The reaction mixture was heated at 95°C to denature proteins. Remarkably, incubation of the supernatant with permeabilized Raw264.7 cells led to IRF3 dimerization (Fig. 1D, lane 2). This activity depended on the addition of DNA to the cytoplasmic extracts. Other DNA, including poly[dA:dT], poly[dG:dC] and ISD, also stimulated the generation of the STING activator in L929-shSTING cytoplasmic extracts, whereas poly[I:C] and single-stranded RNA had no activity (Fig. 1D). Similar results were obtained with permeabilized THP1 cells (fig. S1F). Knockdown of STING in the permeabilized THP1 cells abolished IRF3 activation by the heat-resistant factor generated by DNA transfected into L929 cells or DNA added to L929 cytosolic extracts (Fig. 1E). Control experiments showed that the knockdown of STING inhibited the activation of IRF3 and induction of IFNβ and TNFα in THP1 cells by HT-DNA transfection (fig. S1G and S1H), but IRF3 activation by poly[I:C] transfection or Sendai virus infection, which is known to activate the RIG-I pathway, was unaffected by the STING knockdown (Fig. 1F). We also tested cytoplasmic extracts from several cell lines for their ability to produce the heat-resistant STING activator (Fig. 1G). Incubation of HT-DNA with extracts from primary MEF cells, mouse bone marrow derived macrophages (BMDM) and L929 cells led to generation of the heat-resistant factor that activated IRF3. Human cell extracts from THP1, but not HEK293T, were also able to produce this STING activator. These results are in agreement with our previous finding that primary MEF, BMDM, L929 and THP1 cells, but not HEK293T cells, possessed the STING-dependent, RNA polymerase III –independent, pathway to induce type-I interferons(3). We next purified the heat-resistant STING activator from L929 cell extracts using several chromatographic steps including a STING-Flag affinity purification step (fig. S2, A and B). Previous research has shown that the bacterial molecules cyclic-di-AMP and cyclic-di-GMP bind to STING and induce type-I interferons(8, 9). However, using nano liquid chromatography mass spectrometry (nano-LC-MS), we did not detect MS or MS/MS spectra consistent with those expected of c-di-GMP ([M+H]+=691) or c-di-AMP ([M+H]+=659). Interestingly, in-depth examination of the MS spectra revealed two ions with mass to charge ratios (m/z) of 675.1 (z=1+) and 338.1 (z=2+), which were present in the active fractions but absent in the background spectra (Fig. 2A). These m/z values, despite the low mass accuracy of the mass spectrometer (LTQ), were equivalent to the average calculated m/z values of c-di-GMP and c-di-AMP (675=[691 + 659]/2). This observation suggested that the detected ion was a hybrid of c-di-GMP and c-di-AMP, i.e., cyclic-GMP-AMP (m/z = 675.107, z=1+; m/z = 338.057, z=2+). Collision induced dissociation (CID) fragmentation of this ion (m/z = 338.1, z=2+) revealed several prominent ions with m/z values expected of the product ions of c-GMP-AMP (cGAMP) (Fig. 2B). Importantly, quantitative mass spectrometry using selective reaction monitoring (SRM) showed that the abundance of the ions representing cGAMP in the fractions from a C18 column correlated very well with their IRF3-stimulatory activities (Fig. 2C; fig. S2C). cGAMP has recently been identified in the bacterium Vibro cholera and shown to play a role in bacterial chemotaxis and colonization(10). However, cGAMP has not been reported to exist or function in eukaryotic cells.
Figure 2. Purification and identification of the heat-resistant STING activator.
(A) Full scan nano-LC-MS spectra of active and inactive fractions from the C18 column using LTQ. Arrows indicate an ion at +1 (675.11) and +2 (338.14) charge states present only in the active fraction. (B) Tandem mass (MS2) spectra after CID fragmentation of the ion with m/z =338.14 (z=2) from the MS1 scan shown in (A). Arrows indicate the m/z values of the expected fragmentation patterns of cyclic-GMP-AMP (cGAMP, bottom). Asterisk (*) indicates an ion (m/z=506) that resulted from a neutral loss of a water molecule (18) from the ion with m/z = 524. (C) Fractions (B7-B12) from the C18 column were analyzed for the presence of cGAMP by selective reaction monitoring of the expected ions and for their ability to stimulate IRF3 dimerization. (D) Comparison of the CID MS2 spectra of the purified STING activator and chemically synthesized cGAMP.
To verify the identity of the heat resistant STING activator, we used a high-resolution high-accuracy mass spectrometer (Q Exactive, Thermo) to perform nano-LC-MS analysis. The cell-derived STING activator had m/z of 675.107 (z=1+) and 338.057 (z=2+), which exactly matched the theoretical values of cGAMP (fig. S2D). To further characterize the structure and function of cGAMP, we developed a ten-step single-flask protocol to chemically synthesize cGAMP (fig. S3). The MS/MS spectra of the cell-derived STING activator were identical to those of the chemically synthesized cGAMP (Fig. 2D). These results demonstrate that L929 cells produced cGAMP.
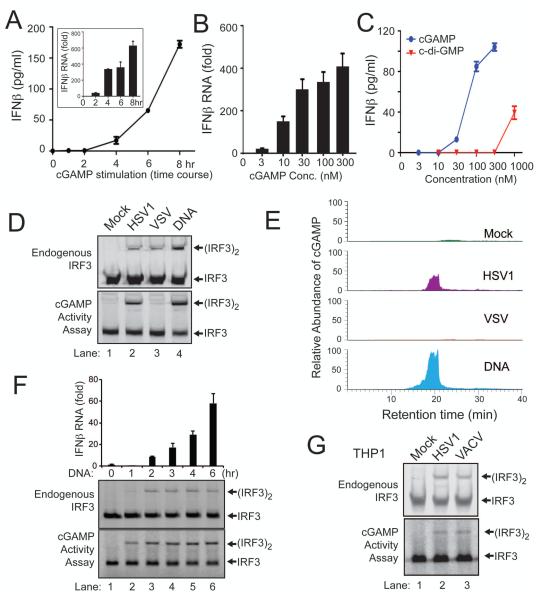
Quantitative RT-PCR and ELISA assays showed that chemically synthesized cGAMP induced IFNβ RNA and protein in L929 cells after introduction into the cells (Fig. 3A). Titration experiments showed that cGAMP induced IFNβ RNA robustly even at concentrations as low as 10 nM (Fig. 3B). In fact, cGAMP was much more potent than c-di-GMP in inducing IFNβ based on ELISA assays (Fig. 3C). cGAMP was also more potent than c-di-GMP and c-di-AMP in activating IRF3 (fig. S4A). To determine if L929 extracts contained enzymes that could synthesize other types of di-nucleotides or oligonucleotides capable of activating IRF3, we tested all four ribonucleotides in various combinations (fig. S4B). ATP and GTP were both necessary and sufficient to support the synthesis of an activator of IRF3, further supporting that L929 contained an enzyme that synthesizes cGAMP from ATP and GTP.
Figure 3. DNA transfection and DNA virus infection induce IFNβ through cGAMP.
(A) Chemically synthesized cGAMP (100 nM) was delivered to digitonin-permeabilized L929 cells for indicated times, then IFNβ RNA and secreted protein were measured by q-RT-PCR (inset) and ELISA, respectively. Unless noted otherwise, the error bars in this and all other panels represent standard errors of the mean (n=3). (B) Similar to (A), except that different concentrations of cGAMP were delivered into L929 cells for 8 hr followed by q-RT-PCR analyses of IFNβ RNA. (C) Similar to (B), except that different concentrations of cGAMP and cdi-GMP were delivered into L929 cells followed by ELISA assays for IFNβ. (D) L929 cells were infected with HSV-1Δ34.5 or VSV-ΔM51-GFP, transfected with HT-DNA, or mock treated. An aliquot of the cell extracts was directly analyzed for IRF3 dimerization (top), whereas another aliquot was heated to denature proteins and the heat-resistant supernatant was assayed for its ability to stimulate IRF3 dimerization in permeabilized Raw264.7 cells (bottom). (E) The heat-resistant supernatant from (D) was fractionated by HPLC using a C18 column, and the presence of cGAMP in the fractions was measured by mass spectrometry using SRM. (F) L929 cells were transfected with 4 μg/ml HT-DNA for the indicated time, then IFNβ RNA was measured by q-RT-PCR and IRF3 dimerization was analyzed by native PAGE. Aliquots of the cell extracts were tested for the presence of cGAMP based on its ability to induce IRF3 dimerization after delivery into Raw264.7 cells. (G) THP1 cells were infected with HSV-1Δ34.5 and Vaccinia virus (VACV) for 6 hr, then the activation of endogenous IRF3 and generation of cGAMP activity were measured as described in (F).
To determine if DNA virus infection leads to the production of cGAMP in cells, we infected L929 cells with HSV-1 lacking ICP34.5, a viral protein known to antagonize interferon production in the infected cells (11). Like DNA transfection, HSV-1ΔICP34.5 infection led to IRF3 activation in L929 cells (Fig. 3D, upper). Cell extracts from the DNA-transfected or virus-infected cells contained a heat-resistant factor that could activate IRF3 in permeabilized Raw264.7 cells (Fig. 3D, lower). As a control, we infected L929 cells with a vesicular stomatitis virus strain, VSV-ΔM51-GFP, an RNA virus known to trigger strong interferon production through the RIG-I pathway (12, 13). In contrast to HSV-1, VSV-infected cells did not contain the heat-resistant IRF3 activator in the same in vitro assay, although VSV infection did induce IRF3 activation in L929 cells (Fig. 3D). The heat resistant factor in HSV-1 infected cells was enriched by reverse phase HPLC and quantified by nano-LC-MS using SRM. DNA transfected or HSV-1 infected cells, but not mock treated or VSV infected cells, produced elevated levels of cGAMP (Fig. 3E). Kinetic experiments showed that, after DNA was transfected into L929 cells, cGAMP was produced before IRF3 dimerization and IFNβ induction could be detected (Fig. 3F). To test if DNA viruses could induce cGAMP production in human cells, we infected THP1 cells with HSV1 or Vaccinia virus (VACV; Fig. 3G). Both viruses induced IRF3 dimerization in the cells. Importantly, both viruses also triggered the production of cGAMP that activated IRF3 (Fig. 3G, lower). Collectively, these results indicate that DNA transfection and DNA virus infections in human and mouse cells produced cGAMP, which led to IRF3 activation.
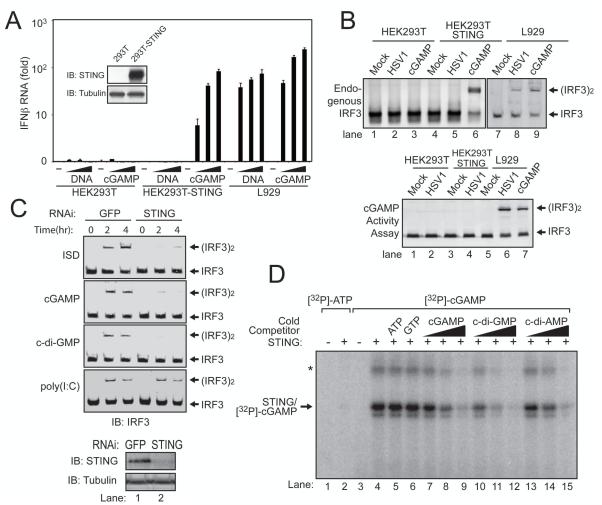
To determine if cGAMP activates IRF3 through STING, we carried out three sets of experiments. First, we established a HEK293T cell line stably expressing STING, stimulated these cells with cGAMP and then measured IFNβ induction by quantitative RT-PCR (Fig. 4A). HEK293T cells did not respond to cGAMP, likely due to a lack of or a very low level of STING expression in these cells. The expression of STING in HEK293T cells rendered a high level of IFNβ induction by cGAMP. However, DNA did not stimulate HEK293T/STING cells to induce IFNβ, consistent with a defect of HEK293T cells in producing cGAMP in response to DNA stimulation. In contrast, L929 cells induced IFNβ in response to stimulation by either cGAMP or DNA. HSV-1 infection induced IRF3 dimerization in L929, but not HEK293T or HEK29T/STING cells (Fig. 4B, upper), suggesting that the production of cGAMP is important for HSV-1 to activate IRF3 in cells. Indeed, extracts from HSV1-infected L929, but not from HEK293T or HEK293T/STING cells, contained the cGAMP activity that led to IRF3 dimerization in permeabilized Raw264.7 cells (Fig. 4B, lower). These results indicate that the expression of STING in HEK293T cells installed the ability of the cells to activate IRF3 and induce IFNβ in response to cGAMP, but was insufficient to install the response to DNA or DNA viruses due to a defect of HEK293T cells in synthesizing cGAMP.
Figure 4. cGAMP binds to STING and activates IRF3 in a STING-dependent manner.
(A) Increasing concentrations of HT-DNA or cGAMP were delivered to indicated cells and the induction of IFNβ was measured by q-RT-PCR. Inset shows immunoblots of STING and β-tubulin in the cell lines. (B) Indicated cell lines were infected with HSV1Δ34.5 or permeabilized with digitionin and then incubated with cGAMP. Activation of endogenous IRF3 was analyzed by native gel electrophoresis (top). Aliquots of the cytosolic extracts were heated to denature proteins, and the supernatant was assayed for its ability to stimulate IRF3 in permeabilized Raw264.7 cells (bottom). (C) cGAMP, c-di-GMP, ISD, or poly[I:C] was delivered into L929 cells stably expressing a shRNA against GFP or STING for the indicated time, followed by analysis of IRF3 dimerization. (D) Recombinant STING protein was incubated with [32P]-ATP or [32P]-cGAMP in the presence or absence of the cold competitors as indicated. After UV crosslinking, the mixtures were resolved by SDS-PAGE followed by autoradiography.
Second, we tested the response of L929 and L929-shSTING cells to cGAMP (Fig. 4C). Similar to ISD and c-di-GMP, cGAMP-induced IRF3 dimerization was dependent on STING. In contrast, poly[I:C] still induced IRF3 dimerization in the absence of STING. These results demonstrate that STING is necessary for cGAMP to activate IRF3.
Finally, we examined whether STING binds to cGAMP directly. Recombinant STING protein containing residues 139-379, which has been shown to bind c-di-GMP(14), was expressed and purified from E. coli and then incubated with 32P-cGAMP followed by UV-induced crosslinking (Fig. 4D). A radiolabelled band corresponding to cross-linked STING - cGAMP complex was detected when both STING and 32P-cGAMP were present. High concentrations of ATP or GTP did not compete with the formation of STING-cGAMP complex. By contrast, the intensity of this band decreased as the concentrations of competing cold cGAMP, c-di-GMP or c-di-AMP increased, suggesting that the cGAMP binding sites on STING might overlap with those that interact with c-di-GMP and c-di-AMP. Indeed, mutations of several residues that were recently shown to participate in the binding of STING to c-di-GMP (14), including S161Y, Y240S and N242A, also impaired the binding of STING to cGAMP (fig. S5). Collectively, these results demonstrate that cGAMP is a ligand that binds to and activates STING.
Cyclic di-nucleotides have been shown to function as bacterial second messengers that regulate a variety of physiological processes, including bacterial motility and biofilm formation(15). A recent report showed that c-di-GMP is produced in the protozoan Dictyostelium and functions as a morphogen to induce stalk cell differentiation(16). In this report, we identified cGAMP as the first cyclic di-nucleotide in metazoa. Moreover, we showed that cGAMP is a potent inducer of type-I interferons. The role of cGAMP is similar to that of cAMP, the best-studied second messenger(17). Like cAMP, which is synthesized by adenylate cyclase upon its activation by upstream ligands, cGAMP is synthesized by a cyclase in response to stimulation by a DNA ligand (18). cAMP binds to and activates protein kinase A and other effector molecules. Similarly, cGAMP binds to and activates STING to trigger the downstream signaling cascades. As an endogenous molecule in mammalian cells, cGAMP may be used in immune therapy or as a vaccine adjuvant.
Supplementary Material
Acknowledgements
We thank Dr. Y. Tanaka for generating HEK293T-STING and L929-shSTING cell lines, Drs. J. Bell, B. Levine and L. Deng for VSV, HSV-1 and VACV, respectively, Dr. R. Debose-Boyd for the PFO plasmid, and Dr. V. Sperandio for the V. cholera strain C6709. The data presented in this paper are tabulated in the main paper and in the supplementary materials. This work was supported by grants from NIH (AI-093967 to Z.J.C. and GM-079554 to C.C). Z. J. C is an investigator of Howard Hughes Medical Institute.
References and Notes
- 1.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic Acid Recognition by the Innate Immune System. Annu Rev Immunol. Apr 5; doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 2.Barber GN. Cytoplasmic DNA innate immune pathways. Immunological reviews. 2011 Sep;243:99. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- 3.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009 Aug 7;138:576. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009 Jul 16; doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka Y, Chen ZJ. STING Specifies IRF3 Phosphorylation by TBK1 in the Cytosolic DNA Signaling Pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossjohn J, et al. Structures of perfringolysin O suggest a pathway for activation of cholesterol-dependent cytolysins. Journal of molecular biology. 2007 Apr 13;367:1227. doi: 10.1016/j.jmb.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proceedings of the National Academy of Sciences of the United States of America. 2009 Dec 8;106:20842. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010 Jun 25;328:1703. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011 Oct 27;478:515. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012 Apr 13;149:358. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mossman KL, Smiley JR. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. Journal of virology. 2002 Feb;76:1995. doi: 10.1128/JVI.76.4.1995-1998.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stojdl DF, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003 Oct;4:263. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 13.Sun Q, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006 May;24:633. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Yin Q, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Molecular cell. 2012 Jun 29;46:735. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr Opin Microbiol. 2009 Apr;12:170. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012 Aug 30;488:680. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blumenthal SA. Earl Sutherland (1915-1975) and the discovery of cyclic AMP. Perspect Biol Med. 2012;55:236. doi: 10.1353/pbm.2012.0017. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type-I interferon pathway. Science. 2012 doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.