Abstract
Immunologic dysfunction, mediated via monocyte activity, has been implicated in the development of HIV-associated neurocognitive disorder (HAND). We hypothesized that transcriptome changes in peripheral blood monocytes relate to neurocognitive functioning in HIV+ individuals, and that such alterations could be useful as biomarkers of worsening HAND.
METHODS
mRNA was isolated from the monocytes of 86 HIV+ adults and analyzed with the Illumina HT-12 v4 Expression BeadChip. Neurocognitive functioning, HAND diagnosis, and other clinical and virologic variables were determined. Data were analyzed using standard expression analysis and weighted gene co-expression network analysis (WGCNA).
RESULTS
Neurocognitive functioning was correlated with multiple gene transcripts in the standard expression analysis. WGCNA identified two nominally significant co-expression modules associated with neurocognitive functioning, which were enriched with genes involved in mitotic processes and translational elongation.
CONCLUSIONS
Multiple modified gene transcripts involved in inflammation, cytoprotection, and neurodegeneration were correlated with neurocognitive functioning. The associations were not strong enough to justify their use as biomarkers of HAND; however, the associations of two co-expression modules with neurocognitive functioning warrants further exploration.
Keywords: HIV-associated neurocognitive disorder, NeuroAIDS, monocyte, IL6R, KEAP1, LRP12, CSNK1A1, WGCNA
INTRODUCTION
Over 32 million individuals are currently infected with HIV-1 worldwide, and 1.2 million within the United States. In most developed countries, the availability of a combination of effective antiretroviral medicines has significantly increased life expectancy. As a chronic disease, HIV has lead to a host of new challenges. Perhaps the most common is HIV-associated neurocognitive disorder (Antinori et al., 2007), which may affect between 40%–70% of infected individuals (McArthur et al., 2005). Prior to the availability of combination anti-retroviral therapy (cART), neurological illness in HIV+ individuals often presented as dementia (i.e., HIV-associated dementia, or HAD), with an acute encephalitis as the frequent underlying cause. A key aspect of the neuropathogenic process of HAD is the phenotypic modification of peripheral blood monocytes and their subsequent migration across the blood brain barrier(Pulliam et al., 1997). CD16+ monocytes are among the first cells to become infected with HIV. This subset of cells expand in HIV and Simian immunodeficiency virus (SIV) infection(Thieblemont et al., 1995, Otani et al., 1998), and also appears to be associated with the pathogenesis of milder HIV-Associated Neurocognitive Disorder (HAND)(Pulliam et al., 1997, Ellery et al., 2007). As the virus gains momentum and the immune system weakens, the subset of CD16+ monocytes increases and cross the blood brain barrier (BBB) as “Trojan Horse” cells with increased frequency. This migration is driven by enhanced chemokine release in the CNS as well as the peripheral immune response (Peluso et al., 1985, Ancuta et al., 2004, Kraft-Terry et al., 2009). Increased traffic of monocytes into the CNS, which is often followed by differentiation into perivascular macrophages (Cosenza et al., 2002), further increases the expression of chemokines such as monocyte chemotactic protein (MCP-1), leading to recruitment of even more monocytes in a feed-forward manner (Persidsky et al., 1999, Persidsky et al., 2000). These processes have also been described in terms of a “push” (peripheral immune activation) and “pull” (a chemokine gradient released from cells within the CNS) mechanisms(Kraft-Terry et al., 2009). In addition to the release of pro-inflammatory cytokines and chemokines, infected monocytes also release viral proteins that are harmful to nearby neurons and other cells (Kedzierska and Crowe, 2002, Glass et al., 1995, Adle-Biassette et al., 1999, Lindl et al., 2007, Kaul and Lipton, 2006, Kraft-Terry et al., 2009). In summary, the crosstalk between the CNS and circulating blood monocytes is thought to be a key mechanism underlying HAND neuropathogenesis. As such, delineating cellular dynamics that occur within monocytes at the early stages of this process could lead to biomarker identification and pharmaceutical intervention.
To date, investigations into the molecular biology of HAND have generally utilized models or tissue representing advanced disease states (e.g., HAD or HIV-encephalitis (HIVE)). Such studies have included transcriptomic and proteomic changes in monocytes, macrophages, brain cells and brain tissue in conjunction with advanced immunosuppression(Pulliam et al., 1997, Pulliam et al., 2004, Gendelman et al., 1997, Carlson et al., 2004, Luo et al., 2003, Wojna et al., 2004, Roberts et al., 2003, Buckner et al., 2011). Gene expression studies have also been widely used to investigate and discover cellular mechanism involved in the pathogenesis advanced HAND and HIVE. Many of these have involved the use of homogenized samples of post-mortem brain tissue (Masliah et al., 2004, Roberts et al., 2003, Roberts et al., 2004, Shapshak et al., 2004a, Gelman et al., 2012), while others have examined specific brain cells in vitro, such as astrocytes or neurons (Shapshak et al., 2004b, Kim et al., 2004, Kolson et al., 2004, Borjabad et al., Shapshak P, 2011), the results of which have led to further insights regarding the neuropathogenesis of HAD and HIVE. However, while studies using brain tissue have been useful in describing cellular alterations associated with HAD, they are of limited utility for defining clinically practical biomarkers. In addition, because most in vivo gene expression studies have examined advanced states of disease, such findings may not generalize to the more pervasive mild HAND. (Brew, 2004, Cysique et al., 2005, Cysique et al., 2004, Robertson et al., 2007, Dore et al., 2003, Sacktor et al., 2002). Currently, in the context of combined antiretroviral therapy (cART), it is thought that HAND develops in a chronic manner, with persistent neuroinflammation and low grade viral replication driven in part by monocytes-derived perivascular macrophages in the CNS(Langford et al., 2003, Yadav and Collman, 2009). Not surprisingly, research targeting monocyte gene expression that utilizes both genome-wide and targeted approaches has yielded important results, implicating an evolving monocyte phenotype characterized by increased chemotactic properties and chronic inflammatory response(Pulliam et al., 2004, Buckner et al., 2011). Despite this, studies thus far have not found an association between monocyte gene transcription and behavioral HAND phenotypes(Sun et al., 2010). However, such studies have had very limited power to detect differences, considering the vast number of comparisons characteristic of transcriptome association studies.
In order to arrive at a biologically meaningful reduction of high dimensional transcriptomic data and to integrate such data with orthogonal genetic and behavioral data, systems biologic data analysis methods such as weighted gene co-expression network analysis (WGCNA) have been developed(Zhang and Horvath, 2005b). Here we used WGCNA to integrate transcriptomic and behavioral data derived from the largest clinical sample to date in an effort to investigate of the neuropathogenesis of mild HAND. We focused our investigation on circulating blood monocytes, as these cells are a primary and early component of this pathogenesis. Our hypothesis was that the WGCNA would identify biological pathways associated with increasing HAND severity.
METHODS
Participants
This study was conducted in accordance with the University of California, Los Angeles Medical Institutional Review Board. Eighty-four (86) participants were recruited from the Multicenter AIDS Cohort Study (MACS) in Los Angeles. MACS participants are seen on a semi-annual basis during which they provide blood and complete comprehensive self-report questionnaires assessing drug use, medication use, and medical co-morbidities. In addition, all participants undergo comprehensive neuropsychological testing and assessment of activities of daily living every two years, from which their HAND status is determined. Forty-nine (57%) participants in the current study were Caucasian/non-Hispanic, 14 (16%) were Caucasian/Hispanic, 8 (9%) were African American/non-Hispanic, and the remaining 15 (17%) considered themselves Other/Hispanic. Additional group characteristics are shown in Tables 1 & 2.
Table 1.
Sample characteristics
| Mean | Median | Standard Deviation | Range | |
|---|---|---|---|---|
| AGE | 52 | 52 | 9.6 | 30–67 |
| GNF* | 49 | 48 | 14 | 23–78 |
| Nadir CD4 | 250 | 260 | 150 | 1–670 |
| Current CD4 | 590 | 580 | 200 | 73–1100 |
| Viral Load | 0.38 | 0 | 1.2 | 0–5.2 |
| CES-D | 14 | 10 | 12 | 0–48 |
| CPE | 1.5 | 1.5 | 0.77 | 0–4.5 |
Global Neurocognitive Functioning
Table 2.
Substance use and HAND status
| N | Percent of Sample | |
|---|---|---|
| Current HAND | ||
| Diagnosis | ||
| Normal | 33 | 38 |
| ANI | 18 | 21 |
| MND | 26 | 30 |
| HAD | 9 | 10 |
| Smoke Tobacco | ||
| Never | 21 | 25 |
| Past | 47 | 54 |
| Current | 18 | 21 |
| Alcohol Use | ||
| None/Rarely | 45 | 51 |
| Monthly | 19 | 23 |
| Weekly | 10 | 12 |
| Daily | 12 | 14 |
| Cannabis Use | ||
| None/Rarely | 62 | 71 |
| Monthly | 4 | 5 |
| Weekly | 8 | 10 |
| Daily | 12 | 14 |
| Cocaine Use | ||
| None/Rarely | 84 | 98 |
| Monthly | 0 | 0 |
| Weekly | 2 | 2 |
| Daily | 0 | 0 |
| Heroin Use | ||
| None/Rarely | 86 | 100 |
Blood processing, Monocyte Isolation, mRNA extraction, and microarray analysis
24ml of fresh blood was collected from participants. Blood was drawn into three 8 mL Cell Preparation Tubes (CPT) containing sodium citrate. Peripheral blood mononuclear cells (PBMCs) were then isolated through centrifugation within 6 hours of collection (Salazar-Gonzalez et al., 1997). PBMCs were washed with phosphate buffered saline, and then monocytes were isolated through Rosette separation (RosetteSep®; Stem Cell Technologies) according to the manufacturer instructions. Monocytes were then pelleted, lysed, and RNA extracted using the Qiagen RNeasy kit including a DNase treatment to eliminate any potentially confounding genomic DNA contamination(Shay et al., 2003). RNA was stored at −80°C and sent in batches to the Southern California Genotyping Consortium (SCGC) for microarray analysis, which was performed with the Illumina Human HT-12 v4 gene expression BeadChip. The expression data and sample characteristics, including all information required by the MIAME standard, are available from our web site http://labs.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/HIVMonocyteExpression/as well as from the NCBI Gene Expression Omnibus (GEO).
Variables Included in the Gene Expression Analysis
Neurocognitive functioning
Participants completed a comprehensive battery of neuropsychological tests as part of the standard MACS protocol. This includes measures of working memory, learning, memory, executive functioning, motor functioning, and information processing speed. T-scores were calculated using normative data derived from the HIV-seronegative MACS cohort, with demographic corrections for age, education, and ethnicity. For this study, we calculated a Global Neurocognitive Functioning score based on the average of the six domain T-scores.
HAND Severity
HAND status was determined via an algorithm developed by MACS investigators. The algorithm is based on neurocognitive test performance and self-reported deficits in activities of daily living(Lawton and Brody, 1969) in accordance with current research criteria(Antinori et al., 2007). Participants are rated as normal, mild (i.e. Asymptomatic Neurocognitive Impairment-ANI), moderate (Minor Neurocognitive Disorder-MND), or severely impaired (HIV-Associated Dementia-HAD).
CNS Penetration Effectiveness (CPE)
The effects of antiretroviral medications on neurocognitive functioning remains unclear(Tozzi et al., 2007, Cysique et al., 2009, Marra et al., 2003). We sought to determine the impact of antiretroviral regimen on monocyte gene expression. We employed the CPE, a score that is based on the pharmacokinetic and pharmacodynamic characteristics of antiretroviral medications(Letendre et al., 2008). CPE scores for the regimen reported at the time of neurocognitive testing were calculated. Higher scores indicate a regimen with increased penetration of the blood brain barrier.
Substance Use
In this study we considered the effects of alcohol, marijuana, and stimulant (cocaine and/or methamphetamine) use on gene expression. MACS participants complete a substance use questionnaire every six months. Drug and alcohol use is recorded in terms of frequency during the last six months and average amount used each time. Participants were considered active users of alcohol, stimulants, or marijuana if they report daily or weekly use, and non-users if they report monthly or less use in the six months preceding the visit.
Depression
Depression was determined with the Center for Epidemiologic Studies Depression Scale (CES-D)(Radloff, 1977). Scores on the CES-D are entered as a continuous variable, with higher scores indicating greater degree of depression.
Virologic measures
The percentage of lymphocytes that were CD4+ T-cells was determined by flow cytometry. HIV viral load was determined via either the COBAS TaqMan HIV-1 Test, Version 2.0 or Roche Amplicor HIV-1 MONITOR Test, Version 1.5. Both tests quantify HIV-1 RNA based on in vitro amplification of the highly conserved HIV-1 gag gene. Nadir CD4 was obtained either by self-reports or, for those who seroconverted during the course of the study, their lowest CD4 according to study records.
Statistical Analysis
Gene array data was processed in Illumina BeadStudio software and log2-transformed to stabilize variance. The normalized data were batch-corrected using the ComBat R function (Johnson et al., 2007). Potential outliers were removed both within each batch and, after batch correction, in the combined data. Outlier identification was based on scaled connectivity of each sample in a sample network (Oldham et al., 2012) based on Euclidean distance. An apparent technical sample-to-sample variation that remained was then removed by quantile normalization (Bolstad et al., 2003). Low-expressed probe sets were removed. The expression data were then adjusted for age by regressing gene expression on age using a linear model and retaining the residuals as age-adjusted expression values.
Differential expression analysis
The relationship between each gene and continuous or ordinal clinical variables (neurocognitive domain and global scores, HAND rating, CPE, CES-D, and substance use variables) and virologic variables (nadir CD4, current CD4, and current viral load) were assessed using Pearson correlation. The Pearson correlation test is appropriate since it only assumes that one of the quantitative variables (here gene expression) follows a normal distribution. For the single dichotomous clinical variable (HAND diagnosis) we used the Mann-Whitney U test (a special case of the Kruskal-Wallis test). To adjust for multiple testing we report the local False Discovery Rate (FDR) q-value estimates calculated using code from the qvalue R package(Storey et al., 2004). Results of these differential expression analyses were used in two ways. First, sets of top identified genes for each phenotype (i.e., clinical and virologic variable) were tested for enrichment in known gene ontology (GO) terms(Ashburner et al., 2000). Second, the clinical/virologic variable - gene relationship values (i.e., correlations) for all genes were retained and converted into a color vectors for visual assessment of disease-gene relationships in the network analysis.
Weighted Gene Co-Expression Network Analysis (WGCNA)
Here we employ for the first time WGCNA in the analysis of HIV monocyte transcriptome data. The WGCNA starts out from the over 36,000 assayed probe set, identifies modules of co-expressed genes, and relates these modules to clinical variables and gene ontology information(Zhang and Horvath, 2005b, Langfelder and Horvath, 2008, Horvath et al., 2006). Because gene modules may correspond to biological pathways, focusing the analysis on modules (and their highly connected intramodular hub genes) amounts to a biologically meaningful data reduction scheme. Highly correlated module genes are represented and summarized by the module eigengene, or ME(Langfelder and Horvath, 2007), as described below. By correlating each gene with the ME we defined a measure of module membership which quantifies how close (i.e., correlated) a gene is to a given module. Module membership measures allow one to annotate all genes on the array and to screen for highly connected intramodular hub genes, which may have particular relevance to disease (Voineagu et al., 2011, Horvath et al., 2006, Ghazalpour et al., 2006). As described below, we use functional enrichment analysis with regard to known gene ontologies to understand the biological significance of the ME and to identify putative disease pathways.
In the following, we provide more details on the weighted correlation network construction. The network construction starts by calculating robust correlations between all genes across all relevant samples. A signed “hybrid” network is constructed by setting adjacency equal to the 5th power of the robust correlation when the correlation is positive, and zero otherwise. Thus, the adjacency takes on values between 0 and 1 and is 0 whenever the corresponding correlation is negative. The power 5 was chosen using the Scale-Free Topology Criterion (Zhang and Horvath, 2005a). We chose the signed network because it keeps tracks of the sign of the correlation and results in modules comprised of positively correlated genes. Recent studies have shown that signed co-expression networks can yield modules with more coherent functional annotation and lead to better identification of small modules(Mason et al., 2009). Topological overlap (TO), a more biologically meaningful measure of node interconnectedness (similarity), was then calculated as described previously (Zhang and Horvath, 2005b, Oldham et al., 2006, Ravasz et al., 2002, Yip and Horvath, 2007). Next, genes were clustered using average-linkage hierarchical clustering with pairwise dissimilarity equal 1 – TO. Modules correspond to branches of the resulting hierarchical clustering tree and were identified using the Dynamic Tree Cut algorithm (Langfelder et al., 2008). This procedure results in modules of highly correlated genes for which a summary expression profile can be defined. We use the module eigengene (ME), defined as the first right-singular vector of the standardized expression matrix (Zhang and Horvath, 2005a). The ME is the mathematically optimal summary of the module expression profiles since it captures the maximum amount of variation (Langfelder and Horvath, 2007, Horvath and Dong, 2008). The MEs were then correlated to the clinical and virologic variables in order to identify modules related to the clinical and virologic variables. To further enhance data interpretation, we used functional enrichment in GO terms and the function userListEnrichment from the WGCNA package to find enrichment for cell type markers and other brain-related categories(Miller et al., 2011).
All data, analysis code, and additional details can be found at our webpage: http://labs.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/HIVMonocyteExpression/.
RESULTS
Standard differential gene expression analysis of clinical variables
The relationship between individual gene expression and the clinical and virologic variables was examined via correlation analysis. Table 3 lists associations that remained statistically significant after correcting for multiple comparisons using False Discovery Rate(Benjamini et al., 2001). We used a q-value cutoff of 0.1, as suggested by Benjamini and Yekutieli (Benjamini and Yekutieli, 2005). Only Global Neurocognitive Functioning showed significant associations with gene transcripts. The full list of associations between genes and variables is provided in Supplemental File A.
Table 3.
Significant associations between gene expression and Global Neurocognitive Functioning
| Gene Name (HGNC Approved Gene Symbol) | R | q-value |
|---|---|---|
| Chromosome 3 open reading frame 17 (C3orf17) | −.472 | 0.03 |
| Cytoplasmic FMRP-Interacting Protein-2 (CYFIP2) | .475 | 0.03 |
| Testis-Derived Transcript (TES) | .478 | 0.03 |
| Prostate androgen-regulated mucin-like protein (PARM1) | .483 | 0.03 |
| Inositol Hexaphosphate Kinase 1 (IHPK1) | −.474 | 0.03 |
| GPI-Anchored Membrane Protein-1 (CAPRIN1) | −.468 | 0.03 |
| Hypoxia Up-regulated-1 (HYOU1) | .451 | 0.05 |
| Interleukin 6-Receptor (IL6R) | −.435 | 0.07 |
| GPN-loop GTPase-2 (GPN2) | −.437 | 0.07 |
| Pumilio, Drosophila, Homolog Of, 1 (Pum1) | −.436 | 0.07 |
| Casein Kinase I, Alpha-1 (CSNK1A1) | −.437 | 0.07 |
| Vibrio Polysaccharide (VPS) | .431 | 0.07 |
| Low density lipoprotein receptor-related protein 12 (LRP12) | −.425 | 0.08 |
| Mannose-1-phosphate guanyltransferase beta (GMPPB) | 425 | 0.08 |
| Nodal Modulator-1 (NOMO1) | .426 | 0.08 |
| Kelch-Like ECH-Associated Protein-1 (KEAP1) | −.419 | 0.09 |
Detailed enrichment analysis of top genes related to clinical variables
To further understand monocyte gene expression pathways related to HAND and the other variables, we examined enriched GO categories based on the top gene associations identified in the differential expression analysis. After applying corrections for multiple comparisons, no gene ontology categories were found to be significantly associated with the variables.
Weighted Gene Co-Expression Network Analysis
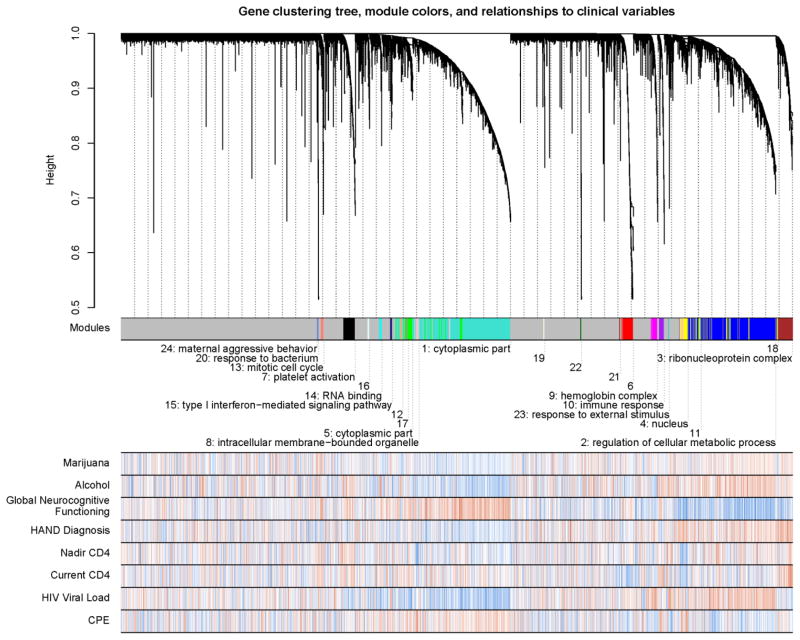
We next applied WGCNA. This approach differs from the standard differential expression analysis described above because it identifies biological meaningful networks before examining their association with the variables of interest. As such, WGCNA makes significantly fewer comparisons, thereby reducing the chance of Type I error. The network analysis (see Methods) identified twenty-four co-expression modules. The gene clustering dendogram, modules, and a heatmap of gene-variable associations are presented in Figure 1. Multiple modules are highly enriched in existing GO terms (Table 4), which suggests their functional interpretation.
Figure 1.
The gene clustering dendograms (top) shows that co-expressed genes tend to cluster into modules corresponding to major biological divisions (cell type, molecular function, etc), as well as disease-relevant changes. The dendrogram results from hierarchical clustering and groups genes into distinct modules. The y-axis corresponds to dissimilarity determined by the extent of topological overlap (1-TO). Top color band (Module label): dynamic tree cutting was used to identify highly parsimonious module definitions, generally dividing modules at significant branch points in the dendrogram. Labels below the module band indicate numeric module labels (1–24) and highest enriched GO terms that suggest the functional interpretation of the modules (modules without a GO term were not significantly enriched in any GO term). The remaining rows indicate the strength of associations between the clinical variables (listed on the left) and individual probes. Red indicates a positive correlation, blue indicates negative correlation.
Table 4.
Enrichment of gene modules in selected GO terms(Ashburner et al., 2000). For each module, the table lists its size (number of probe sets), GO term, enrichment p-value, Bonferroni-corrected enrichment p-value, and number of genes from the module that are included in the listed GO term. For each module we only list the top few terms. Modules not listed in this table were not significantly enriched in any GO terms (after Bonferroni correction)
| Module | Size | Term (Ontology) | p | Bonferroni | Genes in term |
|---|---|---|---|---|---|
| 1 | 4812 | cytoplasmic part (CC) | 6.90E-43 | 8.50E-39 | 1450 |
| 1 | 4812 | mitochondrion (CC) | 2.90E-40 | 3.60E-36 | 464 |
| 1 | 4812 | catalytic activity (MF) | 7.40E-32 | 9.20E-28 | 1189 |
| 2 | 4018 | regulation of cellular metabolic process (BP) | 3.10E-29 | 3.80E-25 | 735 |
| 2 | 4018 | nucleus (CC) | 6.90E-27 | 8.50E-23 | 945 |
| 3 | 806 | ribonucleoprotein complex (CC) | 4.90E-15 | 6.20E-11 | 37 |
| 3 | 806 | mRNA metabolic process (BP) | 5.60E-15 | 7.00E-11 | 39 |
| 4 | 788 | nucleus (CC) | 1.70E-20 | 2.20E-16 | 258 |
| 4 | 788 | RNA metabolic process (BP) | 9.30E-15 | 1.20E-10 | 174 |
| 4 | 788 | membrane-bounded organelle (CC) | 2.80E-14 | 3.50E-10 | 334 |
| 5 | 770 | cytoplasmic part (CC) | 2.60E-09 | 3.30E-05 | 196 |
| 6 | 640 | zinc ion binding (MF) | 8.00E-05 | 0.99 | 44 |
| 7 | 631 | platelet activation (BP) | 2.00E-18 | 2.50E-14 | 32 |
| 7 | 631 | blood coagulation (BP) | 8.90E-17 | 1.10E-12 | 46 |
| 8 | 624 | intracellular membrane-bounded organelle (CC) | 2.30E-13 | 2.80E-09 | 281 |
| 8 | 624 | nucleus (CC) | 6.00E-11 | 7.40E-07 | 195 |
| 9 | 349 | hemoglobin complex (CC) | 1.70E-15 | 2.10E-11 | 9 |
| 10 | 271 | immune response (BP) | 1.40E-19 | 1.80E-15 | 44 |
| 11 | 250 | translational elongation (BP) | 2.50E-05 | 0.31 | 6 |
| 11 | 250 | mRNA metabolic process (BP) | 1.00E-04 | 1 | 12 |
| 12 | 159 | mitochondrial respiratory chain (CC) | 1.00E-05 | 0.12 | 5 |
| 12 | 159 | respiratory chain (CC) | 1.50E-05 | 0.19 | 5 |
| 13 | 150 | mitotic cell cycle (BP) | 7.90E-17 | 9.80E-13 | 28 |
| 14 | 119 | RNA binding (MF) | 8.40E-07 | 0.011 | 14 |
| 14 | 119 | ribonucleoprotein complex (CC) | 2.30E-05 | 0.29 | 10 |
| 15 | 102 | type I interferon-mediated signaling pathway (BP) | 2.50E-27 | 3.10E-23 | 17 |
| 15 | 102 | innate immune response (BP) | 4.40E-27 | 5.40E-23 | 29 |
| 15 | 102 | cytokine-mediated signaling pathway (BP) | 1.20E-23 | 1.50E-19 | 23 |
| 16 | 87 | interleukin-5-mediated signaling pathway (BP) | 8.60E-06 | 0.11 | 2 |
| 20 | 50 | response to bacterium (BP) | 6.40E-19 | 7.90E-15 | 15 |
| 20 | 50 | extracellular region (CC) | 2.00E-13 | 2.40E-09 | 21 |
| 20 | 50 | defense response to fungus (BP) | 1.20E-12 | 1.50E-08 | 6 |
| 23 | 28 | response to external stimulus (BP) | 1.10E-07 | 0.0014 | 8 |
| 24 | 28 | maternal aggressive behavior (BP) | 4.00E-06 | 0.049 | 2 |
CC (Cellular Compartment), MF (Molecular Function) and BP (Biological Process) are the three ontologies used to classify GO terms, as described in (Ashburner et al., 2000).
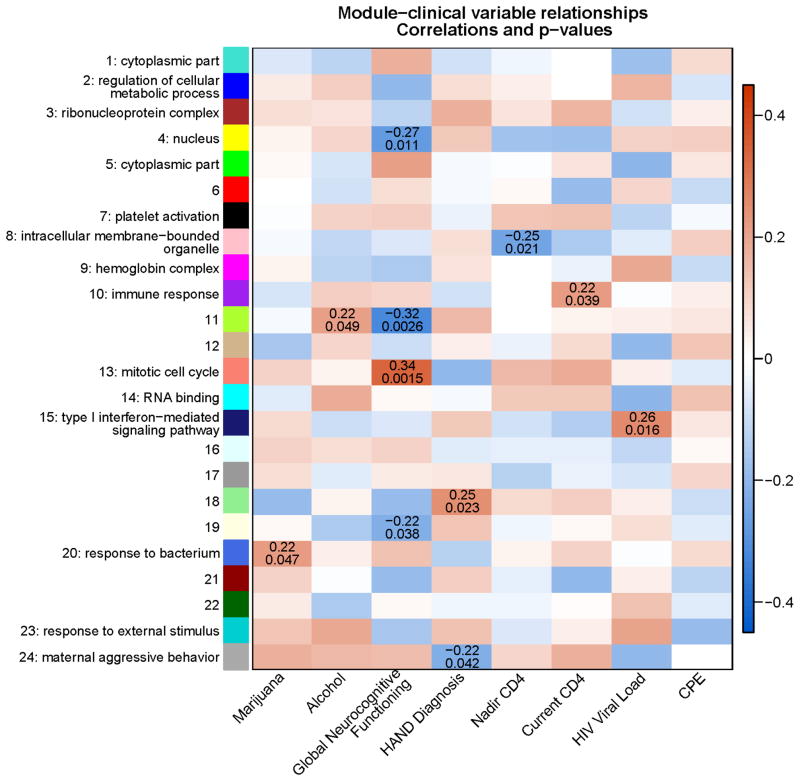
Correlations between Module Eigengenes and Clinical Variables
Clinical and virologic variables were correlated to module eigengenes. Figure 2 shows in grid format the correlations and adjusted p-values for the module/variable associations. Gene ontology analysis was conducted for each module, reflected in their respective labels. Thirteen correlations reached the nominal .05 threshold; however, after applying FDR correction only two associations reached a suggestive 1q-value of <.20. Module 13 (Mitotic Cell Cycle) was positively correlated with Global Neurocognitive Functioning (R=.34; p=.001, q=.16). This module largely consisted of GO categories involved in mitosis. Module 11 was negatively correlated with Global Neurocognitive Functioning (R= −.32; p=.003, q=.16).
Figure 2.
Tombstone chart showing correlations and p-values between modules and clinical variables. After correcting for multiple testing using FDR, module 13 (Mitotic Cell Cycle) was positively correlated with Global Neurocognitive Functioning and module 11 (No clear gene ontology) was negatively correlated with Global Neurocognitive Functioning at the suggestive level of q=0.14. Other associations with nominal p-values of less than 0.05 are shown, although these did not survive FDR correction.
Association of HAND and Global Neurocognitive Functioning with Transcripts Implicated in Previous Studies
As a supplemental analysis, we examined expression levels of genes implicated in HAND in previous studies (Table 5). None of the genes was found to be significantly correlated with HAND or Global Neurocognitive Functioning.
Table 5.
Associations of previously implicated mRNA transcripts with Global Neurocognitive Functioning and HAND
| Global Neurocognitive Functioning | HAND Diagnosis | |||
|---|---|---|---|---|
|
| ||||
| Gene Product (HGNC Approved Gene Symbol) | R | q-value* | R | q-value* |
| Osteopontin (SPP1)(Buckner et al., 2011) | .051 | .68 | .011 | .997 |
| Monocyte Chemotactic Protein 1 (CCL2)(Pulliam et al., 2004) | −.036 | .71 | −.019 | .997 |
| Chemokine-CC Motif Receptor 5 (CCR5)(Pulliam et al., 2004) | −.078 | .63 | −.033 | .997 |
| Enolase-2 (ENO2)(Buckner et al., 2011) | −.143 | .49 | .132 | .997 |
| Ubiquitin-like Modifier ISG15 (ISG15)(Roberts et al., 2003) | .054 | .62 | .066 | .997 |
| Interferon-Gamma-Inducible Protein-10 (CXCL10)(Pulliam et al., 2011) | −.119 | .54 | .198 | .997 |
| Interleukin 6 (IL6)(Yang et al., 2009) | −.063 | .66 | .046 | .997 |
| Sialoadhesin (SIGLEC1)(Pulliam et al., 2004) | −.01 | .92 | .182 | .997 |
Power considerations
While our sample size (n=84) is unprecedented in regards to the study of monocyte expression in relation to HAND, it only allows us to detect moderate to large correlations as can be seen by the following power calculations. At an uncorrected significance threshold of 0.05, a sample size of n=80 achieves 78% power or 96% power if the true underlying correlation is 0.30 or 0.40, respectively. We caution the reader that the uncorrected significance threshold of alpha=0.05 is not appropriate in the context of high dimensional gene expression data. If we had found only 10 co-expression modules, then one could justify a Bonferroni correction that adjusts for 10 comparisons resulting in a significance threshold of alpha=0.005. At this significance level, n=80 subjects would lead to a power of 82% to detect an absolute correlation of 0.40 or higher. Given that we found 24 coexpression modules, these considerations indicate that our study was not sufficiently powered to detect correlations lower than 0.40 between modules and a quantitative clinical outcome.
DISCUSSION
HAND pathogenesis in the current era of widespread cART use and relative virologic control is likely due in part to chronic immunologic over-reactivity and neuroinflammation fueled by an influx of monocytes into the CNS. Delineating the molecular changes that occur in monocytes in relation to neurobehavioral dysfunction may lead to the identification of clinically relevant biomarkers, and further illuminate the biological pathways involved in HAND. While an altered monocyte phenotype was previously identified as being associated with HAD, studies of monocyte transcription changes associated with more mild HIV-related neurocognitive impairment has thus far yielded negative results(Sun et al., 2010). In the current study, we utilized a fairly large sample, a monocyte extraction method with high purity, and a statistical approach allowing for a systems-wide understanding of transcription changes while minimizing the risk of type-I error inherent in gene expression studies.
A standard differential expression analysis, in which we correlated mRNA levels with the clinical variables, identified Global Neurocognitive Functioning as the sole variable with significant associations following correction for multiple comparisons. Interestingly, many of the genes associated with Global Neurocognitive Functioning have either previously been implicated in NeuroAIDS or other neurodegenerative disease, or are involved in pathways relevant to HAND neuropathogenesis. The unexpected finding is that most of those previous studies used brain tissue/cells. The current study found that interleukin-6 receptor (IL-6R) expression was negatively correlated with Global Neurocognitive Functioning. That is, better neurocognitive functioning was associated with lower IL-6R mRNA levels. Increased levels of IL-6R have long been implicated in central nervous system pathology in general, including multiple sclerosis (Padberg et al., 1999) and head trauma (Hans et al., 1999), consistent with our findings. However, it is this receptor’s ligand, IL-6, that has primarily been associated with NeuroAIDS. Yoshioka et al (1994) reported increased expression of IL-6 in endothelial cells in lumbosacral sensory dorsal root ganglia from patients who died with AIDS vs. HIV- controls, leading to the conclusion that IL-6 expression was associated with neuronal degeneration(Yoshioka et al., 1994). More recently, plasma levels of IL-6 mRNA were found to be weakly correlated with memory and learning ability in HIV+ adults(Pedersen et al., 2013). Others have reported associations between depression and IL-6 mRNA in HIV+ cases, but not neurocognitive functioning, suggesting that depression may be a moderating variable(Warriner et al., 2010). However, in our analysis, IL-6 and IL-6R mRNA had very weak correlations with depression, memory, and learning. Further, Pulliam et al found that PBMC-derived soluble IL-6 levels were significantly higher in a very small group of patients with HIV-associated dementia(Pulliam et al., 1996). As such, it is plausible that the decreased expression of IL-6 receptors associated with better neurocognitive functioning reflects decreased IL-6 activity in these monocytes. It should be noted that an early study implicated a possibly neuroprotective and regenerative role for IL-6 that may explain the variable findings over the years for this cytokine(Yoshioka et al., 1995). This is supported by a recent study showing that IL-6 protected against NMDA-induced neurotoxicity when a functioning IL-6 receptor was present(Fang et al., 2012).
Additional genes found to be associated with Global Neurocognitive Functioning warrant attention. Casein kinase 1-alpha-1 (CSNK1A1) is one of a family of casein kinases previously implicated in neurodegenerative disease(Perez et al., 2011). Expression of this gene was negatively correlated with Global Neurocognitive Functioning. Hypoxia up-regulated-1 (HYOU1) has been shown in numerous studies to play a protective role in various cells types, including astrocytes, neurons, and macrophages(Janeczko, 1991, Kuwabara et al., 1996, Ozawa et al., 2001, Tamatani et al., 2001, Zhao et al., 2010). Consistent with this, expression of this gene was positively correlated with Global Neurocognitive Functioning. Low density lipoprotein receptor-related protein-12 (LRP12), an apolipoprotein-E receptor that mediates the endocytosis and degradation of amyloid precursor protein(Kounnas et al., 1995), was negatively correlated with Global Neurocognitive Functioning. Activation of this receptor by Tat has been shown to mediate HIV-induced neuropathology(Liu et al., 2000). This gene has also been linked to an increased risk of Alzheimer’s disease(Kang et al., 1997, Bian et al., 2005). Kelch-like ECH-Associated protein-1 (KEAP1) was negatively correlated with Global Neurocognitive Functioning. The transcription factor Nrf2, which enhances cytoprotective gene expression following pathological insults, is sequestered by KEAP1 in cytoplasm, where it is subsequently degraded by proteosomes(Itoh et al., 1999). KEAP1 is inactivated under oxidative stress, leading to Nrf2 activation of antioxidant and detoxifying genes(Motohashi and Yamamoto, 2004). Consistent with this, KEAP1 antibodies have been immunostained in the lesions of numerous neurodegenerative diseases, including Dementia with Lewy bodies, Parkinson’s disease, Alzheimer’s disease, and amyotrophic lateral sclerosis(Tanji et al., 2013). As such, some have begun examining molecular agents that inhibit the KEAP1/Nrf2 interaction as a treatment for neurodegenerative diseases(Zhao et al., 2011); our findings provide initial support for the inclusion of HAND.
The module-centric analysis afforded by WGCNA allowed us to greatly reduce the number of statistical tests: only 216 hypothesis tests resulted from relating 24 modules to 9 clinical variables. Two of these 216 tests resulted in statistically significant findings following FDR correction. GO analysis of the significant modules indicates that mitotic cell cycle was positively correlated with Global Neurocognitive Functioning, whereas translational elongation was negatively correlated. Interestingly, mitotic cell cycle was negatively correlated with pre-mortem neurocognitive functioning in our examination of frontal grey matter from deceased cases in the National NeuroAIDS Tissue Consortium(Morgello et al., 2001, Levine et al., 2013). Those cases died with advanced illness, some including HIV-encephalitis. With regards to translational elongation, this GO category shares genes with viral transcription and viral infectious cycle GO categories. One possible explanation is that greater transcription activity and viral replication within monocytes is associated with neurocognitive impairment.
In relation to previous studies of monocyte gene expression in HIV+ individuals, our findings provided little if any validation. One possible reason for this is that these past studies used monocytes of a specific CD-receptor phenotype. Pulliam et al (2004) isolated CD14+ monocytes from 23 HIV+ individuals with high or low viral loads, as well as HIV-seronegative controls, and used DNA microarrays to characterize differentially regulated genes in the cells(Pulliam et al., 2004). Monocytes from individuals with high viral loads (>10,000 copies) increased expression of CD16, CCR5, MCP-1, and sialoadhesin, suggesting an inflammatory phenotype. However, they noted that the monocytes are not activated in the “classical sense”; that is, they did not produce a number of proinflammatory cytokines common in the pre-cART era (e.g., IL-6, TNF-alpha), and they showed a transcription profile consistent with chemotactic properties. Specifically, among individuals with high viral loads, they found evidence of increased propensity for chemotaxis characterized by an increase in CCR5 and MCP-1, leading the authors to conclude that the circulating monocyte has evolved in the cART era to a chronic inflammatory state with additional chemotactic features, essentially a mature monocyte/macrophage hybrid. In our analysis, we did not observe any associations between gene expression and viral load; however, the range of viral loads within our sample was more restricted than that of Pulliam et al (2004)(Pulliam et al., 2004), and, as already mentioned, we did not focus our analysis on CD14+ monocytes. In a second study, Buckner et al (2011) examined dynamic transcription changes in monocytes using an in vitro culture model(Buckner et al., 2011). Focusing on a specific monocyte subpopulation (CD14+CD16+CD11b+Mac387+, derived by infecting healthy donor cells with HIV), these investigators found these monocytes to be adept at crossing a laboratory model of the blood brain barrier. Gene expression analysis revealed upregulation of chemotactic and metastasis-related genes, as opposed to inflammatory genes. In addition, Buckner et al. described dynamic changes as the monocytes matured into macrophages, including an increase in the expression of enolase 2, followed by a decrease once the cell was fully differentiated. Osteopontin was also observed to have increased expression in the maturing monocytes. This important in vitro study demonstrated that the dynamic changes in monocyte transcription may provide clues concerning biological processes necessary for neuropathogenesis. While our in vivo analysis did not identify any of the genes observed by Buckner et al., we did not isolate this particular subpopulation of monocytes.
Finally, in an attempt to link global monocyte transcription changes to HAND, Sun et al examined whether or not CD14+ monocyte gene expression and other peripheral factors (CD4, ApoE genotype, viral load, lipopolysaccharide and soluble CD14) were associated with neurocognitive impairment in a group of 44 HIV+ individuals on cART, as well as 11 HIV–seronegative controls(Sun et al., 2010). Monocyte gene expression was not correlated with neurocognitive impairment. More recently, the same group focused their analysis on neurophysiological measures rather than HAND(Pulliam et al., 2011). Specifically, they examined whether peripheral immune activation was associated with brain metabolite concentrations, as measured by magnetic resonance spectroscopy. Thirty-five HIV+ persons on cART and 8 HIV-seronegative adults were examined. Approximately half of the HIV+ sample was considered to have mild neurocognitive impairment. Absolute concentrations of brain metabolites in the frontal white matter, anterior cingulate cortex, and basal ganglia were determined and then examined in relation to the monocyte transcriptome and a global neurocognitive measure. Among the HIV+ participants, an interferon-alpha induced activation transcriptome phenotype was strongly correlated with N-acetyl-aspartate in the frontal white matter. Notably, Interferon-gamma inducible protein-10 (IP-10) was strongly correlated with plasma protein levels, and plasma IP-10 was inversely correlated with N-acetylaspartate in the anterior cingulated cortex. This study is particularly notable as it is the first to connect transcription changes with putative HIV-related neurophysiological changes. This tactic of focusing on more intermediate phenotypes, which may be more reliably measured as compared to the clinical syndrome of HAND, may hold the greatest promise for elucidating the neuropathogenesis of NeuroAIDS. While our findings did not indicate an association between IP-10 and HAND, we again point out that we did not isolate monocytes of a specific phenotype.
A number of methodological factors limit the extent to which our findings may be generalized. Firstly, the sample was largely homogeneous concerning disease severity. As such, the variation in neurocognitive functioning was small. We are currently recruiting participants from the National NeuroAIDS Tissue Consortium, a cohort with considerable variation with regards to HAND spectrum diagnoses and neurocognitive functioning(Morgello et al., 2001). Secondly, while ours is the largest sample thus far in HAND-related transcriptome studies, it is still relatively underpowered to detect true differences in gene expression (as can be seen from our power studies). Finally, we did not employ an HIV-seronegative comparison group since our main interest was to study HAND development among HIV+ individuals. However, since Global Neurocognitive Functioning was the sole variable with significant findings, the significant associations described here will need to be re-examined in a seronegative sample to ensure that any associations are indeed HIV-related, as opposed to being a general biological association regardless of infection.
In summary, our analysis of global monocyte gene expression revealed interesting associations of a number of highly relevant genes with neurocognitive functioning, but did not reveal significant associations with the diagnosis of HAND. Our finding of elevated levels of IL6R, CSNK1A1, LRP12, and KEAP1 mRNA in association with greater neurocognitive impairment is consistent with past studies and may provide justification for exploring these proteins and their related pathways further; however, the associations were not adequately robust to justify their use as biomarkers of HAND. A modest association between two co-expression modules and neurocognitive functioning may also be worthy of further exploration.
Highlights.
This study used standard differential gene expression analysis and WGCNA
Expression of several genes was genes associated with neurocognitive functioning
Two biological processes were associated with neurocognitive functioning
No associations were found with HAND diagnosis, CD4, viral load, or substance use
Acknowledgments
Our deepest gratitude goes to the participants and staff of the Multicenter AIDS Cohort Study in Los Angeles. This study was primarily funded by the National Institute for Drug Abuse grant R01DA030913 (Levine & Horvath). The Los Angeles site of the Multicenter AIDS Cohort Study is funded the National Institute of Allergy and Infectious Disease grant U01-AI-35040 (Detels). The research described was also supported by NIH/National Center for Advancing Translational Science (NCATS) UCLA CTSI grant number UL1TR000124. We would also like to acknowledge the technical assistance of Jesse Hogan and the UCLA Virology Core Laboratory which is part of the NIH-funded UCLA Center for AIDS Research grant AI-28697 (Zack).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew J. Levine, Email: ajlevine@mednet.ucla.edu, Department of Neurology, National Neurological AIDS Bank, David Geffen School of Medicine at the University of California, Los Angeles.
Steve Horvath, Email: shorvath@mednet.ucla.edu, Department of Human Genetics, David Geffen School of Medicine at the University of California, Los Angeles and Department of Biostatistics, University of California, Los Angeles
Eric N. Miller, Email: emiller@ucla.edu, Department of Psychiatry and Biobehavioral Science, David Geffen School of Medicine at the University of California, Los Angeles
Elyse J. Singer, Email: esinger@ucla.edu, Department of Neurology, National Neurological AIDS Bank, David Geffen School of Medicine at the University of California, Los Angeles
Paul Shapshak, Email: pshapshak@gmail.com, Department of Medicine (Division of Infectious Disease & International Medicine) and, Department of Psychiatry & Behavioral Medicine, University of South Florida, Morsani College of Medicine, Tampa
Gayle C. Baldwin, Email: gbaldwin@ucla.edu, Department of Medicine, David Geffen School of Medicine at the University of California, Los Angeles
Otoniel Martínez-Maza, Email: omartinez@mednet.ucla.edu, Departments of Obstetrics & Gynecology and Department Microbiology, Immunology & Molecular Genetics, David Geffen School of Medicine at the University of California, Los Angeles, Department of Epidemiology, UCLA Fielding School of Public Health
Mallory D. Witt, Email: mwitt@labiomed.org, David Geffen School of Medicine at the University of California, Los Angeles, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center
Peter Langfelder, Email: Peter.langfelder@gmail.com, Department of Human Genetics, David Geffen School of Medicine at the University of California, Los Angeles
References
- ADLE-BIASSETTE H, CHRETIEN F, WINGERTSMANN L, HERY C, EREAU T, SCARAVILLI F, TARDIEU M, GRAY F. Neuronal apoptosis does not correlate with dementia in HIV infection but is related to microglial activation and axonal damage. Neuropathol Appl Neurobiol. 1999;25:123–33. doi: 10.1046/j.1365-2990.1999.00167.x. [DOI] [PubMed] [Google Scholar]
- ANCUTA P, MOSES A, GABUZDA D. Transendothelial migration of CD16+ monocytes in response to fractalkine under constitutive and inflammatory conditions. Immunobiology. 2004;209:11–20. doi: 10.1016/j.imbio.2004.04.001. [DOI] [PubMed] [Google Scholar]
- ANTINORI A, ARENDT G, BECKER JT, BREW BJ, BYRD DA, CHERNER M, CLIFFORD DB, CINQUE P, EPSTEIN LG, GOODKIN K, GISSLEN M, GRANT I, HEATON RK, JOSEPH J, MARDER K, MARRA CM, MCARTHUR JC, NUNN M, PRICE RW, PULLIAM L, ROBERTSON KR, SACKTOR N, VALCOUR V, WOJNA VE. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69:1789–99. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASHBURNER M, BALL CA, BLAKE JA, BOTSTEIN D, BUTLER H, CHERRY JM, DAVIS AP, DOLINSKI K, DWIGHT SS, EPPIG JT, HARRIS MA, HILL DP, ISSEL-TARVER L, KASARSKIS A, LEWIS S, MATESE JC, RICHARDSON JE, RINGWALD M, RUBIN GM, SHERLOCK G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25n9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BENJAMINI Y, DRAI D, ELMER G, KAFKAFI N, GOLANI I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- BENJAMINI Y, YEKUTIELI D. Quantitative trait Loci analysis using the false discovery rate. Genetics. 2005;171:783–90. doi: 10.1534/genetics.104.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIAN L, YANG JD, GUO TW, DUAN Y, QIN W, SUN Y, FENG GY, HE L. Association study of the A2M and LRP1 Genes with Alzheimer disease in the Han Chinese. Biol Psychiatry. 2005;58:731–7. doi: 10.1016/j.biopsych.2005.05.013. [DOI] [PubMed] [Google Scholar]
- BOLSTAD BM, IRIZARRY RA, ASTRAND M, SPEED TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- BORJABAD A, BROOKS AI, VOLSKY DJ. Gene expression profiles of HIV-1-infected glia and brain: toward better understanding of the role of astrocytes in HIV-1-associated neurocognitive disorders. J Neuroimmune Pharmacol. 5:44–62. doi: 10.1007/s11481-009-9167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BREW BJ. Evidence for a change in AIDS dementia complex in the era of highly active antiretroviral therapy and the possibility of new forms of AIDS dementia complex. Aids. 2004;18(Suppl 1):S75–8. [PubMed] [Google Scholar]
- BUCKNER CM, CALDERON TM, WILLAMS DW, BELBIN TJ, BERMAN JW. Characterization of monocyte maturation/differentiation that facilitates their transmigration across the blood-brain barrier and infection by HIV: implications for NeuroAIDS. Cell Immunol. 2011;267:109–23. doi: 10.1016/j.cellimm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARLSON KA, CIBOROWSKI P, SCHELLPEPER CN, BISKUP TM, SHEN RF, LUO X, DESTACHE CJ, GENDELMAN HE. Proteomic fingerprinting of HIV-1-infected human monocyte-derived macrophages: a preliminary report. J Neuroimmunol. 2004;147:35–42. doi: 10.1016/j.jneuroim.2003.10.039. [DOI] [PubMed] [Google Scholar]
- COSENZA MA, ZHAO ML, SI Q, LEE SC. Human brain parenchymal microglia express CD14 and CD45 and are productively infected by HIV-1 in HIV-1 encephalitis. Brain Pathol. 2002;12:442–55. doi: 10.1111/j.1750-3639.2002.tb00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CYSIQUE LA, BREW BJ, HALMAN M, CATALAN J, SACKTOR N, PRICE RW, BROWN S, ATKINSON JH, CLIFFORD DB, SIMPSON D, TORRES G, HALL C, POWER C, MARDER K, MCARTHUR JC, SYMONDS W, ROMERO C. Undetectable cerebrospinal fluid HIV RNA and beta-2 microglobulin do not indicate inactive AIDS dementia complex in highly active antiretroviral therapy-treated patients. J Acquir Immune Defic Syndr. 2005;39:426–9. doi: 10.1097/01.qai.0000165799.59322.f5. [DOI] [PubMed] [Google Scholar]
- CYSIQUE LA, MARUFF P, BREW BJ. Prevalence and pattern of neuropsychological impairment in human immunodeficiency virus-infected/acquired immunodeficiency syndrome (HIV/AIDS) patients across pre- and post-highly active antiretroviral therapy eras: a combined study of two cohorts. J Neurovirol. 2004;10:350–7. doi: 10.1080/13550280490521078. [DOI] [PubMed] [Google Scholar]
- CYSIQUE LA, VAIDA F, LETENDRE S, GIBSON S, CHERNER M, WOODS SP, MCCUTCHAN JA, HEATON RK, ELLIS RJ. Dynamics of cognitive change in impaired HIV-positive patients initiating antiretroviral therapy. Neurology. 2009;73:342–8. doi: 10.1212/WNL.0b013e3181ab2b3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DORE GJ, MCDONALD A, LI Y, KALDOR JM, BREW BJ. Marked improvement in survival following AIDS dementia complex in the era of highly active antiretroviral therapy. Aids. 2003;17:1539–45. doi: 10.1097/00002030-200307040-00015. [DOI] [PubMed] [Google Scholar]
- ELLERY PJ, TIPPETT E, CHIU YL, PAUKOVICS G, CAMERON PU, SOLOMON A, LEWIN SR, GORRY PR, JAWOROWSKI A, GREENE WC, SONZA S, CROWE SM. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol. 2007;178:6581–9. doi: 10.4049/jimmunol.178.10.6581. [DOI] [PubMed] [Google Scholar]
- FANG XX, JIANG XL, HAN XH, PENG YP, QIU YH. Neuroprotection of Interleukin-6 Against NMDA-induced Neurotoxicity is Mediated by JAK/STAT3, MAPK/ERK, and PI3K/AKT Signaling Pathways. Cell Mol Neurobiol. 2012 doi: 10.1007/s10571-012-9891-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GELMAN BB, CHEN T, LISINICCHIA JG, SOUKUP VM, CARMICAL JR, STARKEY JM, MASLIAH E, COMMINS DL, BRANDT D, GRANT I, SINGER EJ, LEVINE AJ, MILLER J, WINKLER JM, FOX HS, LUXON BA. The National NeuroAIDS Tissue Consortium Brain Gene Array: Two Types of HIV-Associated Neurocognitive Impairment. PLoS One. 2012;7:e46178. doi: 10.1371/journal.pone.0046178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GENDELMAN HE, PERSIDSKY Y, GHORPADE A, LIMOGES J, STINS M, FIALA M, MORRISETT R. The neuropathogenesis of the AIDS dementia complex. Aids. 1997;11(Suppl A):S35–45. [PubMed] [Google Scholar]
- GHAZALPOUR A, DOSS S, ZHANG B, WANG S, PLAISIER C, CASTELLANOS R, BROZELL A, SCHADT EE, DRAKE TA, LUSIS AJ, HORVATH S. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GLASS JD, FEDOR H, WESSELINGH SL, MCARTHUR JC. Immunocytochemical quantitation of human immunodeficiency virus in the brain: correlations with dementia. Ann Neurol. 1995;38:755–62. doi: 10.1002/ana.410380510. [DOI] [PubMed] [Google Scholar]
- HANS VH, KOSSMANN T, JOLLER H, OTTO V, MORGANTI-KOSSMANN MC. Interleukin-6 and its soluble receptor in serum and cerebrospinal fluid after cerebral trauma. Neuroreport. 1999;10:409–12. doi: 10.1097/00001756-199902050-00036. [DOI] [PubMed] [Google Scholar]
- HORVATH S, DONG J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORVATH S, ZHANG B, CARLSON M, LU KV, ZHU S, FELCIANO RM, LAURANCE MF, ZHAO W, QI S, CHEN Z, LEE Y, SCHECK AC, LIAU LM, WU H, GESCHWIND DH, FEBBO PG, KORNBLUM HI, CLOUGHESY TF, NELSON SF, MISCHEL PS. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103:17402–7. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ITOH K, WAKABAYASHI N, KATOH Y, ISHII T, IGARASHI K, ENGEL JD, YAMAMOTO M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANECZKO K. The proliferative response of S-100 protein-positive glial cells to injury in the neonatal rat brain. Brain Res. 1991;564:86–90. doi: 10.1016/0006-8993(91)91355-5. [DOI] [PubMed] [Google Scholar]
- JOHNSON WE, RABINOVIC A, LI C. Adjusting batch effects in microarray expression data using Empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- KANG DE, SAITOH T, CHEN X, XIA Y, MASLIAH E, HANSEN LA, THOMAS RG, THAL LJ, KATZMAN R. Genetic association of the low-density lipoprotein receptor-related protein gene (LRP), an apolipoprotein E receptor, with late-onset Alzheimer’s disease. Neurology. 1997;49:56–61. doi: 10.1212/wnl.49.1.56. [DOI] [PubMed] [Google Scholar]
- KAUL M, LIPTON SA. Mechanisms of neuronal injury and death in HIV-1 associated dementia. Curr HIV Res. 2006;4:307–18. doi: 10.2174/157016206777709384. [DOI] [PubMed] [Google Scholar]
- KEDZIERSKA K, CROWE SM. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr Med Chem. 2002;9:1893–903. doi: 10.2174/0929867023368935. [DOI] [PubMed] [Google Scholar]
- KIM SY, LI J, BENTSMAN G, BROOKS AI, VOLSKY DJ. Microarray analysis of changes in cellular gene expression induced by productive infection of primary human astrocytes: implications for HAD. J Neuroimmunol. 2004;157:17–26. doi: 10.1016/j.jneuroim.2004.08.032. [DOI] [PubMed] [Google Scholar]
- KOLSON DL, SABNEKAR P, BAYBIS M, CRINO PB. Gene expression in TUNEL-positive neurons in human immunodeficiency virus-infected brain. J Neurovirol. 2004;10(Suppl 1):102–7. doi: 10.1080/753312760. [DOI] [PubMed] [Google Scholar]
- KOUNNAS MZ, MOIR RD, REBECK GW, BUSH AI, ARGRAVES WS, TANZI RE, HYMAN BT, STRICKLAND DK. LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell. 1995;82:331–40. doi: 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- KRAFT-TERRY SD, BUCH SJ, FOX HS, GENDELMAN HE. A coat of many colors: neuroimmune crosstalk in human immunodeficiency virus infection. Neuron. 2009;64:133–45. doi: 10.1016/j.neuron.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUWABARA K, MATSUMOTO M, IKEDA J, HORI O, OGAWA S, MAEDA Y, KITAGAWA K, IMUTA N, KINOSHITA T, STERN DM, YANAGI H, KAMADA T. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J Biol Chem. 1996;271:5025–32. doi: 10.1074/jbc.271.9.5025. [DOI] [PubMed] [Google Scholar]
- LANGFELDER P, HORVATH S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANGFELDER P, ZHANG B, HORVATH S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–20. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- LANGFORD TD, LETENDRE SL, LARREA GJ, MASLIAH E. Changing patterns in the neuropathogenesis of HIV during the HAART era. Brain Pathol. 2003;13:195–210. doi: 10.1111/j.1750-3639.2003.tb00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAWTON MP, BRODY EM. Assessment of older people: self-maintaining and instrumental activities of daily living. Gerontologist. 1969;9:179–86. [PubMed] [Google Scholar]
- LETENDRE S, MARQUIE-BECK J, CAPPARELLI E, BEST B, CLIFFORD D, COLLIER AC, GELMAN BB, MCARTHUR JC, MCCUTCHAN JA, MORGELLO S, SIMPSON D, GRANT I, ELLIS RJ. Validation of the CNS Penetration-Effectiveness rank for quantifying antiretroviral penetration into the central nervous system. Arch Neurol. 2008;65:65–70. doi: 10.1001/archneurol.2007.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVINE AJ, MILLER JA, SHAPSHAK P, GELMAN B, SINGER EJ, HINKIN CH, COMMINS D, MORGELLO S, GRANT I, HORVATH S. Systems analysis of human brain gene expression: mechanisms for HIV-associated neurocognitive impairment and common pathways with Alzheimer’s disease. BMC Med Genomics. 2013;6:4. doi: 10.1186/1755-8794-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDL KA, AKAY C, WANG Y, WHITE MG, JORDAN-SCIUTTO KL. Expression of the endoplasmic reticulum stress response marker, BiP, in the central nervous system of HIV-positive individuals. Neuropathol Appl Neurobiol. 2007;33:658–69. doi: 10.1111/j.1365-2990.2007.00866.x. [DOI] [PubMed] [Google Scholar]
- LIU Y, JONES M, HINGTGEN CM, BU G, LARIBEE N, TANZI RE, MOIR RD, NATH A, HE JJ. Uptake of HIV-1 tat protein mediated by low-density lipoprotein receptor-related protein disrupts the neuronal metabolic balance of the receptor ligands. Nat Med. 2000;6:1380–7. doi: 10.1038/82199. [DOI] [PubMed] [Google Scholar]
- LUO X, CARLSON KA, WOJNA V, MAYO R, BISKUP TM, STONER J, ANDERSON J, GENDELMAN HE, MELENDEZ LM. Macrophage proteomic fingerprinting predicts HIV-1-associated cognitive impairment. Neurology. 2003;60:1931–7. doi: 10.1212/01.wnl.0000064396.54554.26. [DOI] [PubMed] [Google Scholar]
- MARRA CM, LOCKHART D, ZUNT JR, PERRIN M, COOMBS RW, COLLIER AC. Changes in CSF and plasma HIV-1 RNA and cognition after starting potent antiretroviral therapy. Neurology. 2003;60:1388–90. doi: 10.1212/01.wnl.0000058768.73358.1a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASLIAH E, ROBERTS ES, LANGFORD D, EVERALL I, CREWS L, ADAME A, ROCKENSTEIN E, FOX HS. Patterns of gene dysregulation in the frontal cortex of patients with HIV encephalitis. J Neuroimmunol. 2004;157:163–75. doi: 10.1016/j.jneuroim.2004.08.026. [DOI] [PubMed] [Google Scholar]
- MASON MJ, FAN G, PLATH K, ZHOU Q, HORVATH S. Signed weighted gene co-expression network analysis of transcriptional regulation in murine embryonic stem cells. BMC Genomics. 2009;10:327. doi: 10.1186/1471-2164-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCARTHUR JC, BREW BJ, NATH A. Neurological complications of HIV infection. Lancet Neurol. 2005;4:543–55. doi: 10.1016/S1474-4422(05)70165-4. [DOI] [PubMed] [Google Scholar]
- MILLER JA, CAI C, LANGFELDER P, GESCHWIND DH, KURIAN SM, SALOMON DR, HORVATH S. Strategies for aggregating gene expression data: The collapseRows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORGELLO S, GELMAN BB, KOZLOWSKI PB, VINTERS HV, MASLIAH E, CORNFORD M, CAVERT W, MARRA C, GRANT I, SINGER EJ. The National NeuroAIDS Tissue Consortium: a new paradigm in brain banking with an emphasis on infectious disease. Neuropathol Appl Neurobiol. 2001;27:326–35. doi: 10.1046/j.0305-1846.2001.00334.x. [DOI] [PubMed] [Google Scholar]
- MOTOHASHI H, YAMAMOTO M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–57. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- OLDHAM MC, HORVATH S, GESCHWIND DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103:17973–8. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLDHAM MC, LANGFELDER P, HORVATH S. Network methods for describing sample relationships in genomic datasets: application to Huntington’s disease. BMC Syst Biol. 2012;6:63. doi: 10.1186/1752-0509-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OTANI I, AKARI H, NAM KH, MORI K, SUZUKI E, SHIBATA H, DOI K, TERAO K, YOSIKAWA Y. Phenotypic changes in peripheral blood monocytes of cynomolgus monkeys acutely infected with simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1998;14:1181–6. doi: 10.1089/aid.1998.14.1181. [DOI] [PubMed] [Google Scholar]
- OZAWA K, KONDO T, HORI O, KITAO Y, STERN DM, EISENMENGER W, OGAWA S, OHSHIMA T. Expression of the oxygen-regulated protein ORP150 accelerates wound healing by modulating intracellular VEGF transport. J Clin Invest. 2001;108:41–50. doi: 10.1172/JCI11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PADBERG F, FENEBERG W, SCHMIDT S, SCHWARZ MJ, KORSCHENHAUSEN D, GREENBERG BD, NOLDE T, MULLER N, TRAPMANN H, KONIG N, MOLLER HJ, HAMPEL H. CSF and serum levels of soluble interleukin-6 receptors (sIL-6R and sgp130), but not of interleukin-6 are altered in multiple sclerosis. J Neuroimmunol. 1999;99:218–23. doi: 10.1016/s0165-5728(99)00120-4. [DOI] [PubMed] [Google Scholar]
- PEDERSEN KK, PEDERSEN M, GAARDBO JC, RONIT A, HARTLING HJ, BRUUNSGAARD H, GERSTOFT J, ULLUM H, NIELSEN SD. Persisting Inflammation and Chronic Immune Activation but Intact Cognitive Function in HIV-infected patients After Long Term Treatment with Combination Antiretroviral Therapy. J Acquir Immune Defic Syndr. 2013 doi: 10.1097/QAI.0b013e318289bced. [DOI] [PubMed] [Google Scholar]
- PELUSO R, HAASE A, STOWRING L, EDWARDS M, VENTURA P. A Trojan Horse mechanism for the spread of visna virus in monocytes. Virology. 1985;147:231–6. doi: 10.1016/0042-6822(85)90246-6. [DOI] [PubMed] [Google Scholar]
- PEREZ DI, GIL C, MARTINEZ A. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med Res Rev. 2011;31:924–54. doi: 10.1002/med.20207. [DOI] [PubMed] [Google Scholar]
- PERSIDSKY Y, GHORPADE A, RASMUSSEN J, LIMOGES J, LIU XJ, STINS M, FIALA M, WAY D, KIM KS, WITTE MH, WEINAND M, CARHART L, GENDELMAN HE. Microglial and astrocyte chemokines regulate monocyte migration through the blood-brain barrier in human immunodeficiency virus-1 encephalitis. Am J Pathol. 1999;155:1599–611. doi: 10.1016/S0002-9440(10)65476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PERSIDSKY Y, ZHENG J, MILLER D, GENDELMAN HE. Mononuclear phagocytes mediate blood-brain barrier compromise and neuronal injury during HIV-1-associated dementia. J Leukoc Biol. 2000;68:413–22. [PubMed] [Google Scholar]
- PULLIAM L, CLARKE JA, MCGRATH MS, MOORE D, MCGUIRE D. Monokine products as predictors of AIDS dementia. AIDS. 1996;10:1495–500. doi: 10.1097/00002030-199611000-00006. [DOI] [PubMed] [Google Scholar]
- PULLIAM L, GASCON R, STUBBLEBINE M, MCGUIRE D, MCGRATH MS. Unique monocyte subset in patients with AIDS dementia. Lancet. 1997;349:692–5. doi: 10.1016/S0140-6736(96)10178-1. [DOI] [PubMed] [Google Scholar]
- PULLIAM L, REMPEL H, SUN B, ABADJIAN L, CALOSING C, MEYERHOFF DJ. A peripheral monocyte interferon phenotype in HIV infection correlates with a decrease in magnetic resonance spectroscopy metabolite concentrations. AIDS. 2011;25:1721–6. doi: 10.1097/QAD.0b013e328349f022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PULLIAM L, SUN B, REMPEL H. Invasive chronic inflammatory monocyte phenotype in subjects with high HIV-1 viral load. J Neuroimmunol. 2004;157:93–8. doi: 10.1016/j.jneuroim.2004.08.039. [DOI] [PubMed] [Google Scholar]
- RADLOFF LS. The CES-D scale: A self-report depression scale for research in the general population. Applied Psychological Measurement. 1977;1:385–401. [Google Scholar]
- RAVASZ E, SOMERA AL, MONGRU DA, OLTVAI ZN, BARABASI AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–5. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- ROBERTS ES, BURUDI EM, FLYNN C, MADDEN LJ, ROINICK KL, WATRY DD, ZANDONATTI MA, TAFFE MA, FOX HS. Acute SIV infection of the brain leads to upregulation of IL6 and interferon-regulated genes: expression patterns throughout disease progression and impact on neuroAIDS. J Neuroimmunol. 2004;157:81–92. doi: 10.1016/j.jneuroim.2004.08.030. [DOI] [PubMed] [Google Scholar]
- ROBERTS ES, ZANDONATTI MA, WATRY DD, MADDEN LJ, HENRIKSEN SJ, TAFFE MA, FOX HS. Induction of pathogenic sets of genes in macrophages and neurons in NeuroAIDS. Am J Pathol. 2003;162:2041–57. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBERTSON KR, SMURZYNSKI M, PARSONS TD, WU K, BOSCH RJ, WU J, MCARTHUR JC, COLLIER AC, EVANS SR, ELLIS RJ. The prevalence and incidence of neurocognitive impairment in the HAART era. AIDS. 2007;21:1915–21. doi: 10.1097/QAD.0b013e32828e4e27. [DOI] [PubMed] [Google Scholar]
- SACKTOR N, MCDERMOTT MP, MARDER K, SCHIFITTO G, SELNES OA, MCARTHUR JC, STERN Y, ALBERT S, PALUMBO D, KIEBURTZ K, DE MARCAIDA JA, COHEN B, EPSTEIN L. HIV-associated cognitive impairment before and after the advent of combination therapy. J Neurovirol. 2002;8:136–42. doi: 10.1080/13550280290049615. [DOI] [PubMed] [Google Scholar]
- SALAZAR-GONZALEZ JF, MARTINEZ-MAZA O, AZIZ N, KOLBERG JA, YEGHIAZARIAN T, SHEN LP, FAHEY JL. Relationship of plasma HIV-RNA levels and levels of TNF-alpha and immune activation products in HIV infection. Clin Immunol Immunopathol. 1997;84:36–45. doi: 10.1006/clin.1997.4364. [DOI] [PubMed] [Google Scholar]
- SHAPSHAK PDR, DURAN EM, FARINETTI F, MINAGAR A, COMMINS D, RODRIGUEZ H, CHIAPPELLI F, KANGUEANE P, LEVINE AJ, SINGER E, SOMBOONWIT C, SINNOTT JT. Gene expression for HIV-associated dementia and HIV encephalitis in microdissected neurons 1: preliminary analysis. Neurobehavioral HIV Medicine. 2011;3:53–78. [Google Scholar]
- SHAPSHAK P, DUNCAN R, MINAGAR A, RODRIGUEZ DE LA VEGA P, STEWART RV, GOODKIN K. Elevated expression of IFN-gamma in the HIV-1 infected brain. Front Biosci. 2004a;9:1073–81. doi: 10.2741/1271. [DOI] [PubMed] [Google Scholar]
- SHAPSHAK P, DUNCAN R, TORRES-MUNOZ JE, DURAN EM, MINAGAR A, PETITO CK. Analytic approaches to differential gene expression in AIDS versus control brains. Front Biosci. 2004b;9:2935–46. doi: 10.2741/1449. [DOI] [PubMed] [Google Scholar]
- SHAY AH, CHOI R, WHITTAKER K, SALEHI K, KITCHEN CM, TASHKIN DP, ROTH MD, BALDWIN GC. Impairment of antimicrobial activity and nitric oxide production in alveolar macrophages from smokers of marijuana and cocaine. J Infect Dis. 2003;187:700–4. doi: 10.1086/368370. [DOI] [PubMed] [Google Scholar]
- STOREY JD, TAYLOR JE, SIEGMUND D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2004;66:187–205. [Google Scholar]
- SUN B, ABADJIAN L, REMPEL H, CALOSING C, ROTHLIND J, PULLIAM L. Peripheral biomarkers do not correlate with cognitive impairment in highly active antiretroviral therapy-treated subjects with human immunodeficiency virus type 1 infection. J Neurovirol. 2010;16:115–24. doi: 10.3109/13550280903559789. [DOI] [PubMed] [Google Scholar]
- TAMATANI M, MATSUYAMA T, YAMAGUCHI A, MITSUDA N, TSUKAMOTO Y, TANIGUCHI M, CHE YH, OZAWA K, HORI O, NISHIMURA H, YAMASHITA A, OKABE M, YANAGI H, STERN DM, OGAWA S, TOHYAMA M. ORP150 protects against hypoxia/ischemia-induced neuronal death. Nat Med. 2001;7:317–23. doi: 10.1038/85463. [DOI] [PubMed] [Google Scholar]
- TANJI K, MARUYAMA A, ODAGIRI S, MORI F, ITOH K, KAKITA A, TAKAHASHI H, WAKABAYASHI K. Keap1 is localized in neuronal and glial cytoplasmic inclusions in various neurodegenerative diseases. J Neuropathol Exp Neurol. 2013;72:18–28. doi: 10.1097/NEN.0b013e31827b5713. [DOI] [PubMed] [Google Scholar]
- THIEBLEMONT N, WEISS L, SADEGHI HM, ESTCOURT C, HAEFFNER-CAVAILLON N. CD14lowCD16high: a cytokine-producing monocyte subset which expands during human immunodeficiency virus infection. Eur J Immunol. 1995;25:3418–24. doi: 10.1002/eji.1830251232. [DOI] [PubMed] [Google Scholar]
- TOZZI V, BALESTRA P, BELLAGAMBA R, CORPOLONGO A, SALVATORI MF, VISCO-COMANDINI U, VLASSI C, GIULIANELLI M, GALGANI S, ANTINORI A, NARCISO P. Persistence of neuropsychologic deficits despite long-term highly active antiretroviral therapy in patients with HIV-related neurocognitive impairment: prevalence and risk factors. J Acquir Immune Defic Syndr. 2007;45:174–82. doi: 10.1097/QAI.0b013e318042e1ee. [DOI] [PubMed] [Google Scholar]
- VOINEAGU I, WANG X, JOHNSTON P, LOWE JK, TIAN Y, HORVATH S, MILL J, CANTOR RM, BLENCOWE BJ, GESCHWIND DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–4. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARRINER EM, ROURKE SB, ROURKE BP, RUBENSTEIN S, MILLIKIN C, BUCHANAN L, CONNELLY P, HYRCZA M, OSTROWSKI M, DER S, GOUGH K. Immune activation and neuropsychiatric symptoms in HIV infection. J Neuropsychiatry Clin Neurosci. 2010;22:321–8. doi: 10.1176/jnp.2010.22.3.321. [DOI] [PubMed] [Google Scholar]
- WOJNA V, CARLSON KA, LUO X, MAYO R, MELENDEZ LM, KRAISELBURD E, GENDELMAN HE. Proteomic fingerprinting of human immunodeficiency virus type 1-associated dementia from patient monocyte-derived macrophages: A case study. J Neurovirol. 2004;10(Suppl 1):74–81. doi: 10.1080/753312756. [DOI] [PubMed] [Google Scholar]
- YADAV A, COLLMAN RG. CNS inflammation and macrophage/microglial biology associated with HIV-1 infection. J Neuroimmune Pharmacol. 2009;4:430–47. doi: 10.1007/s11481-009-9174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YIP AM, HORVATH S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics. 2007;8:22. doi: 10.1186/1471-2105-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOSHIOKA M, BRADLEY WG, SHAPSHAK P, NAGANO I, STEWART RV, XIN KQ, SRIVASTAVA AK, NAKAMURA S. Role of immune activation and cytokine expression in HIV-1-associated neurologic diseases. Adv Neuroimmunol. 1995;5:335–58. doi: 10.1016/0960-5428(95)00012-q. [DOI] [PubMed] [Google Scholar]
- YOSHIOKA M, SHAPSHAK P, SRIVASTAVA AK, STEWART RV, NELSON SJ, BRADLEY WG, BERGER JR, RHODES RH, SUN NC, NAKAMURA S. Expression of HIV-1 and interleukin-6 in lumbosacral dorsal root ganglia of patients with AIDS. Neurology. 1994;44:1120–30. doi: 10.1212/wnl.44.6.1120. [DOI] [PubMed] [Google Scholar]
- ZHANG B, HORVATH S. A general framework for weighted gene co-expression network analysis. Statistical Applications in Genetics and Molecular Biology. 2005a:4. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- ZHANG B, HORVATH S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005b;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- ZHAO J, REDELL JB, MOORE AN, DASH PK. A novel strategy to activate cytoprotective genes in the injured brain. Biochem Biophys Res Commun. 2011;407:501–6. doi: 10.1016/j.bbrc.2011.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO L, ROSALES C, SEBURN K, RON D, ACKERMAN SL. Alteration of the unfolded protein response modifies neurodegeneration in a mouse model of Marinesco-Sjogren syndrome. Hum Mol Genet. 2010;19:25–35. doi: 10.1093/hmg/ddp464. [DOI] [PMC free article] [PubMed] [Google Scholar]