Abstract
ING4, a new member of the ING (inhibitor of growth) family of tumour suppressor genes, has been found to be deleted or down-regulated in gliomas, breast tumours, and head and neck squamous cell carcinomas. The goal of the present study was to investigate whether the expression and alternative splicing of ING4 transcripts are involved in the initiation and progression of stomach adenocarcinoma. ING4 mRNA and protein expression was examined in gastric adenocarcinoma tissues and human gastric adenocarcinoma cell lines by RT-PCR, real-time RT-PCR, tissue microarray immunohistochemistry, and western blot analysis. Alterations in ING4 transcripts were determined through sequence analysis of ING4 cDNA. Our data showed that ING4 mRNA and protein were dramatically reduced in stomach adenocarcinoma cell lines and tissues, and significantly less in female than in male patients. We also found that reduced ING4 mRNA expression correlated with the stage of the tumour. Interestingly, by sequence analysis, we discovered five novel aberrantly spliced variant forms of ING4 v1 and ING4 v2. These variants cause a codon frame-shift and, eventually, deletion of the NLS or PHD domain contributing to the mislocalization of p53 and/or HAT/HDAC complexes and, subsequently, altered gene expression in gastric adenocarcinoma. These results suggest that attenuated and aberrant ING4 expression may be involved in the initiation and progression of stomach adenocarcinoma.
Keywords: cancer, stomach, adenocarcinoma, tumour suppressor, mRNA expression, protein expression, splice variants
Introduction
Worldwide, stomach cancer is the third and fifth most common malignancy in males and females, respectively [1], with the highest risk populations in Asian countries [2]. The incidence rate is significantly related to age and is approximately twice as high in males than in females [1]. To aid in the development of a more effective therapy for stomach cancer, it is crucial to elucidate the molecular defects that lead to disease progression.
ING4 is a tumour suppressor protein that has been implicated in apoptosis, cell cycle arrest, gene transcription, DNA repair, and other biological events [3]. In gliomas, a significant decrease in ING4 mRNA correlates with tumour grade [4]. ING4 protein is low in gliomas but enriched in normal brain tissues [4]. Reduced ING4 mRNA and allelic loss are further observed in head and neck squamous cell carcinomas [5]. In addition, deletion of the ING4 locus occurs in breast cancer cell lines and primary breast tumours [6]. Inactivating mutations in ING4 transcripts are also found in other human cancer cell lines [6]. Furthermore, ING4 overexpression results in reduced S-phase cells, and p53-dependent apoptosis [7]. Presumably, ING4 also interacts with NF-κB and inhibits brain tumour angiogenesis by repressing NF-κB-responsive gene transcription [4]. ING4 also suppresses the loss of contact inhibition induced by MYC [6].
Here we analysed the potential involvement of ING4 in the development of gastric adenocarcinoma. For the first time, we propose that reduced ING4 expression may be involved in the initiation and progression of gastric adenocarcinoma. We also give evidence for the existence of novel aberrantly spliced variant forms of ING4 v1 and ING4 v2 that possibly alter ING4 function in gastric adenocarcinoma.
Materials and methods
Tissues and cell lines
All tumour and normal surgical (stomach) or autopsy (brain) specimens used in this study represented excess pathological/normal materials obtained in accordance with procedures approved by the Human Ethics Review Board at the First and Second Affiliated Hospitals of Harbin Medical University (Harbin, China) and Beijing Friendship Hospital, Affiliate of Capital University of Medical Sciences (Beijing, China). Surgical tissues were obtained with written consent from patients. The normal and tumour states of specimens were confirmed by examination of haematoxylin and eosin (H&E)-stained histology sections by pathologists at the same hospitals.
Gastric adenocarcinoma cell lines (poorly differentiated MGC-803 and BGC-823; moderately differentiated SGC-7901; and undifferentiated HGC-27) and human embryonic kidney (HEK) 293 cells were from Shanghai Institute of Cell Biology (Shanghai, China). Cell lines were maintained in RPMI 1640 medium containing 10% fetal calf serum.
Reverse transcription PCR (RT-PCR)
Total RNA was isolated using Trizol (Invitrogen). cDNA synthesis was performed using the First-Strand cDNA Synthesis Kit (Promega). β-actin was used as an internal PCR control. The primers were ING4 : 5′-ATGGCTGCGGGGATGTATTTGGAAC-3′ and 5′-CTATTTCTTCTTCCGTTCTTGGGAGCAG-3′ [4]; β-actin: 5′-ACTCTTCCAGCCTTCCTTCC-3′ and 5′-CATACTCCTGCTTGCTGATCC-3′. The PCR products were subjected to electrophoresis and bands were visualized using the Alpha Innotech gel documentation system, utilizing a Multilmage™ Light Cabinet. Images were analysed using Alpha Innotech (version 1.2.0.1) software.
Densitometry was performed using the Scion Image program. ING4 mRNA in tumour and normal specimens was designated as T and N, respectively (calculated as ING4/β-actin densitometric values). T/N indicates the relative ING4 mRNA levels in tumour specimens. Statistical analysis was performed using the t-test. A p value of less than 0.05 was considered significant.
Real-time RT-PCR
Real-time RT-PCR was performed using Taqman TM′ technology and analysed using an ABI 7700 Sequence Detector (Gene Core Bio Technologies, Shanghai, China). Specific ING4 primers (5′-CAAGGAATTTGGTGACGACAAG-3′ and 5′-TCCAGCCGCCGAATGT-3′) and ING4 hybridization probes (FAM-TTTGTCCACCATCTCATAGGTCTGCATG G-TAMRA) were also synthesized by Gene Core Bio Technologies. Normalization was based on β-actin PCR products, which were also quantified by real-time RT-PCR. The real-time RT-PCR results were analysed using the Light Cycler analysis software. Real-time RT-PCR using SYBR Green was performed using a Hot Start Fluorescent PCR Core Reagent Kit (Bio Basic Inc, Markham, Ontario, Canada). Statistical analysis was performed using the t-test. A p value of less than 0.05 was considered significant.
ING4 transcript analysis
Full-length ING4 cDNA sequences from nine gastric adenocarcinoma samples were sub-cloned into a pCR 2.1-TOPO vector (Invitrogen), transformed into E. coli Top10 competent cells (Invitrogen). Positive clones were selected and sequenced (Invitrogen Biological Engineering, Shanghai, China). At least five clones of each cDNA sample were sequenced.
Tissue microarray (TMA) and immunohistochemistry
The TMA and paired normal and tumour samples were taken from different patients. The TMA samples (80 tumour and 40 normal tissues) were from patients at the Beijing Friendship Hospital. The 40 paired normal and tumour tissues were from patients at the First and Second Affiliated Hospitals of Harbin Medical University. The paired tissues were snap-frozen in liquid nitrogen immediately following collection and stored at −80 °C.
Tissue sections, including the TMA samples, were deparaffinized in xylene and rehydrated in graded alcohol. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide (10 min). Antigen was retrieved by autoclaving in EDTA buffer (2 min). Tissues were then incubated in rabbit serum (20 min) to reduce non-specific staining. ING4 goat poly-clonal antibody (Abcam) was applied overnight (4 °C). Immunoreactivity was detected using the Streptavidin/Peroxidase Histostain™ -Plus Kit (Zymed) and DAB Kit (Zhongshan Goldenbridge Biotechnology, Beijing, China). Human brain and normal stomach tissues incubated with normal goat IgG were used as positive and negative controls, respectively.
Western blot analysis
Tissues were homogenized (in 50 mM Tris–HCl, pH 8.0, containing 150 mM NaCl, 0.02% sodium azide, 0.1% sodium dodecyl sulphate, 1% Nonidet P-40, 1 mM phenylmethylsulphonyl fluoride, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin A) and then sonicated, vortexed, and centrifuged at 12 000 g for 10 min at 4 °C. Supernatants were analysed by SDS-PAGE and western blotting using ING4 (T-15) and β-actin antibodies (Santa Cruz Biotech). Immunoreactive bands were detected using the ECL system (New England Biolabs, Guelph, Ontario, Canada).
Results
ING4 mRNA and protein expression is reduced in gastric adenocarcinoma cell lines and tissues
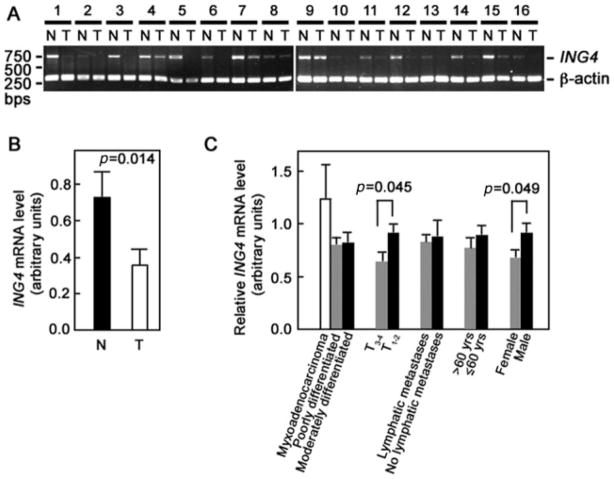
ING4 expression has previously been analysed in various tumour tissues but not in gastric adenocarcinoma. Initially, we analysed 40 paired tumour and adjacent normal tissues for ING4 expression by RT-PCR. Figure 1A shows the general pattern of reduced ING4 mRNA expression in tumour compared with adjacent normal tissues. Reduced ING4 mRNA expression was found in 75% (30/40) of tumour tissues examined. To confirm these findings, we performed real-time RT-PCR on 13 representative paired samples. As shown in Figure 1B, the average level of ING4 mRNA in gastric adenocarcinoma tissues was 0.34 ± 0.07, compared with 0.71 ± 0.13 in normal adjacent tissues (p = 0.014), and thus, ING4 levels of the tumour tissues were, on average, about half of those in adjacent normal tissues.
Figure 1.
ING4 mRNA is significantly reduced in gastric adenocarcinoma tissues compared with normal tissues. (A) Forty paired tumour and adjacent normal tissues were analysed for ING4 expression by RT-PCR. Two gels showing 16 representative paired tumour (T) and normal (N) tissues are shown. β-actin was used as internal loading control. (B) Average ING4 mRNA levels in 13 randomly picked paired tumour (T) and normal (N) samples were determined by real-time RT-PCR. (C) Analysis of relative ING4 mRNA level (T/N) according to gender (27 males, 13 females), age of patient (≤60 years, n = 23; >60 years, n = 17), lymphatic metastasis (lymphatic metastasis, n = 13; no lymphatic metastasis, n = 27), tumour stage (T1 – 2, n = 29; T3 – 4, n = 11), and extent of differentiation of the gastric adenocarcinoma (moderately differentiated, n = 10; poorly differentiated, n = 26; myxoadenocarcinoma, n = 4). Densitometry of PCR products was performed to analyse ING4 levels. Statistical analysis was performed by the t-test. p value <0.05 was considered significant
We then sought to determine whether ING4 mRNA expression correlates with patient gender and age, lymphatic metastasis, tumour stage, and extent of differentiation of the adenocarcinoma. As shown in Figure 1C, the relative ING4 mRNA level (T/N) in tumour tissues was significantly lower in female than in male patients (p = 0.049). The ING4 mRNA level also correlated with depth of tumour penetration. In other words, ING4 levels in tumour stages 3 and 4 (T3 – 4) were significantly lower than those in tumour stages 1 and 2 (T1 – 2) (p = 0.045). Conversely, ING4 levels in tumour tissues did not correlate with the patient’s age, lymphatic metastasis, or extent of differentiation of the adenocarcinoma. Furthermore, we did not observe significant differences in ING4 levels in the intestinal and diffuse (0.8115 ± 0.09124 versus 0.8478 ± 0.07712, respectively; p = 0.802) types of adenocarcinomas, indicating that ING4 levels do not correlate with the type of gastric adenocarcinoma.
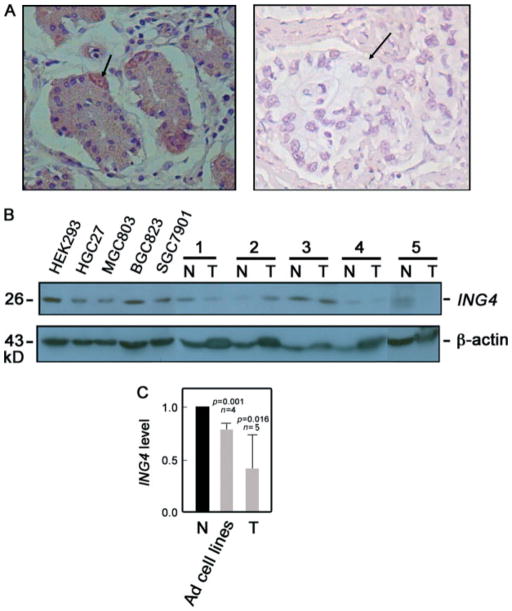
We then proceeded to examine ING4 protein expression by immunohistochemistry. Of the 40 patients examined, we found weak ING4 staining (0 to +) in 72.5% (29/40) of the tumour tissues and in 20% (8/40) of the adjacent normal tissues (p = 0.000; data not shown). Thus, 52.5% more of the tumour tissues than the normal tissues showed weak ING4 staining. Generally, we found stronger ING4 staining in the nucleus than in the cytoplasm, and staining was observed in almost all normal tissues (as in Figure 2A, left panel), while reduced or lack of staining was mostly noted in tumour tissues (as in Figure 2A, right panel). Strong ING4 staining was also seen in parietal cells (Figure 2A, left panel, arrow).
Figure 2.
ING4 protein expression is reduced in gastric adenocarcinoma cell lines and tissues. (A) ING4 expression in representative paired tumour (right panel) and normal adjacent (left panel) tissues was analysed by immunohistochemistry. Original magnification: 400 ×. Arrows indicate a stomach parietal cell (left panel) and a stomach adenocarcinoma nest (right panel). (B) Representative adenocarcinoma cell lines (HGC27, MGC803, BGC823, and SGC7901) and paired tumour (T) and normal (N) adjacent tissues (n = 5) were examined for ING4 protein expression by western blot analysis. HEK293 cells served as a positive control. β-actin blot was used for internal loading control. The 29 kD ING4 band detected in gastric adenocarcinoma tissues and cell lines is consistent with the possibility that the epitope recognized by the ING4 (T-15) antibody lies in an internal region of ING4 v1 and ING4 v2 but is absent or masked in the aberrantly spliced variant forms. (C) Analysis of the ING4/β-actin ratio following densitometric analysis of ING4 immunoblot. Values of normal tissues were normalized to 1. Statistical analysis was performed by the t-test tissue
To support our immunohistochemistry data, we examined representative normal and adenocarcinoma cell lines and tissues by western blotting (Figure 2B). Consistent with our results above, densitometry of blots and analysis of the ING4/β-actin ratio (Figure 2C) revealed that stomach adenocarcinoma cell lines and tissues express lower ING4 protein levels compared with normal tissues.
To evaluate ING4 protein expression more thoroughly in gastric adenocarcinoma, we utilized the microarray–immunohistochemistry method to analyse 80 tumour and 40 normal tissues further. Consistent with our RT-PCR/real-time RT-PCR results, Table 1 shows that most of the tumour tissues had negligible or low ING4 levels. In fact, out of 120 tumour tissues, 99 (82.5%) showed weak ING4 signals. Conversely, of the 80 normal tissues, 30 (37.5%) showed weak ING4 signals. Therefore 45% more of the tumour than normal tissues showed reduced ING4 expression. It is striking that ING4 was negligible in 43 (35.8%) of the tumour tissues but in only 3 (3.8%) of the normal tissues, suggesting that lack of ING4 may indeed be involved in the initiation and progression of gastric adenocarcinoma.
Table 1.
Analysis of ING4 expression in normal stomach and gastric adenocarcinoma tissues
p < 0.05.
p values were calculated by the Mann–Whitney test.
We then further examined potential correlations between ING4 protein expression and patient gender or age, and extent of differentiation of the adenocarcinoma. Table 2 shows that about 80% of affected males (26/31) and females (73/89) have reduced ING4 expression. Among them, however, 54.8% of females have negligible levels of ING4 as opposed to 29.2% in males. Female patients are therefore almost twice as likely to lack ING4. Although a higher percentage of patients 60 years old and younger (48/64) seemed to show reduced ING4 levels (0 to +) compared with patients older than 60 (31/56), the difference was statistically insignificant. Similarly, differences in ING4 expression in poorly, moderately, and well-differentiated tumour cells were not statistically significant. Therefore, as with the mRNA levels, ING4 protein levels did not correlate with age or extent of differentiation of the stomach adenocarcinoma. In addition, no significant difference (p = 0.112) in ING4 protein levels was observed in the intestinal and diffuse types of stomach adenocarcinomas (data not shown).
Table 2.
ING4 expression by gender, age, and extent of gastric adenocarcinoma differentiation by TMA analysis (n = 120)
| Variable | N |
ING4
|
p | ||||
|---|---|---|---|---|---|---|---|
| 0 | + | ++ | +++ | ||||
| Gender | Male | 31 | 17 | 9 | 5 | 0 | |
| Female | 89 | 26 | 47 | 15 | 1 | 0.043*† | |
| Age, years | ≤60 | 64 | 20 | 28 | 8 | 0 | |
| >60 | 56 | 11 | 20 | 4 | 3 | 0.689† | |
| Differentiation | Poorly differentiated | 66 | 29 | 30 | 6 | 1 | |
| Moderately differentiated | 37 | 11 | 17 | 9 | 0 | ||
| Well differentiated | 13 | 3 | 8 | 2 | 0 | 0.125‡ | |
p < 0.05.
p values were calculated by the Mann–Whitney test.
p values were calculated by the Kruskal–Wallis test.
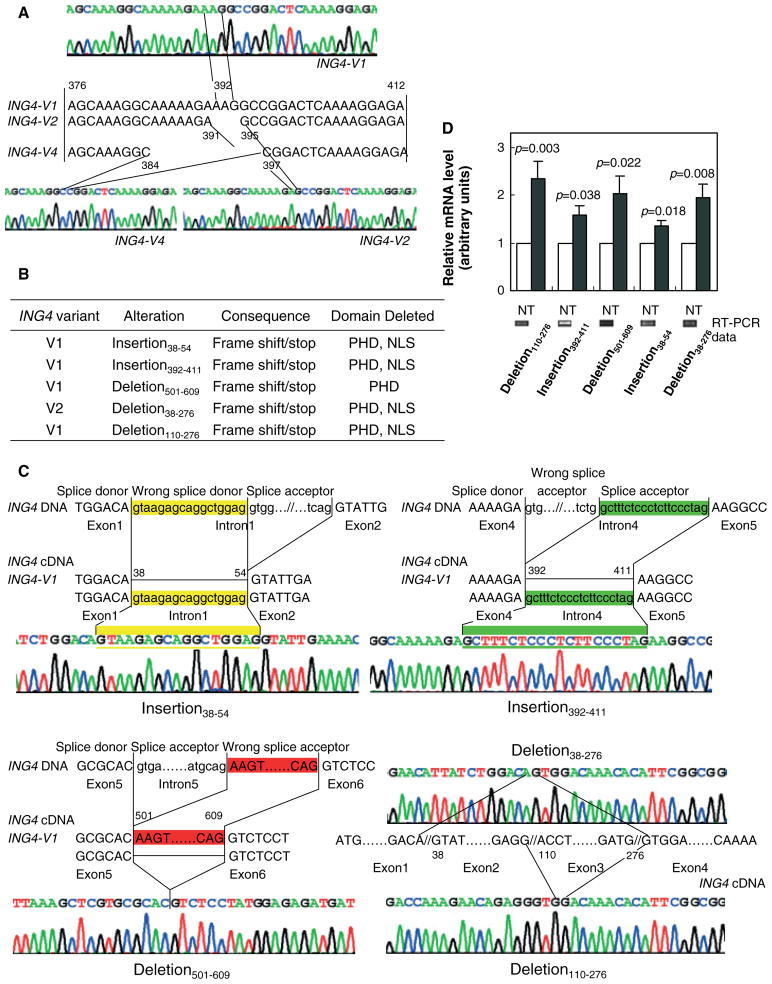
ING4 transcript analysis revealed novel spliced variants of ING4 v1 and ING4 v2
Alterations of ING4 transcripts in gastric adenocarcinoma have not yet been elucidated. We therefore examined all the coding regions of ING4 for potential alterations at the transcriptional level in nine representative gastric adenocarcinoma tissues and cell lines by clone sequencing. Our data indicated that ING4 v1, ING4 v2, and ING4 v4, but not ING4 v3, were present in all of the adenocarcinoma samples examined and their sequence integrity was well maintained (Figure 3A). Interestingly, we discovered four novel spliced variants of ING4 v1 (two insertions and two deletions) (Figures 3B and 3C). We also found a novel deletion in ING4 v2. A summary of these alterations, their consequences, and deleted domains are presented in Figures 3B and 4. Specifically, in ING4 v1, we found insertion at positions 38–54, reserving the head of the first intron (Figure 3C), and another insertion at positions 392–411, reserving the end of the fourth intron (Figure 3C). These insertions cause a frame-shift, deleting the bipartite nuclear localization signal (NLS) domain and the c-terminal plant homeo-domain (PHD) zinc-finger domain (Figure 4). We further found deletion at positions 501–609 in ING4 v1 (Figure 3C), deleting the head of the sixth exon, which may cause ING4 products to lose the PHD zinc-finger domain (Figure 4).
Figure 3.
Five novel aberrantly spliced variant forms of ING4 v1 and ING4 v2 were found in gastric adenocarcinoma tissues. (A) ING4 v1, ING4 v2, and ING4 v4 (but not ING4 v3) were detected in stomach adenocarcinoma cell lines and tissues. The partial sequence maps of ING4 v1, v2 and v4, and cDNA sequences of the variable region (exon 4) of the three ING4-spliced variants are shown. (B) List of the five novel ING4 v1 and ING4 v2 insertions and/or deletions. PHD and NLS stand for the c-terminal plant homeo-domain zinc-finger domain and the bipartite nuclear localization signal domain, respectively. (C) Schematic representations of 38–54 (yellow; upper left panel) and 392–411 (green; upper right panel) insertions, and 501–609 (red; lower left panel) deletion in ING4 v1. Upper- and lowercase letters represent exon and intron sequences of ING4 genomic DNA, respectively. Splice donor, wrong splice donor causing the alteration, and splice acceptor are noted. ING4 v1 and ING4 v2 sequences are also shown. Schematic representations of deletion (38–276) of ING4 v2 and deletion (110–276) of ING4 v1 (lower right panel). Sequence maps of ING4 exons 1, 2, 3, and 4 indicate that stomach adenocarcinoma tissues harbour deletion of nucleotides 38–276 which include the second exon and the third exon of ING4 v2, and deletion of nucleotides 110–276 which include the third exon of ING4 v1 in the stomach adenocarcinoma cell line MGC-803. (D) The mRNA levels of the ING4 variants in normal (N) and tumour (T) tissues. Deletion 110–276 (T, n = 15; N, n = 13). Insertion 392–411 (T, n = 8; N, n = 11). Deletion 501–609 (T, n = 12; N, n = 11). Insertion 38–54(N, n = 7; N, n = 7). Deletion 38–276 (N, n = 10; T, n = 9). The values for normal tissues were normalized to 1. Representative RT-PCR gels for normal tissues are also shown
Figure 4.
Amino acid sequence alignment of the aberrantly spliced variant forms of ING4 v1 (A) and ING4 v2 (B) found in gastric adenocarcinoma tissues. Uppercase letters represent sequence identity, while lowercase letters in red indicate sequence variations due to a frame-shift in translation. (C) Diagrammatic representation of the aberrantly spliced variant forms of ING4 v1 and ING4 v2. Different conserved domains are indicated by different coloured boxes, including a leucine zipper-like motif (LZL); the nuclear localization sequence (NLS); and the plant homeo-domain (PHD). The red box indicates the region where sequence variations occur due to a frame-shift in translation
In ING4 v2, we found deletion at positions 38–276, deleting the whole second and third exons, which results in truncation of ING4, and deletion of both NLS and PHD zinc-finger domains (Figure 4). These four alterations were found singly or in combination (ie 38–54 insertion and 501–609 deletion) in three out of the nine tumour tissues examined. In addition, in MGC803 adenocarcinoma cells, we observed deletion at positions 110–276 in ING4 v1, deleting the whole third exon, which results in ING4 truncation, and subsequently, deletion of both NLS and PHD zinc-finger domains as well (Figure 4). The ING4 variants were also detected in normal gastric tissues but at considerably lower levels than in tumour tissues (Figure 3D). Up-regulation of ING4 variants did not correlate with patient clinical parameters. Sequence analysis of ten adenocarcinoma tissues did not show ING4 gene mutation (data not shown).
Discussion
The tumour suppressor ING4 has been shown to be deleted or down-regulated in gliomas, breast tumours, and head and neck squamous cell carcinomas [5]. For example, in head and neck squamous cell carcinomas, ING4 mRNA is reduced in 76% of primary tumours [5]. In glioblastomas, ING4 levels are up to six times lower than in normal brain tissues [4]. Here, we found that ING4 mRNA was likewise significantly lower in gastric adenocarcinoma tissues than in normal stomach tissues. Reduced ING4 mRNA level correlated with depth of penetration of the tumour tissue. Thus, ING4 levels in advanced tumour stages 3 and 4 (T3 – 4) are lower than those in earlier tumour stages 1 and 2 (T1– 2), which in turn have ING4 levels lower than those in normal tissues. Therefore, the pattern of ING4 expression in normal and T1 – 2 and T3 – 4 gastric adenocarcinoma is consistent with that in normal, glioma, and glioblastoma tissues.
Worldwide, males are more predominantly affected than females by adenocarcinoma at any site in the stomach [8]. This has been associated with oestrogen-regulated progression of gastric adenocarcinoma. To some extent, increased lifetime exposure to oestrogen and/or progesterone may protect women from gastric cancer [9]. Indeed, there are α- and β-type oestrogen receptors (ERs) in normal and cancerous gastric mucosa [10]. Supposedly, ING1b stimulates oestrogen-induced ER-transcriptional activity in a dose-dependent manner, and this activation is consistent with ING1b function in chromatin remodelling [11]. Here, we found stronger ING4 staining in parietal cells than in any other cells in the normal gastric tissue. Interestingly, gastric parietal cells can synthesize and secrete oestrogen into the portal vein [12]. We then hypothesized that ING4 might also be involved in ER signalling that possibly protects women from gastric cancer. However, we found that ING4 levels in tumour tissues of female patients were significantly lower than those in male patients. Thus, ING4 may not play a significant role in ER signalling, which seems to reduce gastric cancer incidence in women. Furthermore, it appears that ING4 is not responsible for the higher incidence of male gastric adenocarcinoma.
Inactivating mutations in ING4 transcripts have been found in various human cancer cell lines. These mutations were localized in the NLS and PHD finger domains, or cause the loss of NLS and PHD finger domains [6]. The PHD zinc-finger domain, a functionally conserved structural domain of ING4, can interact with histone acetyltransferase (HAT) and histone deacetylase (HDAC) complexes, which are involved in gene transcriptional regulation [13]. Conversely, the NLS domain is necessary for the effect of ING4 on p53-mediated apoptosis [14]. Certainly, overexpression of ING4 causes a decreased population of S-phase cells, and p53-dependent apoptosis associated with p21 transcriptional up-regulation [7]. However, ING4 also participates in apoptosis induced by the RUNX3 tumour suppressor in MKN-1 gastric adenocarcinoma cells [15]. Therefore, in addition to its tumour-suppressing activity, ING4 may cooperate with other tumour suppressor proteins to induce tumour cell apoptosis in gastric cancer.
ING4, which has been mapped to chromosome 12p13.31, possesses hybridization deletion at the ING4 locus in 10–20% of breast cancer cell lines and primary breast tumours, and in 66% of head and neck squamous cell carcinomas [5,6]. Deletion and missense mutations in ING4 transcripts have also been detected in several tumour cell lines [6], with the most frequent mutation identified as ING4 v4, a variant that is also detectable in normal cell lines and tissues [4,16,17]. Initial studies showed the presence of both ING4 v1 (ING4) and ING4 v4 in the T47D breast cancer cell line [16], but the four identified variants, ING4 v1, ING4 v2, ING4 v3, and ING4 v4, are also present in both normal and tumour tissues, and the percentages of each variant are similar in adult normal tissues [17,18]. In separate studies, the splice variants, ING4 v2, ING4 v3, and ING4 v4, which lack the full NLS domain, were shown to have attenuated ING4 function [17]. These variants were localized in the cytoplasm, supporting the important role of NLS in nuclear localization of ING4 [17].
Here, we found that ING4 v1, ING4 v2, and ING4 v4 were present in stomach adenocarcinoma tissues and that their sequence integrity was maintained (Figure 3A). However, we also discovered four novel ING4 v1 and ING4 v2 spliced variants (insertion and/or deletion) in these tissues. In ING4 v1, we found insertions at positions 38–54 and 392–411, reserving the head of the first intron and the end of the fourth intron, respectively. These insertions cause a frame-shift, deleting the bipartite NLS domain and the c-terminal PHD zinc-finger domain. ING4 v1 deletion at positions 110–276 and 501–609 deletes the third exon and the head of the sixth exon, respectively, causing the ING4 products to lose the PHD zinc-finger domain. We also found ING4 v2 deletion at positions 38–276, deleting the whole second and third exons. This results in truncated ING4 products with deleted NLS and PHD zinc-finger domains. The fact that these alterations result in frame-shift, stop translation, and, eventually, NLS domain (which targets p53) or PHD domain (which targets HAT/HDAC complexes and subsequently regulates gene transcription) deletion suggests that these ING4 variants could contribute to p53 mislocalization and/or HAT/HDAC complexes, and subsequently, altered gene expression in stomach adenocarcinoma.
In separate studies, newly identified ING4 variants, designated as ING4-DEx2, -DEx3, and -DEx6A and -DEx6B, were found to lack exons 2, 3, and 6 (entirely or in part), respectively [16]. All variants were detected in normal human thyroid, brain, and lung tissues but ING4-DEx2 and -DEx6B were also detected in tumour tissues. Aberrant alternative splicing caused by such mutations, which create or disrupt splice sites, correlates with loss of tumour suppressor activity [19]. Potentially, the novel spliced ING4 v1 and ING4 v2 variants in gastric adenocarcinoma result from ING4 mutations that subsequently cause altered ING4 function. It appears that reduced ING4 expression combined with attenuated ING4 function due to insertions and/or deletions promotes the initiation and progression of gastric adenocarcinoma.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No 30 572 092), the Natural Science Foundation of Heilongjiang Province (No LC07C11), and a Research Project of MOE (No 208 035). K-Y Lee is an Alberta Heritage Foundation for Medical Research Senior Scholar.
Footnotes
No conflicts of interest were declared.
References
- 1.Forman D, Burley VJ. Gastric cancer: global pattern of the disease and an overview of environmental risk factors. Best Pract Res Clin Gastroenterol. 2006;20:633–649. doi: 10.1016/j.bpg.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Ngoan LT, Yoshimura T. Pattern and time trends of stomach cancer in Asia from 1950–99. Asian Pac J Cancer Prev. 2002;3:47–54. [PubMed] [Google Scholar]
- 3.Russell M, Berardi P, Gong W, Riabowol K. Grow-ING, Age-ING and Die-ING: ING proteins link cancer, senescence and apoptosis. Exp Cell Res. 2006;312:951–961. doi: 10.1016/j.yexcr.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, et al. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature. 2004;428:328–332. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- 5.Gunduz M, Nagatsuka H, Demircan K, Gunduz E, Cengiz B, Ouchida M, et al. Frequent deletion and down-regulation of ING4, a candidate tumor suppressor gene at 12p13, in head and neck squamous cell carcinomas. Gene. 2005;356:109–117. doi: 10.1016/j.gene.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Kim S, Chin K, Gray JW, Bishop JM. A screen for genes that suppress loss of contact inhibition: identification of ING4 as a candidate tumor suppressor gene in human cancer. Proc Natl Acad Sci U S A. 2004;101:16251–16256. doi: 10.1073/pnas.0407158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373–2378. [PubMed] [Google Scholar]
- 8.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 9.Lindblad M, Ye W, Rubio C, Lagergren J. Estrogen and risk of gastric cancer: a protective effect in a nationwide cohort study of patients with prostate cancer in Sweden. Cancer Epidemiol Biomarkers Prev. 2004;13:2203–2207. [PubMed] [Google Scholar]
- 10.Takano N, Iizuka N, Hazama S, Yoshino S, Tangoku A, Oka M. Expression of estrogen receptor-alpha and -beta mRNAs in human gastric cancer. Cancer Lett. 2002;176:129–135. doi: 10.1016/s0304-3835(01)00739-x. [DOI] [PubMed] [Google Scholar]
- 11.Toyama T, Iwase H, Yamashita H, Hara Y, Sugiura H, Zhang Z, et al. p33(ING1b) stimulates the transcriptional activity of the estrogen receptor alpha via its activation function (AF) 2 domain. J Steroid Biochem Mol Biol. 2003;87:57–63. doi: 10.1016/s0960-0760(03)00388-1. [DOI] [PubMed] [Google Scholar]
- 12.Ueyama T, Shirasawa N, Numazawa M, Yamada K, Shelan-gouski M, Ito T, et al. Gastric parietal cells: potent endocrine role in secreting estrogen as a possible regulator of gastro-hepatic axis. Endocrinology. 2002;143:3162–3170. doi: 10.1210/endo.143.8.8974. [DOI] [PubMed] [Google Scholar]
- 13.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Wang KS, Wang ZQ, Xu LS, Wang QW, Chen F, et al. Nuclear localization signal of ING4 plays a key role in its binding to p53. Biochem Biophys Res Commun. 2005;331:1032–1038. doi: 10.1016/j.bbrc.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 15.Nagahama Y, Ishimaru M, Osaki M, Inoue T, Maeda A, Nakada C, et al. Apoptotic pathway induced by transduction of RUNX3 in the human gastric carcinoma cell line MKN-1. Cancer Sci. 2008;99:23–30. doi: 10.1111/j.1349-7006.2007.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raho G, Miranda C, Tamborini E, Pierotti MA, Greco A. Detection of novel mRNA splice variants of human ING4 tumor suppressor gene. Oncogene. 2007;26:5247–5257. doi: 10.1038/sj.onc.1210335. [DOI] [PubMed] [Google Scholar]
- 17.Unoki M, Shen JC, Zheng ZM, Harris CC. Novel splice variants of ING4 and their possible roles in the regulation of cell growth and motility. J Biol Chem. 2006;281:34677–34686. doi: 10.1074/jbc.M606296200. [DOI] [PubMed] [Google Scholar]
- 18.Tsai KW, Lin WC. Quantitative analysis of wobble splicing indicates that it is not tissue specific. Genomics. 2006;88:855–864. doi: 10.1016/j.ygeno.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]