Abstract
Cathepsin E is an endosomal aspartic proteinase that is predominantly expressed in immune-related cells. Recently, we showed that macrophages derived from cathepsin E-deficient (CatE −/−) mice display accumulation of lysosomal membrane proteins and abnormal membrane trafficking. In this study, we demonstrated that CatE −/− macrophages exhibit abnormalities in autophagy, a bulk degradation system for aggregated proteins and damaged organelles. CatE −/− macrophages showed increased accumulation of autophagy marker proteins such as LC3 and p62, and polyubiquitinated proteins. Cathepsin E deficiency also altered autophagy-related signaling pathways such as those mediated by the mammalian target of rapamycin (mTOR), Akt, and extracellular signal-related kinase (ERK). Furthermore, immunofluorescence microscopy analyses showed that LC3-positive vesicles were merged with acidic compartments in wild-type macrophages, but not in CatE −/− macrophages, indicating inhibition of fusion of autophagosome with lysosomes in CatE −/− cells. Delayed degradation of LC3 protein was also observed under starvation-induced conditions. Since the autophagy system is involved in the degradation of damaged mitochondria, we examined the accumulation of damaged mitochondria in CatE −/− macrophages. Several mitochondrial abnormalities such as decreased intracellular ATP levels, depolarized mitochondrial membrane potential, and decreased mitochondrial oxygen consumption were observed. Such mitochondrial dysfunction likely led to the accompanying oxidative stress. In fact, CatE −/− macrophages showed increased reactive oxygen species (ROS) production and up-regulation of oxidized peroxiredoxin-6, but decreased antioxidant glutathione. These results indicate that cathepsin E deficiency causes autophagy impairment concomitantly with increased aberrant mitochondria as well as increased oxidative stress.
Introduction
Cathepsin E is an intracellular aspartic proteinase that is predominantly distributed in immune-related cells [1–3]. The subcellular localization of cathepsin E is mainly in endosomal compartments in antigen-presenting cells such as macrophages, dendritic cells, and microglia [4–7]. Our previous study has demonstrated that cathepsin E-deficient (CatE −/−) mice spontaneously developed atopic dermatitis-like skin lesions when reared under conventional conditions, but they did not develop these lesions under specific pathogen-free conditions [8]. This finding suggests that this phenotype was triggered by some environmental factor(s), including pathogenic microorganisms. Indeed, we recently reported that CatE −/− mice exhibited increased susceptibility to bacterial infection with Staphylococcus aureus or Porphyromonas gingivalis [9]. However, the precise molecular mechanisms inducing aberrant immune responses in CatE −/− mice remain unclear.
Recently, we reported that peritoneal macrophages from CatE −/− mice display a novel type of lysosomal storage disorder characterized by intracellular accumulation of major lysosomal membrane sialoglycoproteins (LAMP-1 and LAMP-2) [10]. In CatE −/− macrophages, the lysosomal membrane glycoprotein accumulation induced a concomitant elevated lysosomal pH and enhanced extracellular secretion of various lysosomal hydrolases [10,11]. The accumulated membrane proteins in CatE −/− macrophages seem to induce secondary abnormalities in membrane trafficking. In fact, the cell surface levels of Toll-like receptors, chemotactic receptors, and adhesion receptors were significantly lower in CatE −/− macrophages compared with those in wild-type cells [12]. Therefore, we hypothesized that the abnormal membrane trafficking in CatE −/− macrophages influences both extracellular and intracellular events such as cell surface receptor expression and autophagy, respectively.
Autophagy (macro-autophagy) is a process by which the dynamic rearrangement of cellular membranes allows portions of the cytoplasm to be delivered to the lysosomes, where they are degraded [13]. Recent studies have revealed that autophagy is necessary for certain aspects of cellular homeostasis, such as prevention of neurodegeneration [14–16], and for various host defense mechanisms including prevention of tumor progress [17], clearance of pathogenic bacteria and viruses [18], and antigen presentation [19]. Therefore, autophagy is essential for the maintenance of the fundamental cell functions in immune-related cells. Given that a lack of cathepsin E induces various abnormal membrane trafficking events in immune-related cells, it is possible that cathepsin E deficiency causes impairment of autophagy concomitant with the accumulation of toxic proteins or damaged organelles. In this study, we demonstrated that cathepsin E deficiency causes an impaired autophagic flux and an increase in the number of aberrant mitochondria in macrophages.
Materials and Methods
Antibodies to mouse LC3 and beclin-1 were purchased from Medical & Biology Laboratories (Nagoya, Japan). The antibody to p62/SQSTM1 was purchased from Progen Biotechnique (Heidelberg, Germany). The antibody to ubiquitin-protein conjugates was obtained from Biomol International, PA, USA. Antibodies to mammalian target of rapamycin (mTOR), p-mTOR(Ser2448), Akt, p-Akt(Ser473), extracellular signal-related kinase (ERK), and p-ERK (Tyr204, Ser473), S6K, p-S6K (Thr 389), adenosine monophosphate kinase (AMPK)α, p-AMPKα (Thr172), were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies to actin were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The ATP assay kit (CellTiter-Gio Luminescent cell viability assay) was purchased from Promega (Madison, WI, USA). Cell counting kit-8 (CCK-8) was from Dojindo (Kumamoto, Japan). Baf and 3-MA were purchased from Wako Pure Chemicals (Tokyo, Japan). LysoTracker Red DND-99 and Alexa Fluor 488 goat anti-rabbit IgG were purchased from Molecular Probes/Invitrogen (Eugene, Oregon, USA). FCCP was from Sigma-Aldrich Inc (Tokyo, Japan).
Animals
Wild-type and CatE −/− mice with the C57BL/6 genetic background were used as described previously [10]. All animals were maintained according to the guidelines of the Japanese Pharmacological Society. The animals and all experiments were approved by the Animal Research Committees of the Graduate School of Biomedical Sciences, Nagasaki University (Permit Number: 1205080984-2). All surgical procedures were performed under ether anesthesia, and every effort was made to minimize suffering.
Preparation of peritoneal macrophages
Thioglycolate-elicited peritoneal macrophages were isolated from mice as described previously [10]. Briefly, 8- to 14-week-old mice were peritoneally injected with 4.05% thioglycolate (2 mL/mouse). Three and a half days later, peritoneal exudate cells were isolated from the peritoneal cavity by washing with phosphate-buffered saline (PBS). The cells were incubated in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), penicillin (50 units/mL), and streptomycin (50 µg/mL) at 37°C with 5% CO2. After incubation for 2 h, non-adherent cells were removed by washing 3 times with Ca2+/Mg2+-free PBS. Peritoneal macrophages isolated as adherent MAC-2-positive cells were obtained at a purity of greater than 95%.
Preparation of cell lysates
For the preparation of cell lysates, the cells were washed twice with PBS, removed from the plates with a rubber scraper, and centrifuged at 300 ×g for 5 min. The precipitated cells were resuspended in PBS containing 0.1% Triton X-100, and then subjected to sonication for 1 min at 4°C followed by centrifugation at 100,000 ×g for 1 h. The supernatant fraction was referred to as the cell lysate. For sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) or immunoblot analyses, the supernatant was precipitated with TCA at a final concentration of 5% and centrifuged at 12,000 ×g for 15 min after incubation on ice for 15 min. After washing with ice-cold acetone and evaporating with air, the pellets were suspended in the buffer for SDS-PAGE. The cells were washed twice with PBS, removed from the plates by pipetting, and then subjected to centrifugation at 300 × g for 5 min. The precipitated cells were suspended in PBS containing 0.05% Triton X-100, sonicated 3 times for 5 s at 4°C, and subjected to centrifugation at 120,000 × g for 30 min at 4°C. The supernatant was used as the cell lysate.
Gel electrophoresis and western blot analysis
SDS-PAGE and western blot were performed as described previously [20]. The quantification of the immunoreactive bands was analyzed by LAS-1000 and Image Gauge software (Fuji Photo Film Co., Ltd., Tokyo, Japan)
Bulk protein degradation assay
The bulk protein degradation assay was performed according to the method described by Fujita et al. [21] with some modifications. Briefly, cells (1 × 106 cells) were seeded in 35 mm culture dishes and incubated overnight. The medium were substituted with labeling medium containing [14C] leucine (0.5 Ci/mL per dish) for 24 h in 2.0 mL of RPMI 1640 medium supplemented with 10% FBS. Cells were then incubated in 2.0 mL of chase medium (DMEM supplemented with 10% FBS and 10 mM unlabeled leucine) and further incubated for 4 h to remove the contribution of short-lived proteins. After a 4-h incubation, the cells were lysed in 1 mL of 10 mM Tris-HCl buffer at pH 7.5 containing 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.02% sodium azide. The cell lysates and the collected media were mixed with trichloroacetic acid (TCA) to a final TCA concentration of 10%. After centrifugation at 12,000 ×g for 5 min, TCA-insoluble cell lysates were dissolved in 1 mL of 1 N NaOH. The radioactivity of the TCA-insoluble cell lysates and the TCA-soluble media was determined by a scintillation counter. The rate of degradation of the bulk long-lived protein was calculated as the total activity of the media divided by that of the cell lysates.
Immunofluorescence microscopy
The cells were grown on glass cover slips and then preincubated with LysoTracker-Red DND-99 for 30 min. After washing with PBS, the cells were fixed with 4.0% paraformaldehyde in PBS for 30 min at room temperature. The fixed cells were then washed with 50 mM NH4Cl in PBS for quenching, and permeabilized with 0.3% Tween-20 in PBS for 10 min. The cells were incubated with 1% bovine serum albumin and 1% normal goat serum in PBS for 3 h and subsequently incubated with anti-LC3 antibody overnight at 4°C, followed by Alexa Fluor 488 goat anti-rabbit IgG as a secondary antibody. The samples were inspected by microscopy using a laser-scanning confocal imaging system (LSM510 META; Carl Zeiss, Co., Ltd).
Determination of mitochondrial membrane potential in macrophages
The mitochondrial membrane potential was measured using 5,5′,6,6′-tetrachloro-1,1′,3,3′ tetraethylbenzimidazolylcarbocyanine iodide (JC-1; Molecular Probes/Invitrogen) according to a published protocol with some modifications. The peritoneal macrophage suspension (2 × 105 cells/100 μL) was incubated on ice for 15 min with 10 μg/mL of JC-1 in PBS containing 2.5% FBS and 0.01% NaN3 (buffer A). After washing with buffer 3 times, flow cytometry analyses were performed using an Epics XL flow cytometer (Beckman Coulter, Brea, CA, USA).
Isolation of mitochondria
Macrophages grown on 100-mm plates were washed twice with the fraction buffer (250 mM sucrose, 5 mM HEPES, 50 mM KCl, 6 mM MgCl2, 1 mM EDTA, pH 7.5) and scraped with soft rubber scrapers. The cells were then homogenized with 3 strokes in a glass Dounce homogenizer. The homogenates were centrifuged at 1,000 ×g for 10 min at 4°C, and the resultant supernatants were centrifuged at 10,000 ×g for 10 min at 4°C, and used as crude mitochondrial preparations.
Two-dimensional electrophoresis and proteomic analyses
Two-dimensional (2-D) electrophoresis and proteomic analyses were performed as described previously (10). Briefly, the cell lysates from macrophages were applied to immobilized pH gradient (IPG) strips (Amersham Biosciences, Piscataway, NJ, USA) and then subjected to isoelectric focusing using a Multiphor II (Amersham Pharmacia Biotech). After isoelectric focusing, the strips were equilibrated for 15 min in 50 mM Tris-HCl, pH 8.8, containing 6 M urea, 30% glycerol, 1% SDS, and 64 mM dithiothreitol and subsequently immersed for 15 min in the same buffer containing 135 mM iodoacetamide instead of dithiothreitol. The strips were then transferred onto 10% SDS-polyacrylamide gels. After electrophoresis, the gels were silver-stained for proteins. Peptide mass mapping was performed by recording the peptide mass fingerprints of typical in-gel digests of the corresponding gel bands by using matrix-assisted laser desorption ionization time-of-flight mass spectrometer (MS; AXIMA-CFR plus, Shimadzu, Tsukuba, Japan) and the subsequent use of the Mascot search engine (Matrix Science, Tokyo, Japan).
Measurement of oxidative burst by macrophages
Oxidative burst production was measured according to the method described previously with some modification [22]. Briefly, the macrophage cell suspension (1×107 cells /mL) was preincubated at 37°C. Zymosan A suspended in PBS (20 mg/mL) was boiled for 10 min and washed. The zymosan suspension was incubated with an equal volume of mouse serum at 37°C for 30 min. The particles were washed twice with PBS and suspended in their original volume in PBS. The cuvette for the reaction mixture, which contained 0.1 mL of freshly diluted luminol solution (0.2 mM), 0.1ml of the macrophage suspension (2×107 cells/mL) and 0.1 mL of the opsonized zymosan (20 mg/mL) was maintained at 37° C in the Luminophotometer TD-4000 (LABO Science Co., Tokyo, Japan). The intensity of light emitted in the cuvette was recorded automatically. The chemiluminescent response corresponded to the peak of the curve.
Measurement of hydrogen peroxide production (H2O2) by macrophages
Determination of H2O2 production in the culture media by macrophages was performed using an Amplex Redassay kit (Molecular Probes).
Measurement of reduced glutathione (GSH)
GSH was spectrophotometrically determined with nicotinamide adenine dinucleotide phosphate (NADPH) at 412 nm by measuring the intensity of color development with DTNB according to the method of described by Beutler et al. [23] with some modification.
Statistical analysis
Quantitative data are presented as mean + standard deviation (SD). The statistical significance of differences among mean values was assessed by Student’s t-test. P values of <0.05 were considered statistically significant.
Results
Cathepsin E deficiency increases autophagy marker proteins in macrophages
To determine whether cathepsin E is involved in autophagy in macrophages, we performed western blot analysis of microtubule-associated protein 1A/1B light chain 3 (LC3), an autophagy marker protein [24]. LC3 is detected as 2 bands by western blotting; one band represents LC3-I, a cytosolic form of approximately 16 kDa, the other band consists of LC3-II, which is conjugated to phosphatidylethanolamine, as a membrane-associated form of approximately 14 kDa [25]. Western blot analysis revealed higher LC3-I and LC3-II protein levels in CatE −/− macrophage lysates than in wild-type macrophage lysates (Figure 1A). Densitometric analysis for the quantification of protein levels showed that LC3-I and LC3-II protein levels were significantly higher in CatE −/− macrophages than in wild-type cells (Figure 1B).
Figure 1. Levels of autophagy-related proteins in wild-type and CatE−/− macrophages.
(A) After a 1-day culture, the cell lysates (100 µg protein for each) derived from wild-type (+/+) and CatE−/− macrophages (−/−) were subjected to SDS-PAGE followed by western blotting with specific antibodies to LC3, p62, ubiquitin, and actin. The data indicate the representative western blotting of five independent experiments. (B) Densitometric analysis for the quantification of each protein in the cell lysate of both cell types. The arbitrary density unit was defined as the relative chemiluminescence intensity per mm2 measured by LAS1000. The data are indicated as the mean ± SD values from 5 independent experiments. *P < 0.05 for the indicated comparisons. (C) The cells were cultured for 2 days, and then treated with the lysosomal inhibitors Baf (0.5 μM) and 3-MA (5 mM) or without for 12 h. After lysis, samples were subjected to SDS-PAGE followed by western blotting with antibodies to LC3 or actin. The data indicate the representative western blotting of 4 independent experiments. (D) Pulse-chase experiments using [14C] leucine were performed to study bulk long-lived proteins. The radioactivity of the TCA-insoluble cell lysates and TCA-soluble media was determined, and was used to calculate as the rate of degradation of the long-lived proteins. The data are shown as the percent of degradation of long-lived proteins from 3 independent experiments. *P < 0.05 for the indicated comparisons.
Besides LC3, p62/SQSTM1 is also used as an autophagy marker, because p62 associates directly with LC3 to promote degradation of the ubiquitinated protein aggregates by autophagy [26]. Consequently, blocking autophagy leads to an increase in protein levels and aggregation of p62 [27,28]. The p62 protein levels in CatE −/− macrophages were markedly higher than those in wild-type cells (Figure 1A and B).
Furthermore, we investigated the levels of polyubiquitinated proteins in CatE −/− macrophages, because it has been reported that blockage of autophagy induces cellular polyubiquitinated protein accumulation [27,28]. The high-molecular-weight (>50 kD) polyubiquitinated protein levels of CatE −/− macrophages were slightly higher than those of wild-type cells, although the difference was not statistically significant (Figure 1 A and B). Thus, the increased levels of autophagy-related proteins LC3 and p62, and polyubiquitinated proteins in CatE −/− macrophages, suggest that cathepsin E deficiency induces abnormalities in autophagy.
Using a different approach, to further confirm whether cathepsin E deficiency inhibits autophagy, we treated macrophages with lysosomotropic agents such as bafilomycin A1 (Baf) and 3-methyladenine (3-MA). Baf is a vacuole-type ATPase inhibitor, whereas 3-MA is a class III phosphatidylinostol 3-kinase inhibitor. Treatment of wild-type and CatE −/− macrophages with Baf or 3-MA induced the accumulation of LC3-I and LC3-II, respectively (Figure 1C). Specifically, the patterns of 3-MA treated wild-type cells mimicked those of CatE −/− cells without the inhibitor (Figure 1C). The Baf treatment significantly increased LC3-II levels in both wild-type cells and CatE −/− cells. These data suggest that cathepsin E deficiency inhibits autophagy (Figure 1C).
CatE−/− macrophages macrophages accumulate long-lived proteins
To further examine the differences between wild-type and CatE −/− macrophages, we performed pulse-chase experiments with [14C] leucine to assess the turnover of long-lived proteins in both cell types with and without Baf treatment (Figure 1D). We compared the radioactivity of the trichloroacetic acid (TCA)-insoluble cell lysate and TCA-soluble medium fractions. The percentage of radio-labeled proteins that were released into the media of CatE −/− macrophages was significantly lower than that of wild-type cells (Figure 1D). Under the conditions used in our study, Baf treatment decreased the release of TCA-soluble radioactivity in both wild-type and CatE −/− macrophages (Figure 1D). These results also suggest that CatE −/− macrophages possess relatively lower autophagic degradation activity.
Cathepsin E deficiency alters autophagy-related signaling
It has been reported that autophagy is regulated by various signaling pathways such as those involving or dependent on mTOR, its substrate p70S6 kinase, Akt, ERK, AMPK [29,30]. Therefore, we analyzed these signaling molecules by western blotting. The protein levels of mTOR, Akt, ERK, and AMPK of both wild-type and CatE −/− macrophages were similar (Figure 2). However, the phosphorylation levels of mTOR, Akt, ERK and AMPK were apparently increased in CatE −/− macrophages compared to wild-type cells (Figure 2). By contrast, the phosphorylation of S6 was decreased in CatE −/− macrophages (Figure 2). These results suggest that the autophagy-related signaling was perturbed in CatE −/− macrophages.
Figure 2. Comparison of Akt/mTOR and ERK signaling pathways in wild-type and CatE −/− macrophages.
(A) The cell lysates (100 µg protein for each) derived from wild-type (+/+) and CatE −/− macrophages (−/−) were subjected to SDS-PAGE followed by western blotting with specific antibodies to mTOR, p-mTOR, Akt, p-Akt(Thr308), p-Akt(Ser473), ERK, p-ERK, S-6, p-S6, AMPK, p-AMPK and actin. The data indicate the representative western blotting of 3 independent experiments. (B) Densitometric analysis for the quantification of each protein in the cell lysate of both cell types. The arbitrary density unit was defined as the relative chemiluminescence intensity per mm2 measured by LAS1000. The data are indicated as the mean ± SD values from 3 independent experiments. *P < 0.05 for the indicated comparisons.
Cathepsin E deficiency impairs fusion of autophagosomes with lysosomes
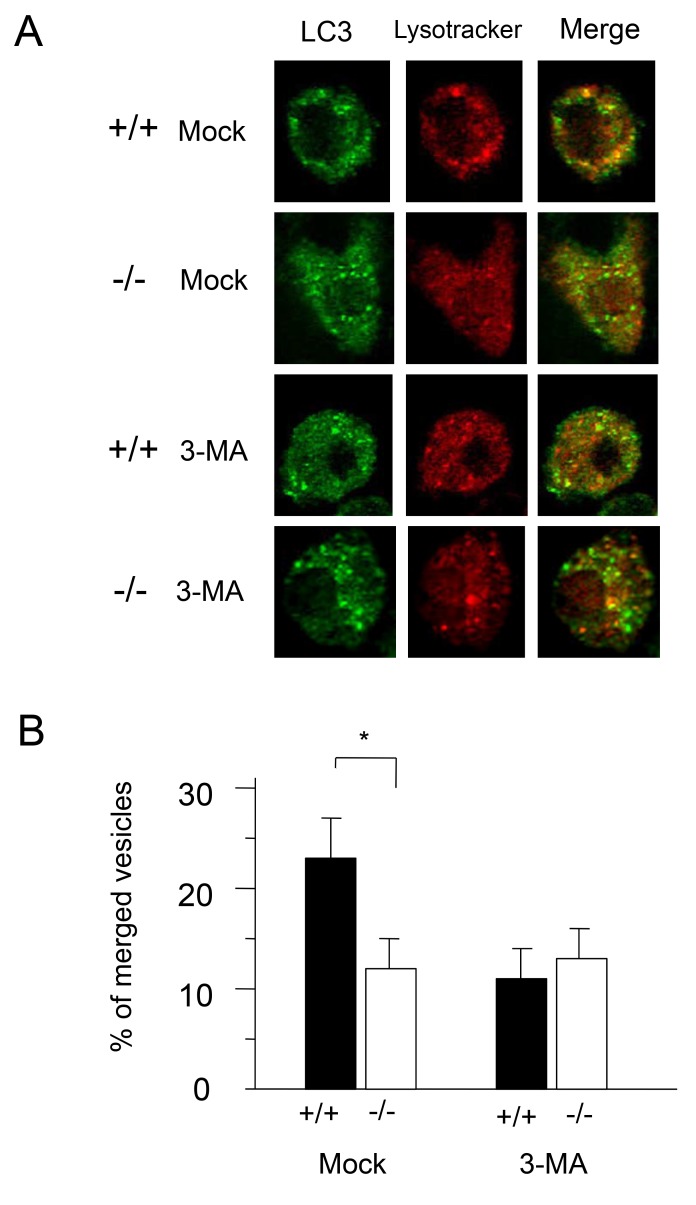
To compare the membrane fusion of autophagosomes with lysosomes in CatE −/− and wild-type macrophages, we analyzed the localization of LC3 and LysoTracker, a fluorescent probe for acidic compartments, such as lysosomes and endosomes by confocal fluorescence microscopy (Figure 3). Upon mock treatment, LC3-positive vesicles in wild-type macrophages were merged with LysoTracker-positive vesicles, whereas those in CatE −/− macrophages did not colocalize with LysoTracker-positive vesicles (Figure 3A). However, when both types of cells were treated with 3-MA, the number of merged vesicles in wild-type cells were apparently decreased, while those in CatE −/− cells remained unaffected (Figure 3A). When the number of LC3-positive vesicles per cell was counted upon mock treatment, the number of those that merged with LysoTracker-positive vesicles was significantly higher in wild-type cells than in CatE −/− cells (Figure 3B). These morphological results also indicate that cathepsin E deficiency inhibits the transport of LC3 to acidic compartments, implying inhibition of autophagy in CatE −/− cells.
Figure 3. Immunofluorescence microscopy of LC3 and LysoTracker in wild-type and CatE -/-macrophages.
(A) Cells on glass cover-slips were preincubated with LysoTracker-Red for 30 min, subsequently fixed, permeabilized with 0.3% Tween-20 in PBS, and allowed to react with anti-LC3 antibody. The cells were then incubated with a fluorescence-labeled secondary antibody and visualized by confocal laser microscopy. (B) Based on the data from immunofluorescence microscopy, the number of LC3- or LysoTracker-positive vesicles per cell was counted. The data are shown as percent merged vesicles per cell and acquired from 3 independent experiments. *P < 0.05 for the indicated comparisons.
Impaired fusion of the autophagosomes with lysosomes was also observed under starvation-induced conditions (Figure 4). When wild-type and CatE −/− macrophages were incubated with a nutrition-depleted medium, LC3-I and LC3-II of CatE −/− macrophages apparently remained, while those of wild-type macrophages gradually disappeared (Figure 4A). After 4 h of incubation, approximately 70 and 60 % of LC3-I and LC3-II, respectively, of CatE −/− macrophages were retained, whereas those of wild-type macrophages decreased to approximately 50% and 35%, respectively (Figure 4B). The results indicate that the degradation of LC3 in CatE −/− macrophages by starvation-induced autophagy was also impaired.
Figure 4. Changes of LC3 in wild-type and CatE −/− macrophages under starvation conditions.
(A) Cultured macrophages derived from wild-type (+/+) and CatE −/− macrophages (−/−) were incubated with Hank’s balanced salt solution for the indicated times (0, 1, 2, 3, and 4). The cell lysates (100 µg protein for each) were subjected to SDS-PAGE followed by western blotting with specific antibodies to LC3. (B) Densitometric analysis for the quantification of each protein in the cell lysate of both cell types. The data was defined as the relative chemiluminescence intensity per mm2 measured by LAS1000. The data are indicated as the mean + SD values from 3 independent experiments. *P < 0.05 for the indicated comparisons.
Cathepsin E deficiency causes mitochondrial abnormalities in macrophages
Autophagy is thought to regulate the turn-over of long-lived proteins and organelles in cells [13]. Given that cathepsin E deficiency inhibits autophagy, degradation of mitochondria may be delayed in CatE −/−cells. To test the accumulation of abnormal mitochondria in CatE −/− macrophages, we measured ATP levels in both cell types. The ATP level in CatE −/− macrophages was significantly lower than that in wild-type cells (Figure 5).
Figure 5. Intracellular ATP levels in wild-type and CatE −/− macrophages.
Macrophages (1 × 105 cells) were cultured on 96-well plates at 37°C for 24 h. After replacement with fresh media, the culture plate was incubated at room temperature (25°C) for 30 min. ATP levels were determined by the manufacturer’s protocol of the CellTiter-Glo assay kit according to the manufacturer’s protocol. The concentration of intracellular ATP was determined by the titration of the control medium without cells plus 0.1−1.0 µM ATP.
We further measured mitochondrial membrane potential with the MitoSensor JC-1, by flow cytometry. JC-1 is a cationic dye, which accumulates in the mitochondria in a potential-dependent manner, and the ratio of green to red fluorescence depends on the membrane potential only [31]. The JC-1 staining pattern of wild-type macrophages exhibited reddish fluorescence similar to that observed in normal mitochondria (Figure 6A). However, the staining pattern of mitochondria in CatE −/− macrophages changed to that of aggregates, indicating depolarization of the mitochondria (Figure 6A). The percentage of depolarized cells in CatE−/− macrophages was significantly increased to approximately 22%, whereas that in wild-type macrophages was approximately 5% (Figure 6B).
Figure 6. Mitochondrial membrane potential of wild-type and CatE −/− macrophages.
(A). Mitochondrial membrane potential was measured with JC-1. A suspension of peritoneal macrophages (2 × 105 cells/100 μL) was incubated on ice for 15 min with JC-1 in PBS containing 2.5% FBS and 0.01% NaN3. After washing, flow cytometric analyses were performed. (B) Comparison of the percentage of gated number of macrophages. Ten thousand cells were analyzed in each sample. The data are indicated as mean ± SD values of data from 4 independent experiments. *P < 0.05 for the indicated comparisons.
Mitochondrial respiration is the most reliable parameter for measuring mitochondrial bioenergetics. Therefore, we measured the oxygen consumption of mitochondrial fractions from wild-type and CatE −/− macrophages by using substrates for mitochondrial complex I (glutamate/malate) or complex II (succinate) (Figure 7). In the presence (state 3) or absence of ADP (state 4), the oxygen consumption of the mitochondria isolated from CatE −/− macrophages was lower than that of wild-type cells (Figure 7A and B). Reduced respiration in mitochondria from CatE −/− macrophages was observed in the presence of carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), a protonophore used for mitochondrial respiration (Figure 7C). Thus, mitochondrial dysfunction was observed in CatE −/− macrophages. However, electron microscopic analysis did not reveal any morphological differences between wild-type and CatE −/− macrophages (data not shown).
Figure 7. Mitochondrial respiration of wild-type and CatE −/− macrophages.
After homogenization, mitochondrial fractions were isolated from macrophages. Respiratory substrate (10 mM glutamate plus 10 mM malate or 5 mM succinate with 10 µM rotenone) was then added and state 4 respiration was measured for 2 min; ADP (1.5 mM) was added, and state 3 respiration was measured for a further 2–4 min. The respiration rates were measured by an oxygraph. The rates were normalized to citrate synthase activity in the same samples. (A) state 3 (in the presence of ADP), (B) state 4 (in the absence of ADP) (C) the uncoupling agent FCCP.
CatE −/− macrophages show an increased oxidative stress
Next, we hypothesized that the abnormal mitochondria in CatE−/− macrophages might affect molecules other than mitochondria, such as cytosolic proteins. To identify proteins that had been changed by cathepsin E deficiency, we examined high resolution 2-D gel electrophoresis within a pI range of 4−7 and a molecular mass range of 15−200 kDa, and compared the proteomic profiles of the cell lysates from both wild-type and CatE −/− macrophages. The 2-D gel electrophoresis analysis detected more than 100 spots. After comparison of the spot volumes, we identified 3 series of protein spots having apparent molecular masses of 38 kDa (pI 6.0−6.7) (a) and 31 kDa (pI 6.0−6.5) (b) that were different in CatE−/− macrophages in comparison to wild-type cells (Figure 8). Proteomic analysis showed that the 38 kDa spot that increased in CatE−/− macrophages was identical with annexin A1. It is known that annexin A1 is a multifunctional protein that is up-regulated by inflammatory and oxidative stimulation (33). In contrast, the two spots of approximately 31 kDa, which increased and decreased, were identified as peroxiredoxin-6. Peroxiredoxin-6 is an antioxidative protein existing in two forms: A reduced form and an oxidized form [32] [33]. To confirm the intracellular conformation of peroxiredoxin-6, we treated wild-type macrophages with hydrogen peroxide (H2O2) analyzed by the 2-D gel electrophoretic profile. Upon H2O2 stimulation, the acidic spot of peroxiredoxin-6 showed an apparent increase, whereas the basic spot a slight decrease (Figure 8B). In addition to these results, various studies have suggested that impairments in the electron transport system in mitochondria are associated with decreased energy production and increased formation of reactive oxygen species (ROS) [34] [35]. Taken together, these results with increased annexin A1 and oxidized peroxiredoxin-6 suggest increased oxidative stress in CatE −/− macrophages.
Figure 8. Two-dimensional gel maps of the cell lysates of macrophages.
(A) Proteome analysis of wild-type and CatE −/− macrophages revealed that almost all the spots located in the areas under pI 6.0–6.7 were identified annexin A1, and those under pI 6.0–6.5 were identified the oxidized and reduced forms of peroxiredoxin-6 respectively. These data indicate the representative 2-D gel maps of 3 independent experiments. (B) Proteomic analysis with wild-type macrophages treated with H2O2 or untreated.
To further test the increased oxidative stress in CatE −/− macrophages, we measured oxidative burst in macrophages by chemiluminescence after stimulation with zymosan (Figure 9A). The relative concentration of superoxide (O2 −) in CatE −/− macrophages was approximately 150% that of wild-type cells (Figure 9A). However, the induced oxidative burst was thought to result from a mixture of O2 − from NADPH oxidase in phagosomes and endogenous O2 − from mitochondrial respiration. Therefore, to exclude ROS production derived from NADPH oxidase, we measured the levels of H2O2 in the extracellular media of macrophages without any stimulation. As shown in Figure 8B, the H2O2 levels were significantly higher in media from CatE −/− macrophages than in those from wild-type cells. We also measured the intracellular levels of glutathione (GSH), which is an antioxidant tripeptide and a useful marker of the redox state [36]. The levels of GSH were markedly diminished in CatE −/− macrophages compared with wild-type cells (Figure 9C). Thus, CatE −/− macrophages apparently exhibited increased oxidative stress in terms of increased H2O2 and decreased GHS levels. Importantly, we found that H2O2 levels in serua from CatE −/− mice were also significantly increased compared with those in wild-type mice (Figure 9D). These results indicate that oxidative stress was increased in CatE −/− cells through an NADPH oxidase-independent pathway including mitochondrial respiration.
Figure 9. Levels of oxidative burst, H2O2 production, and reduced glutathione in wild-type and CatE −/− macrophages.
(A) Levels of oxidative burst by macrophages. Macrophages in cell suspension (1 × 107 cells/mL) were incubated with zymosan suspension at 37 °C for 30 min. Levels of oxidative burst of macrophages were determined by a luminophotometer. (B) The levels of H2O2 production of macrophages. Macrophages (1 × 106 cells/50 μL) were cultured without any stimulation at 37°C for 1 h. Determination of H2O2 production into the culture media by macrophages was performed by an Amplex Redassay kit. (C) The GSH levels of cell lysates of macrophages were assayed with NADPH, measuring the development with DTNB at 412 nm with spectrophotometer. (D) Levels of H2O2 in the serum (50 μL) of mice were determined by an Amplex Redassay kit. The data are the means ± SD of values from 4 independent experiments. *P <0.05, **P <0.01, ***P <0.001.
Discussion
In this study, we demonstrated that cathepsin E deficiency resulted in increased levels of the autophagy marker proteins LC3, p62, and polyubiquitinated proteins. In addition, cathepsin E deficiency caused delayed turnover of long-lived proteins, and impaired fusion of autophagosome to lysosomes in CatE −/− macrophages. These results indicate that cathepsin E deficiency causes impairment of autophagy. However, CatE −/− macrophages display mitochondrial abnormalities such as decreased intracellular ATP levels, depolarized mitochondrial membrane potential, and decreased mitochondrial oxygen consumption. Although 3-MA and Baf have effects on autophagy-lysosome, but these inhibitors are thought to have little, if any, direct inhibitory effect on mitochondrial function. Taken together, we conclude that in macrophages, cathepsin E is essential for eliminating abnormal mitochondria by the autophagy process.
In CatE −/− macrophages, the impaired autophagic flux is probably a secondary event. Our previous study demonstrated that CatE −/− macrophages displayed accumulation of major lysosomal membrane glycoproteins, and followed by elevated lysosomal pH and enhanced soluble lysosomal enzyme secretion [10]. These primary events, including accumulating LAMPs and elevated lysosomal pH presumably causes secondary events related to abnormal membrane trafficking. In fact, CatE −/− macrophages show decreased surface levels of various receptors [9] [12]. Similarly, the accumulation of LC3 proteins and impaired fusion of autophagosomes to lysosomes also result in abnormal membrane trafficking in CatE −/− macrophages. The amount of the membrane form of LC3-II correlates with the number of autophagosomes [24]; hence, the findings that CatE −/− macrophages accumulate both the cytoplasmic LC3-I and the membrane-bound LC3-II imply that both membrane trafficking and degradation are impaired.
The phenotype of CatE −/− macrophages with impaired autophagy is similar to that of cells lacking lysosomal proteins, although cathepsin E is an endosomal proteinase. For example, LC3 protein levels are elevated in neuronal ceroid lipofuscinosis (Batten diseases) [37], multiple sulfate deficiency, mucopolysaccharidosis type IIIA [30], and Niemann-Pick Type C disease [38]. Moreover, accumulation of p62 and polyubiquitinated proteins has been reported in murine cells deficient in autophagy (Atg-5 or Atg-7 deficient mice) [27,28]. Because p62 directly associates with LC3 to promote the degradation of poly-ubiquitinated protein aggregates by autophagy [31], the intracellular p62 accumulation also indicates impaired autophagy. Thus, our findings indicate that the endosomal protease cathepsin E is involved in autophagy-lysosome degradation systems in macrophages.
The impaired autophagic flux in CatE −/− macrophages resulted in various mitochondrial abnormalities. These results presumably imply an accumulation of abnormal mitochondria in CatE −/− macrophages due to decreased mitochondrial degradation and thereby impaired membrane trafficking. This notion is supported by data showing decreased fusion of autophagosomes to lysosomes in CatE −/− macrophages. Similar results were observed in mice deficient in other cathepsins. Koike et al. demonstrated that cathepsin D single-deficient or cathepsins B and L double-deficient mice show the accumulation of LC3 and autophagosomes in the brain [37]. We therefore speculate that the lack of morphological abnormalities in CatE −/− macrophages were due to the amounts of lysosomal proteinases. It is known that professional phagocytic macrophages contain large amounts of lysosomal proteinases [39,40], whereas neurons contain small amounts of lysosomal proteinases. Therefore, it would be difficult to detect the abnormal accumulation of autophagic substrates, including abnormal mitochondria, in macrophages.
Autophagy-related signaling was perturbed in CatE −/− macrophages. Autophagy is regulated by two major signaling pathways in response to starvation: the class I phosphatidylinositol 3-phosphate kinase/Akt/mTOR/S6K/AMPK signaling pathway and the Ras/Raf-1/ ERK/ JNK pathway [41] [42]. The former pathway negatively regulates autophagy, while the latter pathway positively regulates autophagy [41] [42]. In case of CatE −/− macrophages, however, both the mTOR/Akt/AMPK and the ERK pathways were activated, whereas S6K was inactivated. Therefore, the perturbed signaling pathways are likely to correlate with increased oxidative stress in CatE −/− macrophages. In fact, a recent study has reported that there is a relationship between the up-regulation of Akt/mTOR and ERK signaling pathways and the increased ROS activity in other lysosomal storage disorders [43]. Since major intracellular sources of ROS are thought to be derived from dysfunctional mitochondria with impaired respiration [44], the increased ROS production in CatE −/− macrophages is probably due to the accumulation of dysfunctional mitochondria resulting from impaired autophagy.
Conclusion
CatE −/− macrophages display inhibited autophagy, which results in increased numbers of abnormal mitochondria and ROS production, implying that cathepsin E is an essential proteinase for mitophagy in macrophages.
Acknowledgments
We would like to thank Prof. Keiichi I. Nakayama (Kyushu University, Department of Molecular and Cellular Biology, Medical Institute of Bioregulation) for generating the CatE −/− mice.
Funding Statement
This work was supported in part by a Cosmetology Research Foundation Grant and a Grant-in-Aid for Scientific Research (T.T.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yamamoto K, Kawakubo T, Yasukochi A, Tsukuba T (2012) Emerging roles of cathepsin E in host defense mechanisms. Biochim Biophys Acta 1824: 105-112. doi: 10.1016/j.bbapap.2011.05.022. PubMed: 21664991. [DOI] [PubMed] [Google Scholar]
- 2. Sakai H, Saku T, Kato Y, Yamamoto K (1989) Quantitation and immunohistochemical localization of cathepsins E and D in rat tissues and blood cells. Biochim Biophys Acta 991: 367-375. doi: 10.1016/0304-4165(89)90130-X. PubMed: 2655714. [DOI] [PubMed] [Google Scholar]
- 3. Saku T, Sakai H, Tsuda N, Okabe H, Kato Y et al. (1990) Cathepsins D and E in normal, metaplastic, dysplastic, and carcinomatous gastric tissue: an immunohistochemical study. Gut 31: 1250-1255. doi: 10.1136/gut.31.11.1250. PubMed: 2253908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sastradipura DF, Nakanishi H, Tsukuba T, Nishishita K, Sakai H et al. (1998) Identification of cellular compartments involved in processing of cathepsin E in primary cultures of rat microglia. J Neurochem 70: 2045-2056. PubMed: 9572291. [DOI] [PubMed] [Google Scholar]
- 5. Nishioku T, Hashimoto K, Yamashita K, Liou SY, Kagamiishi Y et al. (2002) Involvement of cathepsin E in exogenous antigen processing in primary cultured murine microglia. J Biol Chem 277: 4816-4822. doi: 10.1074/jbc.M108382200. PubMed: 11719510. [DOI] [PubMed] [Google Scholar]
- 6. Bennett K, Levine T, Ellis JS, Peanasky RJ, Samloff IM et al. (1992) Antigen processing for presentation by class II major histocompatibility complex requires cleavage by cathepsin E. Eur J Immunol 22: 1519-1524. doi: 10.1002/eji.1830220626. PubMed: 1601038. [DOI] [PubMed] [Google Scholar]
- 7. Tsukuba T, Ikeda S, Okamoto K, Yasuda Y, Sakai E et al. (2006) Characterization of rat cathepsin E and mutants with changed active-site residues and lacking propeptides and N-glycosylation, expressed in human embryonic kidney 293T cells. FEBS J 273: 219-229. doi: 10.1111/j.1742-4658.2005.05062.x. PubMed: 16367762. [DOI] [PubMed] [Google Scholar]
- 8. Tsukuba T, Okamoto K, Okamoto Y, Yanagawa M, Kohmura K et al. (2003) Association of cathepsin E deficiency with development of atopic dermatitis. J Biochem 134: 893-902. doi: 10.1093/jb/mvg216. PubMed: 14769879. [DOI] [PubMed] [Google Scholar]
- 9. Tsukuba T, Yamamoto S, Yanagawa M, Okamoto K, Okamoto Y et al. (2006) Cathepsin E-deficient mice show increased susceptibility to bacterial infection associated with the decreased expression of multiple cell surface Toll-like receptors. J Biochem 140: 57-66. doi: 10.1093/jb/mvj132. PubMed: 16877769. [DOI] [PubMed] [Google Scholar]
- 10. Yanagawa M, Tsukuba T, Nishioku T, Okamoto Y, Okamoto K et al. (2007) Cathepsin E deficiency induces a novel form of lysosomal storage disorder showing the accumulation of lysosomal membrane sialoglycoproteins and the elevation of lysosomal pH in macrophages. J Biol Chem 282: 1851-1862. PubMed: 17095504. [DOI] [PubMed] [Google Scholar]
- 11. Kakehashi H, Nishioku T, Tsukuba T, Kadowaki T, Nakamura S et al. (2007) Differential regulation of the nature and functions of dendritic cells and macrophages by cathepsin E. J Immunol 179: 5728-5737. PubMed: 17947645. [DOI] [PubMed] [Google Scholar]
- 12. Tsukuba T, Yanagawa M, Okamoto K, Okamoto Y, Yasuda Y et al. (2009) Impaired chemotaxis and cell adhesion due to decrease in several cell-surface receptors in cathepsin E-deficient macrophages. J Biochem 145: 565-573. doi: 10.1093/jb/mvp016. PubMed: 19174547. [DOI] [PubMed] [Google Scholar]
- 13. Mizushima N, Klionsky DJ (2007) Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr 27: 19-40. doi: 10.1146/annurev.nutr.27.061406.093749. PubMed: 17311494. [DOI] [PubMed] [Google Scholar]
- 14. Shibata M, Lu T, Furuya T, Degterev A, Mizushima N et al. (2006) Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem 281: 14474-14485. doi: 10.1074/jbc.M600364200. PubMed: 16522639. [DOI] [PubMed] [Google Scholar]
- 15. Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069-1075. doi: 10.1038/nature06639. PubMed: 18305538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132: 27-42. doi: 10.1016/j.cell.2007.12.018. PubMed: 18191218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levine B (2007) Cell biology: autophagy and cancer. Nature 446: 745-747. doi: 10.1038/446745a. PubMed: 17429391. [DOI] [PubMed] [Google Scholar]
- 18. Schmid D, Münz C (2007) Innate and adaptive immunity through autophagy. Immunity 27: 11-21. doi: 10.1016/j.immuni.2007.07.004. PubMed: 17663981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levine B, Deretic V (2007) Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 7: 767-777. doi: 10.1038/nri2161. PubMed: 17767194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsukuba T, Hori H, Azuma T, Takahashi T, Taggart RT et al. (1993) Isolation and characterization of recombinant human cathepsin E expressed in Chinese hamster ovary cells. J Biol Chem 268: 7276-7282. PubMed: 8463263. [PubMed] [Google Scholar]
- 21. Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A et al. (2008) An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 19: 4651-4659. doi: 10.1091/mbc.E08-03-0312. PubMed: 18768752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kadowaki T, Yoneda M, Okamoto K, Maeda K, Yamamoto K (1994) Purification and characterization of a novel arginine-specific cysteine proteinase (argingipain) involved in the pathogenesis of periodontal disease from the culture supernatant of Porphyromonas gingivalis. J Biol Chem 269: 21371-21378. PubMed: 8063764. [PubMed] [Google Scholar]
- 23. Beutler E, Duron O, Kelly BM (1963) Improved method for the determination of blood glutathione. J Lab Clin Med 61: 882-888. PubMed: 13967893. [PubMed] [Google Scholar]
- 24. Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3: 542-545. PubMed: 17611390. [DOI] [PubMed] [Google Scholar]
- 25. Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E (2005) Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1: 84-91. doi: 10.4161/auto.1.2.1697. PubMed: 16874052. [DOI] [PubMed] [Google Scholar]
- 26. Komatsu M, Waguri S, Koike M, Sou YS, Ueno T et al. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149-1163. doi: 10.1016/j.cell.2007.10.035. PubMed: 18083104. [DOI] [PubMed] [Google Scholar]
- 27. Komatsu M, Waguri S, Chiba T, Murata S, Iwata J et al. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880-884. doi: 10.1038/nature04723. PubMed: 16625205. [DOI] [PubMed] [Google Scholar]
- 28. Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y et al. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885-889. doi: 10.1038/nature04724. PubMed: 16625204. [DOI] [PubMed] [Google Scholar]
- 29. Shinojima N, Yokoyama T, Kondo Y, Kondo S (2007) Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy. Autophagy 3: 635-637. PubMed: 17786026. [DOI] [PubMed] [Google Scholar]
- 30. Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C et al. (2008) A block of autophagy in lysosomal storage disorders. Hum Mol Genet 17: 119-129. PubMed: 17913701. Available online at: PubMed: 17913701 [DOI] [PubMed] [Google Scholar]
- 31. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA et al. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131-24145. doi: 10.1074/jbc.M702824200. PubMed: 17580304. [DOI] [PubMed] [Google Scholar]
- 32. Woo HA, Chae HZ, Hwang SC, Yang KS, Kang SW et al. (2003) Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science 300: 653-656. doi: 10.1126/science.1080273. PubMed: 12714748. [DOI] [PubMed] [Google Scholar]
- 33. Manevich Y, Fisher AB (2005) Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med 38: 1422-1432. doi: 10.1016/j.freeradbiomed.2005.02.011. PubMed: 15890616. [DOI] [PubMed] [Google Scholar]
- 34. Orrenius S, Gogvadze V, Zhivotovsky B (2007) Mitochondrial oxidative stress: implications for cell death. Annu Rev Pharmacol Toxicol 47: 143-183. doi: 10.1146/annurev.pharmtox.47.120505.105122. PubMed: 17029566. [DOI] [PubMed] [Google Scholar]
- 35. Pamplona R, Barja G (2006) Mitochondrial oxidative stress, aging and caloric restriction: the protein and methionine connection. Biochim Biophys Acta 1757: 496-508. doi: 10.1016/j.bbabio.2006.01.009. PubMed: 16574059. [DOI] [PubMed] [Google Scholar]
- 36. McEligot AJ, Yang S, Meyskens FL Jr. (2005) Redox regulation by intrinsic species and extrinsic nutrients in normal and cancer cells. Annu Rev Nutr 25: 261-295. doi: 10.1146/annurev.nutr.25.050304.092633. PubMed: 16011468. [DOI] [PubMed] [Google Scholar]
- 37. Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I et al. (2005) Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Am J Pathol 167: 1713-1728. doi: 10.1016/S0002-9440(10)61253-9. PubMed: 16314482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liao G, Yao Y, Liu J, Yu Z, Cheung S et al. (2007) Cholesterol accumulation is associated with lysosomal dysfunction and autophagic stress in Npc1 -/- mouse brain. Am J Pathol 171: 962-975. doi: 10.2353/ajpath.2007.070052. PubMed: 17631520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES (2005) Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307: 1630-1634. doi: 10.1126/science.1108003. PubMed: 15761154. [DOI] [PubMed] [Google Scholar]
- 40. McCurley N, Mellman I (2010) Monocyte-derived dendritic cells exhibit increased levels of lysosomal proteolysis as compared to other human dendritic cell populations. PLOS ONE 5: e11949. doi: 10.1371/journal.pone.0011949. PubMed: 20689855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meijer AJ, Codogno P (2004) Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol 36: 2445-2462. doi: 10.1016/j.biocel.2004.02.002. PubMed: 15325584. [DOI] [PubMed] [Google Scholar]
- 42. Codogno P, Meijer AJ (2005) Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ 12 Suppl 2: 1509-1518. doi: 10.1038/sj.cdd.4401751. PubMed: 16247498. [DOI] [PubMed] [Google Scholar]
- 43. Vidal-Donet JM, Cárcel-Trullols J, Casanova B, Aguado C, Knecht E (2013) Alterations in ROS activity and lysosomal pH account for distinct patterns of macroautophagy in LINCL and JNCL fibroblasts. PLOS ONE 8: e55526. doi: 10.1371/journal.pone.0055526. PubMed: 23408996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443: 787-795. doi: 10.1038/nature05292. PubMed: 17051205. [DOI] [PubMed] [Google Scholar]