Summary
Cotransins are cyclic heptadepsipeptides that bind the Sec61 translocon to inhibit cotranslational translocation of a subset of secreted and type I transmembrane proteins. The few known cotransin-sensitive substrates are all targeted to the translocon by a cleavable signal sequence, previously shown to be a critical determinant of cotransin sensitivity. By profiling two cotransin variants against a panel of secreted and transmembrane proteins, we demonstrate that cotransin side-chain differences profoundly affect substrate selectivity. Among the most sensitive substrates we identified is the pro-inflammatory cytokine, tumor necrosis factor alpha (TNFα). Like all type II transmembrane proteins, TNFα is targeted to the translocon by its membrane-spanning domain, indicating that a cleavable signal sequence is not strictly required for cotransin sensitivity. Our results thus reveal an unanticipated breadth of translocon substrates whose expression is inhibited by Sec61 modulators.
Introduction
Cotranslational translocation into the endoplasmic reticulum (ER) is an essential, early step in the biogenesis of secreted and transmembrane proteins. Selective delivery of these proteins to the ER is initiated when the first hydrophobic domain, either a cleavable N-terminal signal sequence or an internal transmembrane domain, emerges from the ribosome. This hydrophobic domain is initially captured by the signal recognition particle (SRP), which targets the ribosome-nascent chain complex to the ER membrane via the SRP receptor (Egea et al., 2005; Halic and Beckman, 2005). The nascent chain is then transferred to the Sec61 translocon, a multi-subunit transmembrane protein complex that mediates cotranslational translocation and membrane integration of nearly all secreted and transmembrane proteins (Rapoport, 2007; Mandon, et al., 2009). Once at the translocon, the signal sequence or transmembrane domain is thought to trigger a conformational change in Sec61α, which opens the translocation channel for polypeptide entry into the ER lumen and/or the lipid bilayer (Hegde and Kang, 2008; Plath et al., 1998).
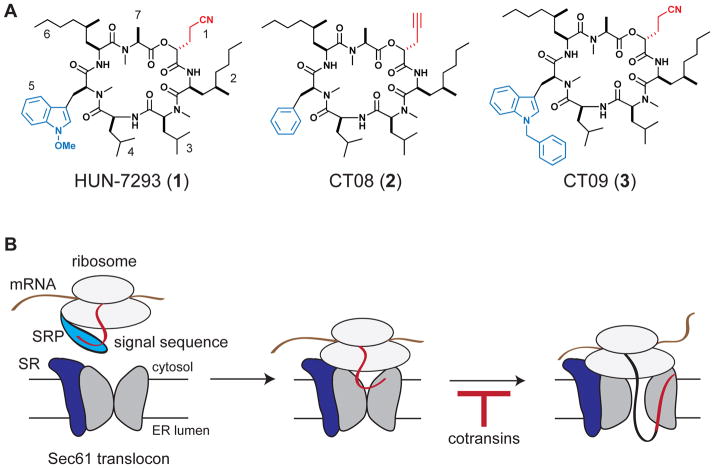
We (Garrison et al., 2005) and another group (Besemer et al., 2005) previously characterized the biological mechanism of “cotransins”, a class of cyclic heptadepsipeptides structurally related to the fungal natural product HUN-7293 (compound 1, Figure 1A). Cotransins potently inhibit the cotranslational translocation of a subset of secreted and transmembrane proteins, resulting in their proteasomal degradation in the cytosol. Mechanistic studies with VCAM (vascular cell adhesion molecule-1) suggested that cotransins inhibit gating of the Sec61 translocon by the VCAM signal sequence, thereby preventing the nascent polypeptide from accessing the ER lumen (Figure 1B). Consistent with this model, a clickable photoaffinity probe related to compound 2 (CT08, Figure 1A) identified the core translocon subunit, Sec61α, as a direct high-affinity target (MacKinnon et al., 2007). Moreover, signal sequence swapping experiments revealed that cotransin sensitivity is determined by the precise identity of the N-terminal signal sequence (Garrison et al., 2005; Besemer et al., 2005). Thus, cotransins appear to selectively disrupt the decisive interaction between Sec61α and a subset of cleavable N-terminal signal sequences.
Figure 1. Cotransins, cyclic depsipeptide inhibitors of cotranslational translocation.
(A) Chemical structures of HUN-7293, CT08, and CT09.
(B) Cotransins inhibit cotranslational translocation of a subset of secreted and transmembrane proteins by preventing signal sequence-dependent opening of the Sec61 translocon. Signal recognition particle (SRP); signal recognition particle receptor (SR).
Given that many proteins with cleavable signal sequences are attractive therapeutic targets, understanding the basis of cotransin selectivity is of considerable interest. To date, only five cotransin-sensitive proteins (VCAM, ICAM-1, E-selectin, P-selectin, and VEGF-A) have been identified, all of which contain cleavable signal sequences lacking any obvious sequence similarity (Garrison et al., 2005; Harant et al., 2007; Boger et al., 1999). Mutagenesis studies have suggested a rough correlation between signal sequence hydrophobicity and cotransin sensitivity, although numerous exceptions to this trend imply a more complex relationship (Harant et al., 2006; Harant et al., 2007). Similarly, little is known about the relationship between cotransin structural elements (e.g., its amino and hydroxy acid side chains) and substrate selectivity/promiscuity. An extensive analysis of HUN-7293 variants revealed that side-chain or backbone modifications can dramatically affect potency (Chen et al., 2002). However, the analysis of only two secretory protein substrates in this study (VCAM and intercellular adhesion molecule-1, ICAM-1) left the issue of selectivity largely unresolved. We therefore sought to define a broader range of cotransin-sensitive secretory and transmembrane substrates, and at the same time, determine whether substrate selectivity can be altered by side-chain modifications.
Results and discussion
In the structure-activity study described above (Chen et al., 2002), most of the HUN-7293 variants were at least 20 times more potent at blocking the expression of VCAM, as compared to ICAM. We were intrigued by the sole exception, a compound we have named CT09 (compound 3, Figure 1A), which had the opposite preference, displaying slightly increased potency against ICAM relative to VCAM. CT09 differs structurally from HUN-7293 by the presence of an N-benzyl indole, which replaces the N-methoxy indole at position 5 (Figure 1A). Although only two secretory proteins were examined, the results of Chen et al. led us to hypothesize that subtle side-chain differences could significantly impact cotransins’ selectivity/promiscuity.
We therefore synthesized CT09 and quantified its effects on the expression of 25 secreted and transmembrane proteins implicated in autoimmune disease, inflammation, and cancer (Table S1). In parallel, we profiled the related cyclic heptadepsipeptide CT08 (compound 2, Figure 1A), which we previously found to inhibit VCAM expression at low nanomolar concentrations (MacKinnon et al, 2007). We quantified endogenously expressed proteins in primary human cells (endothelial, peripheral blood mononuclear, and bronchial epithelial cells) under stimulatory conditions designed to model human pathophysiology (Berg et al., 2010). Protein expression levels were quantified by validated immunoassays (for details, see Experimental Procedures).
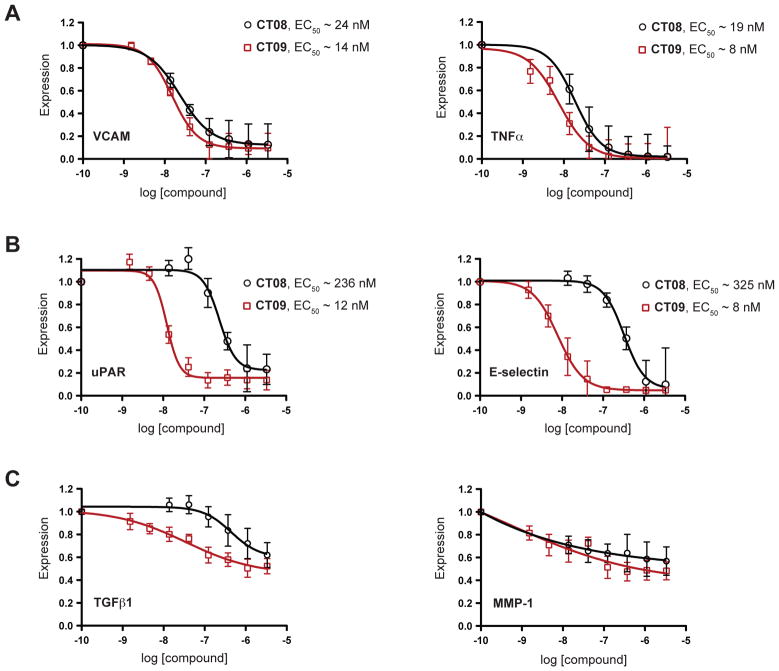
Strikingly, CT08 and CT09 were equipotent at blocking the expression of a subset of proteins but had vastly different potencies toward the majority of proteins tested. Based on their relative sensitivities to CT08 and CT09, the 25 proteins on the panel can be divided into three classes (For the complete dataset, see Figure S1). The first class comprises only two proteins, VCAM and TNFα, both of which were sensitive to low nanomolar concentrations of CT08 or CT09 (Figure 2A). In the second and largest class (72% of the panel) are proteins whose expression was inhibited by CT09 much more potently than CT08 (Figure 2B and S1B). This class is exemplified by the urokinase plasminogen activator receptor, uPAR (20-fold more sensitive to CT09) and the cell adhesion protein, E-Selectin (40-fold more sensitive to CT09). Vascular endothelial growth factor receptor-2 (VEGFR2) provides an extreme example of this selectivity, as it was unaffected by up to 3.3 μM CT08, yet was maximally inhibited by ~100 nM CT09 (>80-fold more sensitive to CT09; Figure S1B). Finally, a third class of proteins (20% of the panel) was only partially affected by micromolar concentrations of either CT08 or CT09 (< 60% inhibition at 3.3 μM; Figure 2C and S1C).
Figure 2. CT08 and CT09 differentially inhibit secretory and transmembrane protein expression.
Primary human cells were stimulated under various pro-inflammatory conditions (see Supplemental Information for details) in the presence of increasing concentrations of CT08 or CT09. After 24 h, expression levels of a total of 25 secreted and membrane proteins (see Table S1 for details) were quantified by cellular immunoassays and normalized to DMSO controls (mean ± SEM; n = 3). EC50 values were estimated by curve-fitting with Prism software (for the complete set of dose-response curves and 95% confidence intervals, along with EC50 values grouped by system, see Figure S1). (A) Class I proteins, VCAM and TNFα, are similarly sensitive to CT08 and CT09. (B) Class II proteins, uPAR and E-selectin, show greater sensitivity to CT09 than CT08. (C) Class III proteins, TGFβ1 and MMP-1, show < 60% inhibition by 3.3 μM CT09 or CT08.
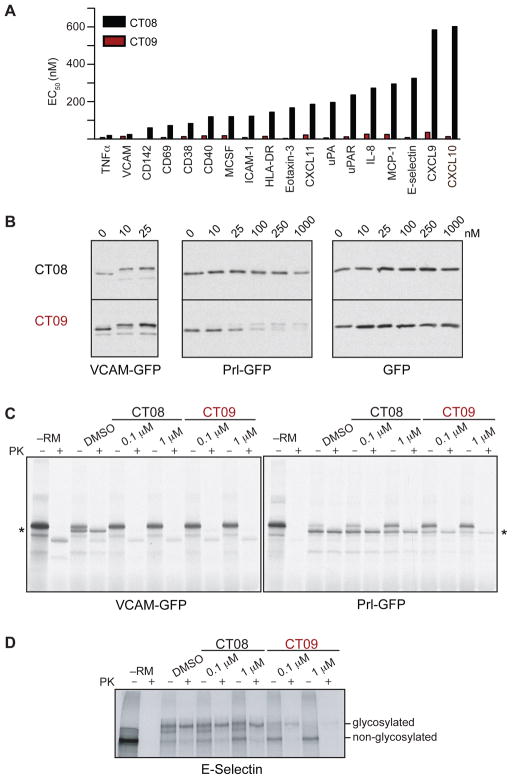
A comparison of CT08 and CT09 EC50 values across the panel reveals two salient features (Figure 3A). First, CT09 is extremely potent, inhibiting the expression of most secreted and transmembrane proteins at concentrations below 100 nM. By contrast, at a concentration of 100 nM, CT08 completely inhibited the expression of only two proteins VCAM and TNFα (Figure 2A); higher concentrations were required to inhibit the expression of most other proteins (Figure 2B and S1B). Second, the data reveal an unconventional structure-activity relationship: side-chain differences between CT08 and CT09 (Figure 1B) impart dramatic potency differences (EC50 ratios) that vary over two orders of magnitude depending on which secretory protein is examined (exemplary CT08:CT09 EC50 ratios: ~1.7 for VCAM; ~20 for uPAR; ~200 for CXCL10; Figure 1B). Moreover, the rank order of protein sensitivity (ranked by increasing EC50) differs between CT08 and CT09. For example, whereas TNFα is the most CT08-sensitive protein (Fig. 3A), there are nine proteins that are equally or more CT09-sensitive than TNFα (Fig. S1B).
Figure 3. CT08 is relatively selective, whereas CT09 is a highly promiscuous inhibitor.
(A) EC50 values for CT08 and CT09 across the panel of 25 proteins were estimated by curve-fitting with Prism. Proteins are listed in order of decreasing sensitivity to CT08. (B) HeLa cells were transfected with plasmids encoding GFP or GFP containing the signal sequence (plus 10 mature domain residues) from VCAM or Prl fused to the N-terminus. Transfected cells were treated with the indicated concentrations of CT08 or CT09 for 20 h prior to cell lysis and Western blot analysis with antibodies to GFP. (C) and (D) mRNA encoding the indicated proteins was translated in reticulocyte lysate in the presence of [35S]-Met, rough ER microsomes (except –RM lanes), and the indicated concentrations of CT08 or CT09. The translated material was left untreated or treated with proteinase K (PK) and analyzed by SDS-PAGE followed by autoradiography. Asterisk = translocated VCAM/Prl-GFP.
We hypothesized that the differential selectivity with which CT08 and CT09 inhibit secretory protein expression arises at the level of cotranslational translocation and that such differences depend primarily on the signal sequences that mediate translocon targeting. To test this hypothesis, we generated constructs containing the signal sequence plus ten mature domain residues of either VCAM or preprolactin (Prl) fused to the N-terminus of GFP; both constructs also contained a C-terminal ER-retention signal (Snapp et al., 2006). Treatment of transfected HeLa cells with nanomolar concentrations of either CT08 or CT09 resulted in an upward shift in the apparent mobility of VCAM-GFP (Fig. 3B), consistent with a lack of signal sequence cleavage due to inhibition of cotranslational translocation. By contrast, expression of the mature, signal-cleaved form of Prl-GFP was relatively insensitive to CT08 (EC50 > 1 μM), yet was potently inhibited by CT09 (EC50 ~ 25 nM, Fig. 3B). The apparent reduced stability of full-length Prl-GFP relative to VCAM-GFP likely derives from the greater hydrophobicity of the Prl signal sequence, recently shown to be an important determinant in the degradation of secretory proteins mislocalized to the cytosol (Hessa et al., 2011). GFP lacking a signal sequence was unaffected by either CT08 or CT09. We confirmed the selectivity of CT08 vs. CT09 by comparing their effects in an in vitro translocation system containing reticulocyte lysates and pancreatic ER microsomes. Consistent with the cell-based experiments, in vitro tanslocation of VCAM-GFP was abolished by low concentrations of both CT08 and CT09, whereas Prl-GFP was significantly more sensitive to CT09 than CT08 (Fig. 3C). These experiments, coupled with the cell-based experiments in Fig. 3B (VCAM-GFP and GFP constructs), also demonstrate that neither CT08 nor CT09 affects translation (see –PK lanes, Fig. 3C). Finally, in vitro translocation assays confirmed that full-length E-selectin is least 10-fold more sensitive to CT09 (EC50 < 100 nM) than CT08 (Fig. 3D), consistent with the cell profiling experiments. We conclude that CT09 broadly inhibits cotranslational translocation and is relatively indiscriminate with respect to signal sequence. By constrast, sensitivity to CT08 is highly signal sequence-dependent.
We further characterized TNFα because of its therapeutic relevance to autoimmune disorders (Palladino et al., 2003) and its potent inhibition by the more selective cotransin, CT08. It was particularly important to corroborate TNFα inhibition in multiple systems, as it represents the first example of a cotransin-sensitive translocon substrate that lacks a cleavable signal sequence. TNFα is cotranslationally inserted into the ER membrane with a type II orientation prior to its delivery to the plasma membrane (Utsumi et al., 1995; Kriegler et al., 1988); it has a cytoplasmic N-terminal tail, followed by a transmembrane-spanning region and an extracellular C-terminal domain that acts as a pro-inflammatory cytokine when liberated by extracellular proteases. In lieu of a cleavable signal sequence, the hydrophobic membrane-spanning domain serves as a ‘signal anchor’ that mediates SRP-dependent targeting, engagement of the Sec61 translocon, and insertion into the lipid bilayer. As such, it is one of four proteins on our panel that relies on an internal signal anchor for cotranslational translocation (the other type II transmembrane proteins, HLA-DR, CD38 and CD69, were affected by higher concentrations of CT08, Figure 3 and S1B).
Because the inhibitory effects of CT08 and CT09 were revealed in a complex cellular system (peripheral blood mononuclear cells stimulated with LPS) in which endogenous TNFα expression is regulated transcriptionally (Collart et al., 1990), post-transcriptionally (Wang et al., 1997; Kontoyiannis et al., 1999) and translationally (Han, et al., 1990), we performed experiments to rule out indirect effects and test whether CT08 blocks TNFα at the level of cotranslational translocation. We focused on CT08, due to its greater selectivity across the panel and thus lower likelihood of inhibiting TNFα expression by indirect mechanisms.
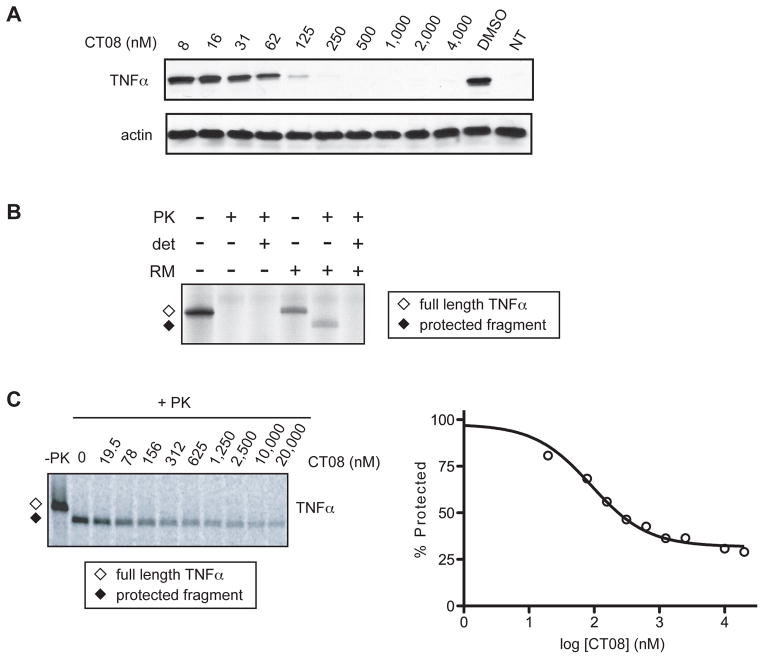
We first expressed TNFα from a cytomegalovirus promoter-driven plasmid in transiently transfected COS-7 cells. In this system, CT08 potently inhibited TNFα expression with an EC50 of less than 60 nM (Figure 4A). As observed with endogenously expressed proteins in primary human cells, the inhibitory effect of CT08 toward ectopically expressed TNFα was similar to its effect on VCAM expressed under identical conditions (MacKinnon et al., 2007). To confirm that CT08 blocks TNFα expression at the level of cotranslational translocation, we moved to the in vitro translocation system. Cotranslational insertion of TNFα into the microsomal membranes was indicated by the formation of a protease-protected fragment consisting of its transmembrane and luminal domains. In the absence of microsomes, or when microsomes were disrupted by detergent, TNFα was completely digested by added protease (Figure 4B). Consistent with the results obtained in transfected cells, cotranslational translocation of wild-type TNFα into ER microsomes was inhibited by nanomolar concentrations of CT08 (Figure 4C).
Figure 4. CT08 inhibits cotranslational translocation of TNFα, a type II transmembrane protein that lacks a cleavable signal sequence.
(A) Expression of TNFα in COS-7 cells treated with CT08. Cell lysates were analyzed by Western blotting. NT = not transfected. (B) mRNA encoding TNFα was translated in the presence of [35S]-cysteine and rough ER microsomes (RM). The translated material was left untreated or treated with proteinase K (PK) in the presence or absence of detergent (det, Triton X-100). Samples were separated by SDS-PAGE and analyzed by autoradiography. The positions of full-length TNFα (open diamond) and its protease-protected fragment (solid diamond) are indicated. (C) In vitro translation/translocation of TNFα, as in Figure 4B. Samples translated in the presence of increasing concentrations of CT08 were digested with proteinase K (+PK), separated by SDS-PAGE, and analyzed by autoradiography and phosphorimaging. A dose-response curve was derived by quantifying the PK-protected TNFα fragment.
The TNFα validation experiments conclusively demonstrate that non-cleavable signal anchors (found in all type II transmembrane proteins and many multi-spanning membrane proteins) can be as susceptible as cleavable signal sequences to cotransin-mediated inhibition. This was unexpected because (1) previous studies indicated that increasing the hydrophobicity of signal sequences leads to decreased cotransin sensitivity (Harant et al., 2006; Harant et al., 2007), and (2) signal anchors, by virtue of their requirement to stably span the lipid bilayer, are universally more hydrophobic than cleavable signal sequences (Martoglio and Dobberstein, 1998; Sakaguchi, et al., 1992). Thus, cotransin sensitivity is not likely based on a single biophysical parameter such as signal sequence/anchor hydrophobicity, raising the possibility that more finely tuned cotransin variants with differing substrate selectivity may be discovered.
Significance
Prior to this work, cotransin-related cyclodepsipeptides had been shown to inhibit the expression of only five secreted and transmembrane proteins, all with cleavable signal sequences lacking any obvious sequence similarity. We made the surprising discovery that structural differences among cotransins (CT08 vs. CT09) can drastically alter the range of inhibited proteins: CT09 inhibited the expression of 20 out of 25 proteins tested at low nanomolar concentrations, whereas CT08 exhibited selectivity for VCAM and TNFα, affecting most of the remaining proteins only at higher concentrations. In the future, it may be possible to refine structure-activity relationships of CT08-like molecules such that even greater selectivity toward TNFα is achieved. Such compounds may provide early leads for small-molecule anti-inflammatory agents that block the functional expression of TNFα.
Supplementary Material
Highlights.
Two cotransin variants were profiled against a panel of secreted/membrane proteins
Cotransin structural differences strongly influence substrate selectivity
TNFα expression is profoundly inhibited by cotransins
Cotransins can block translocation of proteins lacking a cleavable signal sequence
Acknowledgments
This work was supported by the U.S. National Institutes of Health (GM081644 to J.T.; 5F32GM080945 to S.V.M) and the Intramural Research Program of the National Institute of Child Health and Human Development (R.S.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Besemer J, Harant H, Wang S, Oberhauser B, Marquardt K, Foster CA, Schreiner EP, de Vries JE, Dascher-Nadel C, Lindley IJD. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature. 2005;436:290–293. doi: 10.1038/nature03670. [DOI] [PubMed] [Google Scholar]
- Berg EL, Yang J, Melrose J, Nguyen D, Privat S, Rosler E, Kunkel EJ, Ekins S. Chemical target and pathway toxicity mechanisms defined in primary human cell systems. J Pharmacol Toxicol Methods. 2010;61:3–15. doi: 10.1016/j.vascn.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Boger DL, Keim H, Oberhauser B, Schreiner EP, Foster CA. Total synthesis of HUN-7293. J Am Chem Soc. 1999;121:6197–6205. [Google Scholar]
- Chen Y, Bilban M, Foster CA, Boger DL. Solution-phase parallel synthesis of a pharmacophore library of HUN-7293 analogues: A general chemical mutagenesis approach to defining structure–function properties of naturally occurring cyclic (depsi)peptides. J Am Chem Soc. 2002;124:5431–5440. doi: 10.1021/ja020166v. [DOI] [PubMed] [Google Scholar]
- Collart MA, Baeuerle P, Vassali P. Regulation of tumor necrosis factor alpha transcription in macrophages: involvement of four κB-like motifs and of constitutive and inducible forms of NF-κB. Mol. Cell Biol. 1990;10:1498–1506. doi: 10.1128/mcb.10.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Stroud RM, Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Curr Opin Struct Biol. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Garrison JL, Kunkel EJ, Hegde RS, Taunton J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature. 2005;436:285–289. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr Opin Struct Biol. 2005;15:116–125. doi: 10.1016/j.sbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Han J, Brown T, Beutler B. Endotoxin-responsive sequences control cachetin/tumor necrosis factor biosynthesis at the translational level. J Exp Med. 1990;171:465–475. doi: 10.1084/jem.171.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harant H, Lettner N, Hofer L, Oberhauser B, de Vries JE, Lindley IJD. The translocation inhibitor CAM741 interferes with vascular cell adhesion molecule 1 signal peptide insertion at the translocon. J Biol Chem. 2006;281:30492–30502. doi: 10.1074/jbc.M607243200. [DOI] [PubMed] [Google Scholar]
- Harant H, Wolff B, Schreiner EP, Oberhauser B, Hofer L, Lettner N, Maier S, de Vries JE, Lindley IJ. Inhibition of vascular endothelial growth factor cotranslational translocation by the cyclopeptolide CAM741. Mol Pharmacol. 2007;71:1657–1665. doi: 10.1124/mol.107.034249. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Kang SW. The concept of translocational regulaton. J Cell Biol. 2008;182:225–232. doi: 10.1083/jcb.200804157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Hessa T, Sharma A, Mariappan M, Eshleman HD, Gutierrez E, Hegde RS. Protein Targeting and Degradation Pathways are Coupled for Elimination of Mislocalized Proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- Kriegler M, Perez C, DeFay K, Albert I, Lu SD. A novel form of TNF/Catechin is a cell surface cytotoxic transmembrane protein: ramifications for the complex physiology of TNF. Cell. 1988;53:45–53. doi: 10.1016/0092-8674(88)90486-2. [DOI] [PubMed] [Google Scholar]
- MacKinnon AL, Garrison JL, Hegde RS, Taunton J. Photo-leucine incorporation reveals the target of a cyclodepsipeptide inhibitor of cotranslational translocation. J Am Chem Soc. 2007;129:14560–14561. doi: 10.1021/ja076250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon EC, Trueman SF, Gilmore R. Translocation of proteins through the Sec61 and SecYEG channels. Curr Opin Cell Biol. 2009;21:501–570. doi: 10.1016/j.ceb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends in Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- Palladino MA, Bahjat FR, Theodorakis EA, Moldawer LL. Anti-TNF-a therapies: the next generation. Nat Rev Drug Discov. 2003;2:736–746. doi: 10.1038/nrd1175. [DOI] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport T. Signal sequence recognition in posttranslational protein transport across yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Plavec I, Sirenko O, Privat S, Wang Y, Dajee M, Melrose J, Nakao B, Hyptopoulos E, Berg EL, Butcher EC. Method for analyzing signaling networks in complex cellular systems. Proc Natl Acad Sci USA. 2004;101:1223–1228. doi: 10.1073/pnas.0308221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. Protein transport across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M, Tomiyoshi R, Kuroiwa T, Mihara K, Omura T. Functions of signal and signal–anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc Natl Acad Sci USA. 1992;89:16–19. doi: 10.1073/pnas.89.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Mariappan M, Appathurai S, Hedge RS. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol. 2010;619:339–363. doi: 10.1007/978-1-60327-412-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL, Sharma A, Lippincott-Schwartz J, Hegde RS. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc Natl Acad Sci U S A. 2006;103:6536–6541. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi T, Akimaru K, Kawabata Z, Levitan A, Tokunaga T, Tang P, Ide A, Hung MC, Klosteraard J. Human pro-tumor necrosis factor: molecular determinants of membrane translocation, sorting, and maturation. Mol Cell Biol. 1995;15:6398–6405. doi: 10.1128/mcb.15.11.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Ma WJ, Aghajanian C, Spriggs DR. Posttranscriptional regulation of protein expression in human epithelial carcinoma cells by adenine-uridine-rich elements in the 3′-untranslated region of tumor necrosis factor-α messenger RNA. Cancer Res. 1997;57:5426–5433. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.