Abstract
Human monoacylglycerol lipase (hMGL) regulates endocannabinoid signaling primarily by deactivating the lipid messenger 2-arachidonoylglycerol. Agents that carbamylate hMGLs catalytic Ser122 constitute a leading class of therapeutically promising hMGL inhibitors. We have applied peptide-level hydrogen/deuterium exchange mass spectrometry to characterize hMGL’s conformational responses to two potent carbamylating inhibitors, AM6580 (irreversible) and AM6701 (slowly reversible). A dynamic, solvent-exposed lid domain is characteristic of hMGL’s solution conformation. Both hMGL inhibitors restricted backbone enzyme motility in the active-site region and increased substrate binding-pocket solvent exposure. Covalent reaction of AM6580 with hMGL generates a bulkier carbamylated Ser122 residue as compared to the more discrete Ser122 modification by AM6701, a difference reflected in AM6580’s more pronounced effect upon hMGL conformation. We demonstrate that structurally distinct carbamylating hMGL inhibitors generate particular conformational ensembles characterized by regionspecific hMGL dynamics. By demonstrating the distinctive influences of two hMGL inhibitors on enzyme conformation, this study furthers our understanding at the molecular level of the dynamic features of hMGL interaction with small-molecule ligands.
The endocannabinoid system is a ubiquitous signaling network involved in numerous (patho)physiological processes including metabolic control, emotional reactivity, pain sensing, and inflammation.1 Cannabinergic signaling in the mammalian central nervous system (CNS) mainly reflects the full-agonist action of the endogenous lipid mediator, 2- arachidonoylglycerol (2-AG), at the cannabinoid 1 G-protein coupled receptor (CB1R).2 As the serine hydrolase mainly responsible for catalytic 2-AG deactivation in vivo, monoacylglycerol lipase (EC 3.1.1.23) (MGL) is a decisive regulator of 2-AG signaling and, consequently, the production of various neurochemicals in the CNS that underlie synaptic efficiency and influence psychobehavioral status.3,4
Increasing interest has focused on MGL inhibition as a therapeutic modality for enhancing indirectly cannabinergic signaling through elevation of tissue 2-AG tone.5–7 Phenotypic benefits associated with genetic or pharmacological disruption of MGL partly recapitulate those of exogenous CB1R agonists while appearing to pose less risk of the undesirable motor and behavioral effects classically associated with systemic CB1Ragonist application.5,7 Indeed, MGL inhibitors may be an attractive means of augmenting, in a site- and event-specific manner, the increased tissue endocannabinoid levels observed under at least some pathological conditions and considered reflective of an intrinsic adaptive response to restore homeostasis.8–10 The role of downstream products from monoacylglycerol hydrolysis in the etiologies of cancer and inflammation provides additional support for the therapeutic potential of MGL inhibitors.11,12 Although tonic potentiation of tissue 2-AG levels may impair 2-AG-mediated antinociception by desensitizing CB1R,13,14 MGL inhibitors have demonstrated therapeutic benefit in acute preclinical models of inflammation, pain, anxiety, and CNS injury without inciting adverse events.15–18
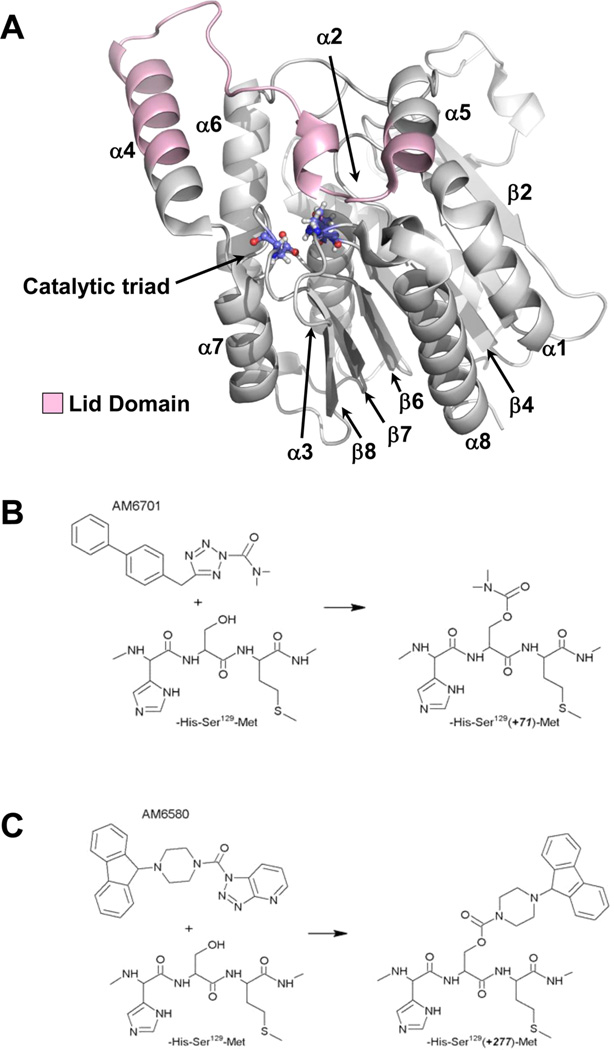
These considerations have promoted the rational design of temporally tuned MGL inhibitors that can potentiate subsets of 2-AG-related signaling circuits to effect a clinically attractive severance of adverse events associated with exogenous CB1R agonists from therapeutic gain. By leveraging the essential role of MGL’s active-site serine nucleophile (Ser122) (Figure 1A) in forming the initial transition state during the catalytic cycle,19 we20–22 and others6,23 have generated several potent, structurally distinct carbamylating inhibitors that inactivate human MGL (hMGL) through covalent enzyme carbamylation at Ser122. Yet optimal translational exploitation of hMGL inhibitors as medicines necessitates a thorough understanding of the inhibitor-interaction profiles that define potent inhibitors with varying hMGL selectivities and durations of action. In this regard, we have utilized two hMGL carbamylating inhibitors, [4-(9H-fluoren-9-yl)piperazin-1-yl][1,2,3]triazolo[4,5-b]-pyridin-1-ylmethanone (AM6580)24 and 5-((biphenyl-4-yl)- methyl)-N,N-dimethyl-2H-tetrazole-2-carboxamide (AM6701) (also known as the 2,5-regioisomer of LY2183240)20,22 (Figure 1B,C) as tool compounds for probing the molecular details of pharmacological hMGL inactivation. Both agents inhibit hMGL in the low nanomolar range through a common mechanism involving rapid carbamylation of Ser122 in the enzyme’s catalytic triad.21,22 Within a 3-h experimental time frame, hMGL inhibition by AM6580 is irreversible, whereas the carbamate group on hMGL Ser122 resulting from enzyme modification byAM6701 begins to be hydrolyzed very slowly such that ≥24 h are required to regenerate catalytically competent hMGL.20,21 This biochemical distinction suggests that the interaction profiles of these two chemically distinct carbamylating inhibitors may impact hMGL conformation in distinctive ways that have contingent implications regarding, for example, the duration and reversibility of hMGL inhibition.
Figure 1.
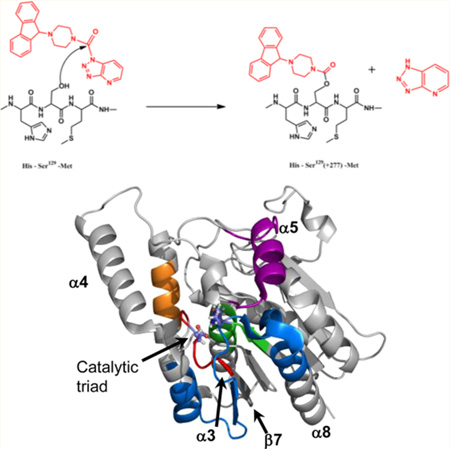
Structure of apo-6-His-hMGL and chemical structures of AM6701 and AM6580 and schematic representation of their carbamylation reaction with hMGL. (A) The catalytic triad (Ser129-Asp246-His276, red/white/blue ball-and-stick depiction), lid domain (residues 158–191, light pink), and secondary structural elements (α-helices 1–8 and β-sheets 2, 4, 6–8) of apo-6-His-hMGL are indicated on the wild-type hMGL crystal structure, PDB ID: 3JW8. (B, C) hMGL inhibitors AM6701 and AM6580 carbamylate the 6-His-hMGL Ser129 active-site nucleophile located between His128 and Met130 in the enzyme’s primary sequence to form the covalent adducts depicted, resulting in theoretical mass increases of +71 and +277, respectively. See also Figure S1 of the Supporting Information.
We report application of peptide-level, solution-phase hydrogen/deuterium exchange mass spectrometry (H/DXMS) to gain detailed insight into the dynamic transitions in hMGL’s solution structure consequent to AM6580 and AM6701 engagement and inhibitor-induced covalent enzyme modification. H/DX-MS involves mass-spectrometry (MS) measurements of backbone amide hydrogens substituted by the heavier deuterium isotope from deuterated water (D2O) at ambient temperature and physiological pH.25,26 The rate of incorporation of deuterium is modulated by the conformational properties of the protein as well as pH and temperature and thus reports on the combined effects of solvent exposure and the hydrogen-bonding characteristic of the protein backbone. Following acid quenching of the exchange reaction, proteolytic digestion with pepsin is used to localize where along the protein backbone the deuterium had been incorporated. Rapid exchange is characteristic of more disordered, solvent-exposed protein regions, whereas slower and more limited exchange indicates a more compact, solvent-shielded, hydrogen-bonded state. Therefore, the difference between deuterium incorporation into apo versus inhibitor-modified hMGL serves as a proxy for the structural impact of the inhibitor’s interaction with the enzyme.
Our results demonstrate that chemically different, small-molecule carbamylating inhibitors generate distinct hMGL conformational ensembles characterized by regionally specific structural responses in the enzyme. To the authors’ best knowledge, these data offer the first direct experimental insight into the molecular dynamics of hMGL in solution upon covalent inhibitor modification. By detailing the hMGL structural excursions characteristic of the hMGL inactive state as induced by two potent, chemically distinct carbamylating inhibitors, the information presented has the potential to aid the design and targeting of new-generation hMGL inhibitors as candidate drugs.
EXPERIMENTAL PROCEDURES
Materials
Standard laboratory chemicals, culture media, isopropyl-β-d-thio-galactopyranoside, lysozyme, and DNase I were purchased from Sigma (St. Louis, MO) and Fisher Chemical (Pittsburgh, PA), unless otherwise specified. SDS– PAGE supplies and Bio Spin P-6 columns were from Bio-Rad (Hercules, CA). AM6580 was synthesized at the Center for Drug Discovery, Northeastern University, by standard routes. 1,2-Deuterium oxide (>99%) was purchased from Cambridge Isotope Laboratories (Andover, MA).
apo-6-His-hMGL Purification and Activity Assay
hMGL was expressed in Escherichia coli with an N-terminal 6-His tag (i.e., apo-6-His-hMGL) and isolated by immobilized metal-affinity chromatography essentially as detailed.27 In brief, five grams (wet-weight) of cells were resuspended in 40 mL of lysis buffer [(200 mM NaCl, 10 mM Na2HPO4, 10 mM NaH2PO4, pH 7.4, containing 1 mM dithiothreitol and 0.05% n-dodecyl-β-d-maltoside and supplemented with lysozyme (0.2 mg/mL) and DNase I (25 µg/mL)] and disrupted on ice by three 1-min sonication cycles, each consisting of 1-s sonication bursts at 50 W power at 5-s intervals (Vibra-Cell 500W, Sonics, Newtown, CT). The cell lysate was centrifuged at 20000g for 25 min at 4 °C. The supernatant was recovered and incubated with 2.0 mL (bed volume) pre-equilibrated BD Talon metalaffinity resin (Clontech, Mountain View, CA) for 1 h at 4 °C with gentle agitation. The suspension was then transferred to a gravity-flow column and allowed to settle. The resin was washed twice with 20 mL of lysis buffer containing 30 mM imidazole. 6-His-tagged protein was eluted in six fractions with 1.0 mL of lysis buffer containing 0.05% n-dodecyl-β-d-maltoside and 300 mM imidazole. Protein samples and molecular mass markers (Bio-Rad) were denatured at 70 °C for 5 min in Laemmli buffer containing 5% β-mercaptoethanol and resolved on SDS-PAGE gels that were stained with Coomassie blue. Enzyme activity was assessed as the hydrolytic production of arachidonic acid from 2-AG with LC quantification.21
Mass Determination of apo-6-His-hMGL and 6-His-hMGL Inhibited by AM6580
Purified apo-6-His-hMGL (3.1 µg, 3 µM) in 200 mM NaCl, 10 mM Na2HPO4, 10 mM NaH2PO4, 5% glycerol, pH 7.4, containing 1 mM dithiothreitol and 0.5% n-dodecyl-β-d-maltoside was incubated for 1 h at room temperature either in the absence of inhibitor or in the presence of AM6580 at an inhibitor/hMGL molar ratio of 4:1. At the start of the incubation and after 1 h, aliquots of each incubation mixture were removed and assayed for enzyme activity to verify that apo-6-His-hMGL was inhibited by AM6580 and was unaffected in the absence of inhibitor. Incubations were terminated by removing excess inhibitor with a Bio-Spin 6 column in the same buffer. For intact mass determination of active apo-6-His-hMGL and enzyme inhibited by AM6580, samples were injected onto a self-packed POROS 20 R2 protein trap (Applied Biosystems, Framingham, MA) and desalted with 1.0 mL of 0.05% aqueous trifluoroacetic acid at a flow rate of 500 µL/min. Protein was eluted into the mass spectrometer from an LC-10ADvp instrument (Shimadzu Scientific, Columbia, MD) using a linear 15–75% acetonitrile gradient over 4 min at 50 µL/min. MS analyses were carried out with an LCT-PremierXE mass spectrometer (Waters Corporation, Milford, MA) under conditions of a standard electrospray source, 3.0 kV capillary voltage, and 35 V cone voltage.
Deuterium Labeling
Deuterium labeling of purified samples of active apo-6-His-hGML and carbamylated enzyme inhibited by AM6701 or AM6580 was initiated with a 15-fold dilution of each enzyme sample (60 pmol of enzyme protein) into a D2O buffer (99.96% D) containing 200 mM NaCl, 10 mM Na2HPO4, 10 mM NaH2PO4, 5% glycerol, 0.02% n-dodecyl-β-d-maltoside, pH 7.4 (pD 7.43). At six specific time points (10 s, 5 min, 15 min, 30 min, 1 h, and 4 h), the labeling reaction was quenched by adding an equal volume of ice-cold acidic quench buffer (200 mM NaCl, 75 mM Na2HPO4, 75 mM NaH2PO4, 5% glycerol, pH 2.46, containing 0.05% n-dodecyl-β-d-maltoside). The samples were then frozen on dry ice and stored at −80 °C until liquid chromatography (LC)-MS analysis.
Pepsin Digestion and MS Analysis
Frozen samples were defrosted quickly and immediately injected into a Waters nanoACQUITY UPLC system with H/DX technology for online digestion and peptide separation, as described previously.28 Peptides originating from pepsin cleavage of apo-6-His-hMGL and 6-His-hMGL reacted with AM6701 or AM6580 were identified from the triplicate analysis of undeuterated control samples using a combination of Waters MSE technology on a Waters Q-TOF Premier instrument and ProteinLynx Global Server 2.5. Triplicate sets of data were collected on three different days for three independent, replicate samples. Common peptides were identified for apo-6-His-hMGL and enzyme carbamylated upon inhibition with either AM6701 or AM6580. All mass spectra were processed with DynamX software (Waters). Relative deuterium levels for each peptide were calculated by subtracting the average mass of a corresponding undeuterated control sample from that of the deuterium-labeled sample for isotopic distributions corresponding to the +1, +2, +3, and +4 charge state of the peptide. The aggregate error of each m/z measurement (i.e., the combined error from all sources including protein preparation, pH, temperature, mass measurement, etc.) was ±0.25, as determined by replicate analyses of peptide standards and prior H/DX-MS data from this experimental setup.29
RESULTS
Recombinant 6-His-hMGL Characterization and Covalent Modification by AM6580
We used our established procedures to overexpress catalytically active, N-terminal hexa-histidine tagged hMGL (6-His-hMGL) in E. coli and to purify the recombinant enzyme by immobilized metal-affinity chromatography. Congruent with prior results,27 SDS-PAGE followed by either Coomassie blue staining or Western blot analysis with anti-5-His antibody demonstrated that the 6-HishMGL used in the current studies was a single monomeric protein whose ~35 kDa molecular mass estimated by SDS-PAGE was consistent with the enzyme’s precisely measured mass of 34178.4 Da (data not shown and Figure S1A of the Supporting Information).
Under the assay conditions specified,22 the 6-His-hMGL preparations used in the present work evidenced an apparent Km of 44 µM and Vmax of 208 µM/min/µg with 2-AG substrate. As previously demonstrated,20–22,24 both AM6701 and AM6580 inhibit the hydrolytic activity of wild-type hMGL and engineered hMGL variants with IC50 values ≤ 88 nM. Prior LC-MS analyses of tryptic peptides generated after the interaction of 6-His-hMGL with AM670127 and hMGL variants retaining the putative Ser122-Asp239-His269 catalytic triad (designated Ser129-Asp246-His276 for 6-His-hMGL) with AM658021 demonstrated directly that both AM6701 and AM6580 are carbamylating agents that covalently modify the enzyme’s catalytic Ser (Figure 1B,C). MS analysis of apo-6-HishMGL before and after incubation with AM6580 is congruent with this finding, since the expected mass increase following active-site Ser129 carbamylation was indeed observed upon apo-6-His-hMGL incubation with this inhibitor (Figure S1 of the Supporting Information). As we have previously observed,21,22 a 4-fold molar excess of either inhibitor relative to apo-enzyme ensured that the carbamylation reaction reached virtual completion.
H/DX-MS Analysis of apo-6-His-hMGL and Inhibitor-Modified Enzyme
Unmodified apo-6-His-hMGL and 6-HishMGL that was covalently carbamylated by either AM6701 or AM6580 were interrogated individually by H/DX-MS to assess the effects that inhibitory carbamylation would have on the enzyme’s solution-phase dynamics. Online pepsin digestion of deuterium-labeled, quenched samples of each state of 6-His-MGL resulted in 103 peptides that covered 100% of the enzyme’s backbone amide hydrogens (Figure S2A of the Supporting Information). Relative deuterium uptake as a function of time for each respective peptide is given in Figure S3 of the Supporting Information for apo-6-His-hMGL and enzyme covalently modified by either AM6701 or AM6580.
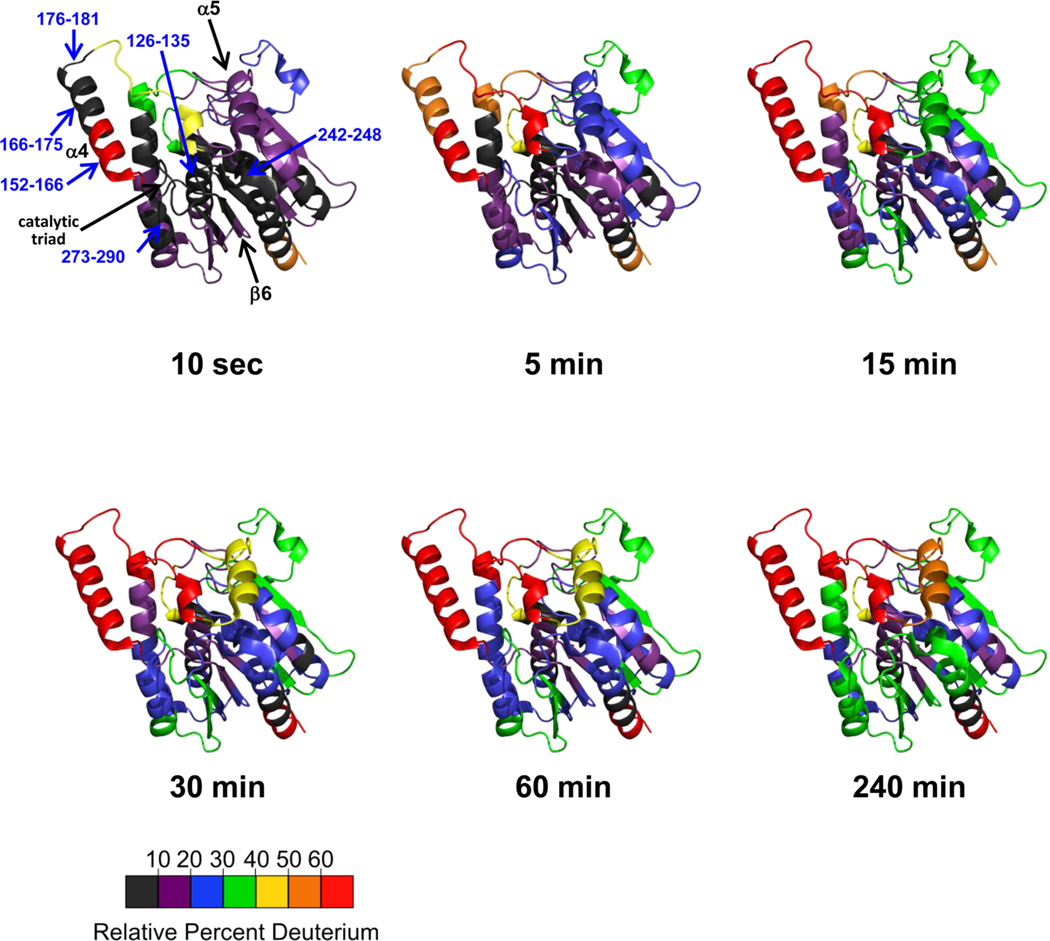
Deuterium incorporation into apo-6-His-hMGL during enzyme incubation periods in D2O ranging from 10 s to 4 h was visualized using the wild-type hMGL X-ray crystal structure, PDB ID: 3JW830 (Figure 2). Consistent with a well-folded and stable protein in solution, we did not observe high levels of deuterium incorporation at acute labeling times in most regions of the enzyme, prominent exceptions being peptides representing the amino- (residues 1–29) and carboxy- (residues 296–310) terminal regions, which showed rapid deuterium uptake (i.e., within 10 s after exposure to D2O) that reached maximal levels minutes thereafter. After 10 s of exposure to deuterium, 38% of the protein backbone was <10% deuterated and 76% of the protein was <20% deuterated (Figure 2 and Figure S3 of the Supporting Information). A core of relatively exchange-resistant regions in apo-6-His-hMGL persisted throughout the labeling time-course (Figure 2). Other apo-6-His-hMGL regions displayed dynamics reflected in steady deuterium incorporation over the 4-h D2O exposure. This phenomenon was especially evident in the lid domain, which covers residues 158–191 and encompasses the C-terminal two-thirds of helix α4, the loop that bridges helices α4 and α5, and the N-terminal turn of helix α5 (Figure 1A).30,31 Certain lid-domain peptides (i.e., peptide 152–166 leading from sheet β6 to helix α4; peptide 166–175 encompassing helix α4; and peptide 176–181 leading from helix α4 to helix α5) had patterns of deuterium uptake distinctive from each other in kinetic and quantitative terms (Figure 2). Peptide 152–166 attained a maximal relative level of deuterium very rapidly (i.e., within 10 s of exposure to D2O), whereas deuterium uptake into lid-domain peptides 166–175 and 176–181 reached maxima more gradually, within the first 10 min of apo-6-HishMGL deuteration, with peptide 166–175 having far more deuterium incorporation relative to peptide 176–181. Conversely, peptides covering the catalytic triad were well protected from exchange. Peptides 126–135, 242–248, and 273–290, which include an amino acid each of the 6-His-hMGL catalytic triad Ser129-Asp246-His276, evidenced maximal deuterium-incorporation levels of only 12%, 38%, and 34%, respectively, after 4 h of D2O exposure (Figure 2). (See Figure S3 of the Supporting Information for the uptake graph of each peptide.)
Figure 2.
Deuteration of apo-6-His-hMGL. Deuterium incorporation into apo-6-His-hMGL is illustrated using the enzyme’s wild-type cartoon structure shown in Figure 1A. Data are presented as relative percent deuterium incorporation using the seven-color scale denoted at 10 s, 5 min, 15 min, 30 min, 60 min, and 240 min of exchange in D2O buffer. The contiguous nonoverlapping peptides used to create this figure are shown in Figure S2B of the Supporting Information, and the relative deuterium incorporation data for each peptide are shown in Figure S3 of the Supporting Information.
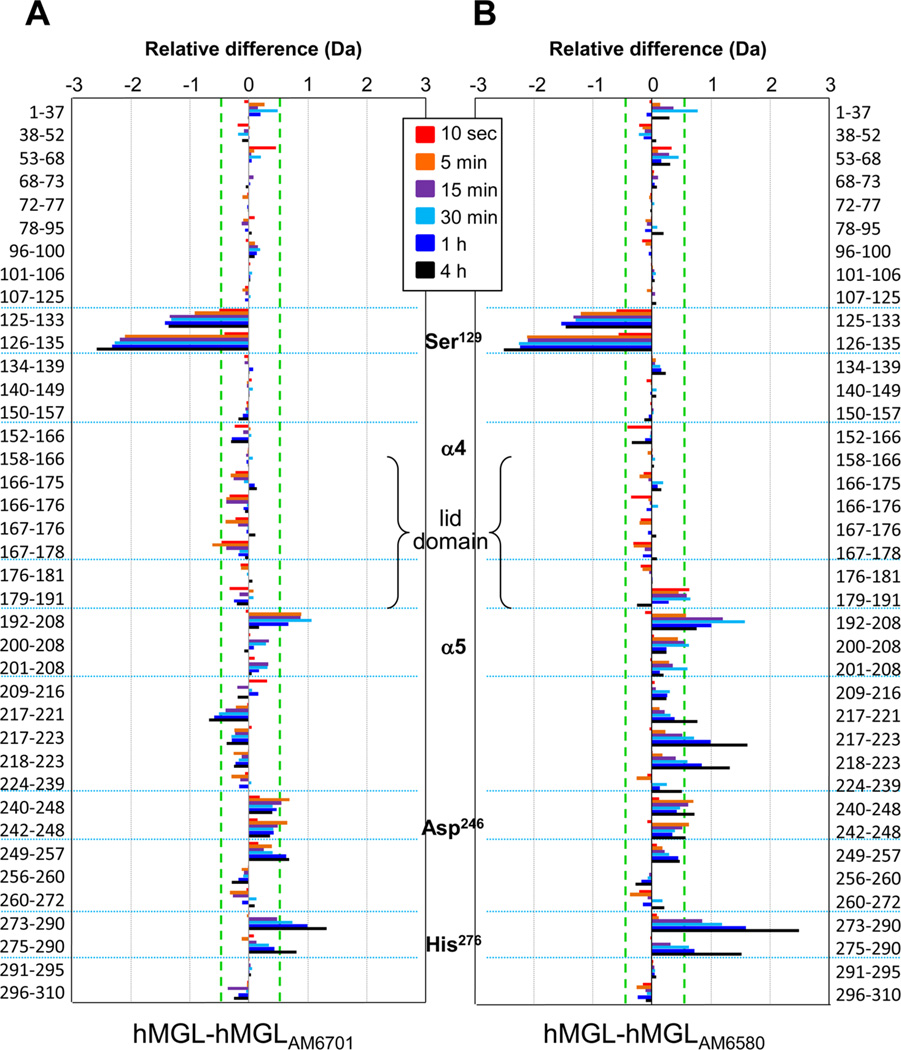
The deuterium exchange into apo-6-His-hMGL, as just described, was then compared to the deuterium exchange into the enzyme after carbamylation by either AM6701 or AM6580. The very gradual reversibility of hMGL inhibition by AM6701 requiring ≥24 h to regenerate catalytically competent hMGL20,21 ensured that virtually all (≥90%) of the enzyme remained inhibited by AM6701 even at the time of the last sampling from D2O incubation for H/DX-MS analysis (i.e., 4 h). For approximately 90% of the enzyme, no differences were observed in deuterium exchange between apo and inhibitor-modified 6-His-hMGL forms. Figure 3 graphically summarizes the differences in deuterium incorporation with time for all individual peptides observed as derived from the corresponding, peptide-level deuterium uptake graphs in Figure S3 of the Supporting Information. In Figure 3, a positive difference signifies increased protection (i.e., less deuterium incorporation) in inhibitor-modified versus apo-6-His-hMGL, whereas a negative difference signifies decreased protection (i.e., more deuterium incorporation) upon inhibitor modification. The aggregate experimental error of each m/z measurement with this experimental setup is ±0.25 Da, as determined by replicate analyses of peptide standards and prior H/DX-MS experiments using this system (see Experimental Procedures).25,26
Figure 3.
Difference plots for peptide-level deuterium uptake by apo-6-His-hMGL vs enzyme covalently modified by carbamylating agent AM6701 or AM6580. Relative difference plots for (A) AM6701-modified and (B) AM6580-modified 6-His-hMGL as compared to the unmodified apo-enzyme are displayed. The differences in deuterium incorporation between the carbamylated and apo enzymes are plotted for the deuterium-exchange time points color-coded, as indicated. The data shown represent the averages of three independent H/DX-MS experiments and are for a subset of the total 6-His-hMGL peptides listed that were common to all three experiments (see also Figure S2B of the Supporting Information). The peptides are listed from N- (top) to C-(bottom) termini at left and right in the figure. Major structural elements (including the beginning of helices α4 and α5 bracketing lid-domain residues 158–191) and the catalytic triad are indicated down the center of the figure. Significant differences in deuterium incorporation (greater/less than 0.5 Da; see Experimental Procedures) fall outside of the green dashed lines. In these graphs, a positive (rightward) difference indicates increased protection (i.e., less deuterium incorporation) in inhibitor-modified 6-His-hMGL, whereas a negative (leftward) difference indicates decreased protection (i.e., more deuterium incorporation) in inhibitor-modified 6-His-hMGL.
Three peptides of the 6-His-hMGL lid domain (i.e., peptides 152–166, 166–175, and 176–181) exemplify regions that were not affected by inhibitor modification of the enzyme. Likewise, the marginal decrease at select sampling times in deuterium uptake by some of the four peptides (i.e., 179–181, 179–191, 182–188, 182–191) encompassed within the terminus of the lid region comprising the distal portion of the loop leading into helix α5 and the initial portion of that helix (Figure S3 of the Supporting Information) upon AM6580-induced enzyme carbamylation was not significant when the overlapping peptide regions were considered in aggregate over the three independent H/DX-MS experiments.
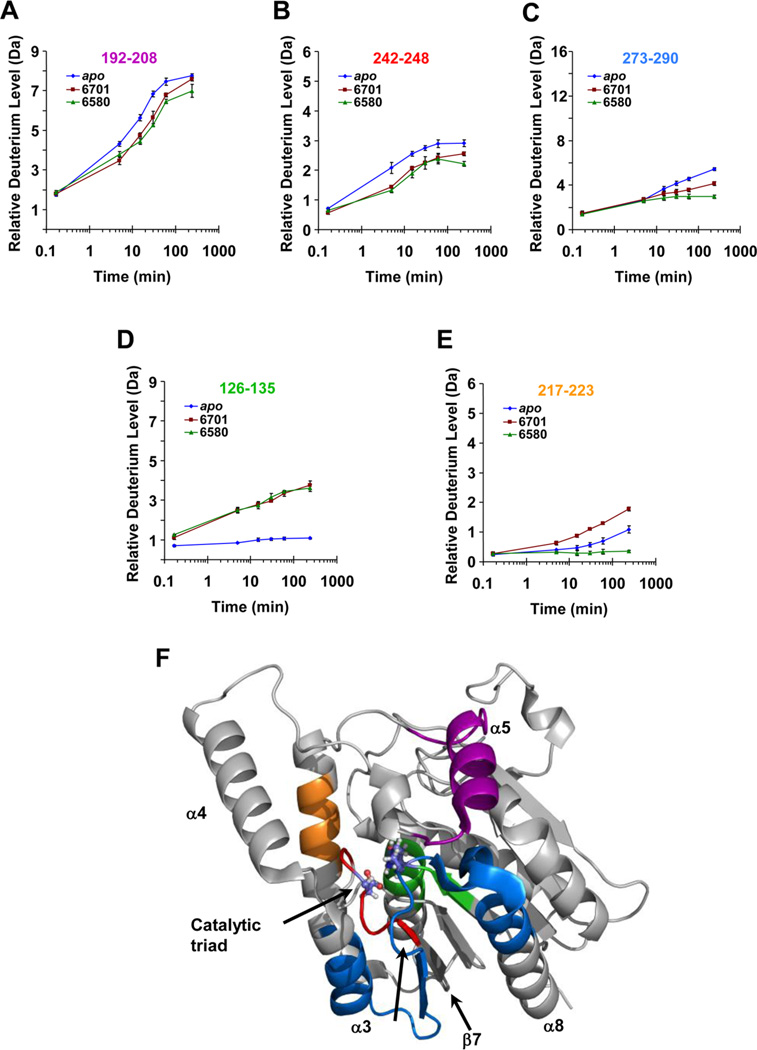
In ~8% of 6-His-hMGL-AM6701, and ~10% of 6-HishMGL-AM6580, deuterium exchange was significantly altered as a consequence of carbamylation (Figures 3 and 4). Carbamylation resulted in a >1 Da decrease (relative to deuterium incorporation in apo-6-His-hMGL) in deuterium uptake into peptide 192–208 (terminal region of helix α5 and loop conjoining helices α5 and α6) for both AM6701-and AM6580-modified forms after 30 min of enzyme exposure to deuterium (Figure 4A). Two peptides that have been localized to the active-site region,30,31 peptides 242–248 and 273–290, showed a decrease in deuterium incorporation upon inhibitor-induced modification of the catalytic Ser, with AM6580 exerting overall the more pronounced protective effect (Figure 4B,C). This protective influence of both AM6701 and AM6580 on some 6-His-hMGL peptides contrasts with the deprotecting effect of these inhibitors on deuterium exchange in other peptides, including peptide 126–135 (at the bottom of the substrate-binding pocket) and peptide 217–223 (helix α6).30,31 In apo-6-His-hMGL, peptide 126–135 was slow to exchange across the entire labeling time-course. Upon modification with AM6701 or AM6580, this peptide became more easily deuterated and showed a consistent increase in incorporation over the 4 h of exposure to deuterium. The relative difference in deuterium incorporated into this peptide reached its maximum difference of 2.25 Da at 4 h (Figure 4D). For peptide 217–223, 6-His-hMGL modification by AM6580 resulted in reduced deuteration, whereas Ser129 carbamylation by AM6701 modestly increased deuterium levels in this peptide (Figure 4E). Figure 4F depicts the localization of these peptides within the wild-type hMGL crystal structure, PDB ID: 3JW8.30
Figure 4.
Regional effects of carbamylating inhibitors AM6701 and AM6580 on H/DX-MS in 6-His-hMGL. (A–E) Plots of relative deuterium incorporation vs time for those specific apo-6-His-hMGL peptides generated by pepsin digestion in which either or both inhibitors elicited a significant difference in deuterium uptake relative to apo-6-His-hMGL. The maximum theoretical amount of deuterium incorporation possible for each respective peptide is designated by the amplitude of the y-axis in each plot. Each data point is the average of three independent experiments, and the error bars represent SD from the mean. apo-6-His-hMGL (blue diamonds) or 6-His-hMGL consequent to Ser129 carbamylation by AM6701 (red squares) or AM6580 (green triangles). (F) Locations of the peptides specified in deuterium incorporation plots (A–E) are color-coded within the wild-type hMGL crystal structure (PDB ID: 3JW8): 192–208, purple; 242–248, red; 273–290, blue; 126–135, green; 217–223, orange. See also Figure S3 of the Supporting Information. For orientation purposes, select α-helices and β-sheets are labeled, and the catalytic-triad residues are depicted in ball-and-stick fashion.
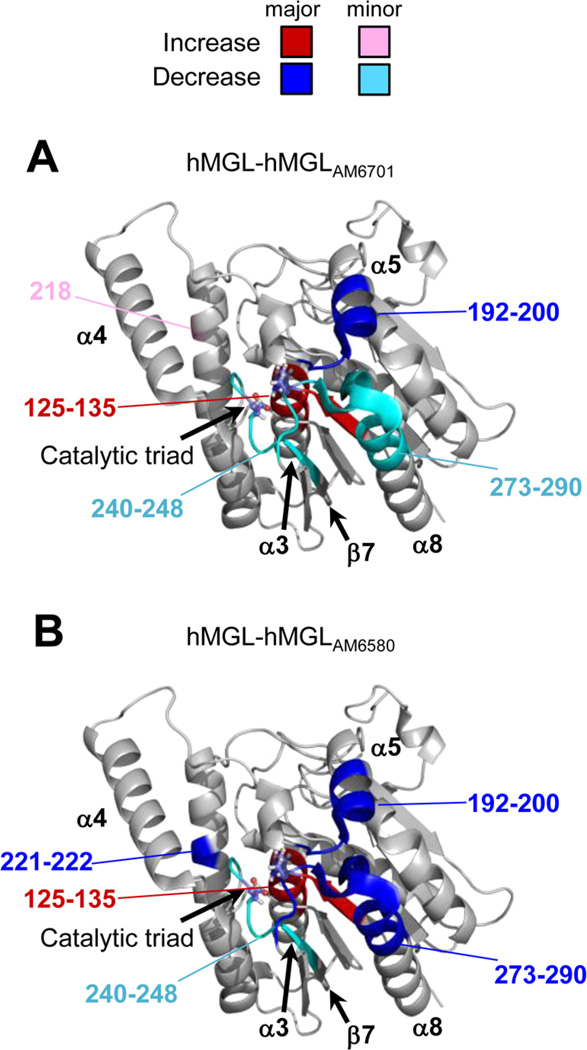
Since there were multiple overlapping peptides in the regions of inhibitor-induced changes in the H/DX-MS profiles of 6-HishMGL (cf. Figure S2 of the Supporting Information), not all the information about the effects of inhibitor binding on enzyme structure was captured in Figure 4, which considers only a select peptide subset. To extract more information, especially regarding the distinctions in hMGL conformational response to the two inhibitors, overlapping peptides were considered, and, when possible, the regions of alteration in deuterium exchange upon inhibitor-hMGL interaction were refined. Figure 5 depicts the refined inhibitor-induced conformational effects and indicates their magnitudes and locations within a wild-type hMGL ribbon structure representation (PDB ID: 3JW8)30 at any time point for both AM6701 (Figure 5A) and AM6580 (Figure 5B). These comparative H/DX-MS data substantiate the significant differences noted above in the structural impact of AM6580-induced carbamylation on the hMGL as compared to that of AM6701 in helix α6 and the N-terminus of helix α8, which resides immediately below lid-domain helix α4. The bulky carbamylating group at 6-His-hMGL Ser129 resulting from the enzyme’s covalent modification by AM6580 elicited more pronounced decreases in deuterium incorporation into helix α6 and the N-terminus of helix α8 than did AM6701, indicating greater shielding of these enzyme regions following AM6580-induced Ser129 carbamylation. Likewise, although both inhibitors increased solvent exposure in the vicinity of the hMGL active site, enhancement of deuterium uptake in this region was more pronounced following 6-His-hMGL Ser129 carbamylation by AM6580 than by AM6701.
Figure 5.
Regional differences in relative deuterium uptake due to 6-His-hMGL carbamylation by inhibitors AM6701 and AM6580. The hMGL crystal structure (PDB ID: 3JW8) is color-coded to indicate the magnitude and locations of the regional intramolecular effects of apo-6-His-hMGL covalent modification by the carbamylating inhibitors AM6701 (A) and AM6580 (B) on relative deuterium uptake by the enzyme. A major inhibitor-induced effect on deuterium uptake (at any exchange-in time point) is considered >1.0 Da; a minor effect (at any exchange-in time point), 0.5–1.0 Da. The regions of major (>1.0 Da) increase and decrease in relative deuterium incorporation due to inhibitor-induced enzyme carbamylation are highlighted in red and blue, respectively. The regions that showed minor (0.5–1.0 Da) increases or decreases in deuterium incorporation are highlighted in pink and teal, respectively. Those regions that showed no significant change in deuterium incorporation are colored gray. For orientation purposes, select α-helices and β-sheets are labeled, and the catalytictriad residues are depicted in ball-and-stick fashion. See also Figure S3 of the Supporting Information.
DISCUSSION
(De)localized backbone and side-chain motions involving active-site and more distant domains contribute to enzyme catalytic function, reaction-rate enhancement, and ligand (substrate, inhibitor) engagement.32 Agents that inhibit catalysis through covalent enzyme modification are associated with unique pharmacological properties that have been therapeutically exploited.33 Accordingly, appreciation of the structural consequences of the interaction between enzymes and small-molecule, active site-targeted inhibitors has proven essential to informing drug-discovery efforts and developing clinically useful therapeutics.34
The present work constitutes the first direct experimental analysis of the hMGL–inhibitor interaction landscape from the standpoint of the enzyme’s conformational dynamics in solution. To examine the consequences of inhibitor engagement on hMGL structure, we utilized as probes AM6580 and AM6701, two potent hMGL inhibitors representative of a leading candidate class of “druggable” hMGL inhibitors, serine-reactive carbamylating agents.5,7,20–23 As carbamylating agents at hMGL’s catalytic Ser122 (i.e., Ser129 for 6-His-hMGL), both AM6580 and AM6701 share the same mode of covalent hMGL inhibition.21,22 However, Ser122 modification by AM6580 leads to a significantly longer-lasting inactivation of hMGL as compared to that elicited by AM6701, which we have demonstrated to be a very slowly reversible hMGL inhibitor.20,21 Arguably, the duration of hMGL inhibition may constitute an important qualifier in applying such agents as potential therapeutics, since prolonged MGL inhibition or genetic ablation in laboratory animals can desensitize CB1R-mediated signaling.13,14 These considerations prompted our selection of AM6580 and AM6701 as hMGL molecular probes for this work.
H/DX-MS has proven to be a seminal technique for direct experimental detection and spatial resolution of enzyme backbone structural changes resulting from chemical modification/ inhibitor interaction.26,35–37 In order to discern the impact of two covalent carbamylating inhibitors on hMGL conformation, we applied the classic, continuous-labeling H/DX-MS approach to catalytically active apo-as well as inhibitor-modified 6-His-hMGL under physiologically relevant conditions. Although the intact mass analysis of apo-6-His-hMGL carbamylated by AM6580 or AM6701 showed the predicted mass increases by MS, we did not observe any peptides containing Ser129 modified with AM6580 or AM6701 (data not shown). This finding was expected on the basis of prior observations that the carbamyl group is lost when a carbamylated peptide derived from pepsin cleavage of an enzyme-carbamyl adduct is subjected to MS-analysis conditions.38,39 Therefore, our 6-His-hMGL-carbamyl adducts likely became destabilized outside of the intact 6-His-hMGL conformation and local enzyme microenvironment under our protein digestion conditions and/or during subsequent peptide MS. Loss of the carbamyl moiety from the peptic peptides in which it had been contained after hydrogen-exchange incubation and quenching of the exchange reaction would not negatively impact the reported deuterium uptake data for those 6-His-hMGL peptides containing Ser129.
The hydrogen-exchange profile of apo-6-His-hMGL established the reference point against which deuterium incorporation into inhibitor-modified enzyme species was compared. Our finding that high-level deuterium incorporation into apo-6-His-hMGL is largely confined to the enzyme’s amino- and carboxy-terminal regions and its lid domain (including helix α4) is consistent with the general nature of hMGL as a member of the α/β-hydrolase superfamily. The canonical α/β-fold core is comprised of eight β-sheets surrounded by six α-helices and a lid domain capping the active site.40–42 Consistent with this principal α/β-hydrolase folding motif and data from a refined hMGL homology model,43 wild-type apo-hMGL and unliganded variants crystallize as compact globular proteins whose ordered, shielded core contrasts with the enzyme’s more exposed lid and unstructured N- and C-terminal regions.30,31 Given this overall hMGL architecture and the ability of H/DX-MS to report on the solvent accessibility and hydrogen bonding of protein amide sites at the peptide level, our data directly demonstrate the greater incorporation of deuterium into the apo-6-His-hMGL lid domain and termini as compared to the rather modest incorporation into the more stable, core α/β-hydrolase fold.
The lid domain of α/β-hydrolases in general, and hMGL in particular, has been considered a dynamic modulator of substrate recruitment/accommodation during the catalytic cycle.3,31,44 Within apo-6-His-hMGL’s lid domain, we have identified two distinct subregions whose pronounced, yet markedly different, patterns of deuterium uptake are indicative of: (a) partial exposure to the solvent environment with a lack of detectable backbone dynamics (peptide 152–166, leading from sheet α6 to and including the beginning of helix α4) and/ or (b) considerable structural dynamics (peptides covering residues 166–191). Among the three published hMGL crystallographic studies,30,31,45 exceptionally large structural variations in the region of the lid domain spanning helix α4 have been noted.45 Upon the basis of the crystal structure of an inhibitor-bound hMGL modified to enhance solubility, a putative ligand-accommodation mode has been inferred whereby substantial structural rearrangement involving helix α4 and the loop connecting helices α4 and α5 would be required for hMGL substrate/inhibitor accommodation.30,45 It has also been speculated that the hMGL lid domain overall — and helix α4 in particular — undergoes conformational change as a mobile flap or cap that transitions the enzyme from an apo- to a liganded form.45 The present H/DX-MS analysis of apo-6-HishMGL provides direct experimental evidence supporting these propositions by demonstrating the dynamic nature of the lid-domain subregion spanning residues 158–181 (i.e., helix α4 and the α4-α5 conjoining loop). Our H/DX-MS data for individual hMGL lid-domain peptides, furthermore, demonstrate the differential degrees of solvent exposure and structural plasticity within the hMGL lid domain, which appear greater than heretofore appreciated from apo-hMGL crystal structures that represent unique states whose static nature often makes it difficult to deduce information on protein dynamics.46
Modification of apo-6-His-hMGL by the pharmacologically related, but structurally different, carbamylating inhibitors AM6580 and AM6701 reduced apo-6-His-hMGL deuterium uptake into peptides 192–208, 242–248, and 273–290. Peptide 192–208 encompasses the terminal region of helix α5 and the loop connecting helices α5 and α6, whereas peptides 242–248 and 273–290 have been localized in the proximity of the active site.30,31 Specifically, peptide 242–248 is part of the apo-6-His-hMGL loop connecting sheet β7 with helix α7 and contains the catalytic-triad residue Asp246, and peptide 273–290 is part of helix α8 and the loop connecting this helix to sheet β8 and contains the His276catalytic-triad residue.30,31 These collective data support conclusion that carbamylation of apo-6-His-hMGL at catalytic Ser129 by either AM6580 or AM6701 restricts the enzyme’s backbone mobility in the substrate-binding pocket, particularly near the active site. In contrast, neither inhibitor influenced deuterium incorporation into prominent lid-domain peptides (including helix α4), indicating that the hMGL lid region largely retains its solution conformation upon enzyme inhibition/covalent modification. This observation suggests that hMGL in solution, even when carbamylated at its catalytic Ser, remains predominantly in a “locked-open” conformation, a scenario that contrasts with the closed state proposed for the crystal structure of an inhibitor-liganded hMGL variant.45 The lid conformation in that crystal structure could have been affected by mutations introduced into the enzyme to increase its solubility for crystallization and/ or may reflect a specific hMGL crystalline state rather than the enzyme’s conformational properties in solution.46 A locked-open conformation for apo- and covalently inhibited hMGL in solution is consistent with our recent experimental demonstration that hMGL engagement with and modification by AM6580 did not alter the solvent exposure of the enzyme’s lid domain when the enzyme is associated with a phospholipid-bilayer nanodisc.47
The apo-6-His-hMGL peptide spanning residues 126–135 contains the catalytic Ser129, and its localization at the bottom of the enzyme’s substrate-binding pocket has been inferred from homology-modeling43 and crystallographic30,31,45 studies. This localization is supported by our observation of a lack of significant deuterium incorporation in this peptide during the 4-h exposure of apo-6-His-hMGL to deuterated solvent, which directly demonstrates the limited solvent accessibility and stable hydrogen-bonding network of the hMGL substrate-binding pocket. In contrast, AM6701 and AM6580 elicited appreciable deuterium incorporation into this same region, suggesting that carbamylation of apo-6-His-hMGL Ser129 by these inhibitors leads to significant stretching/opening of this region of the enzyme’s binding pocket. Since hydrogen exchange reflects both regional solvent accessibility and hydrogen bonding within the protein,48,49 this inhibitor-induced conformational rearrangement of the hMGL substrate-binding pocket may reflect, at least in part, changes in the hydrogen-bond network we have recently identified as a structural correlate to hMGL catalytic activity.20
Although both carbamylating inhibitors studied decreased deuterium uptake into apo-6-His-hMGL peptide 217–223 within helix α6, AM6580 elicited a pronounced shielding effect as compared to AM6701, dramatically abrogating deuterium uptake into this region proximal to the active site. It is tempting to speculate that such extensive hMGL structural responses to carbamylation by AM6580 in this and other regions may contribute to the irreversible nature of hMGL inhibition by AM6580 versus the slow reversibility of hMGL inhibition by AM6701.
CONCLUSION
Within the endocannabinoid signaling system, hMGL is a soluble enzyme and a preferred therapeutic target for a number of psychobehavioral and somatic diseases that represent major unsolved medical problems. Despite the promise of hMGL inhibitors as drugs and recent information on the crystal structures of (modified) hMGL species, the molecular interactions between wild-type hMGL and small-molecule inhibitors remain poorly understood and sparsely documented from an experimental standpoint. The present work embodies the initial application of peptide-level H/DX-MS to interrogate directly the structural effects of two potent inhibitors, AM6701 and AM6580, on the solution conformation of wild-type hMGL. AM6701 and AM6580 are representative of a leading class of hMGL inhibitors, carbamylating agents that covalently modify the enzyme’s active-site Ser122 nucleophile. AM6580 is an irreversible hMGL inhibitor within the time frame that hMGL inhibition by AM6701 slowly reverses to regenerate active enzyme. This approach has enabled us to define and quantify the intramolecular localization and magnitude of the hMGL structural changes associated with inhibitor-induced Ser122 carbamylation. Our data provide the first experimental demonstration that chemically distinct hMGL inhibitors elicit regionally selective changes in hMGL conformation. Major hMGL structural responses to these carbamylating inhibitors include backbone motional restriction in a region proximal to the active site that involves the putative catalytic triad and stretching of the hMGL substrate-binding pocket consequent to inhibitor engagement. The shielding effect of AM6580 within hMGL helices α6 and α8 and the increase in solvent accessibility in the vicinity of the enzyme’s active site were more pronounced than the effects of AM6701 in these regions, this difference in enzyme structural response likely reflective of the more bulky hMGL covalent modification by AM6580 at Ser122. By demonstrating experimentally and for the first time the distinct structural changes that accompany wild-type hMGL covalent modification in solution by two small-molecule carbamylating inhibitors, these results advance our understanding of the hMGL–inhibitor interaction landscape and its dynamic features at the molecular level.
Supplementary Material
Acknowledgments
Funding
This research was supported by National Institutes of Health Grants DA009158 and DA003801 (to A.M.) from the National Institute on Drug Abuse (NIDA), Grants GM086507 and GM101135 (J.R.E.) from the National Institute of General Medical Sciences, and by a research collaboration with Waters Corporation (J.R.E).
ABBREVIATIONS
- 2-AG
2-arachidonoylglycerol
- AM6580
[4-(9H-fluoren-9-yl)-piperazin-1-yl][1,2,3]triazolo[4,5-b]pyridin-1-ylmethanone
- AM6701
5-((biphenyl-4-yl)methyl)-N,N-dimethyl-2H-tetra-zole-2-carboxamide
- CB1R
cannabinoid 1 G-protein coupled receptor 1
- CNS
central nervous system
- H/DX-MS
hydrogen/deuterium exchange mass spectrometry
- LC
liquid chromatography
- 6-His-hMGL
N-terminal hexa-histidine tagged hMGL
- hMGL
human monoacylglycerol lipase
- MGL
monoacylglycerol lipase
- MS
mass spectrometry
Footnotes
ASSOCIATED CONTENT
Supporting Information
MS analysis of apo-6-His-hMGL and 6-His-hMGL inhibited by AM6580 (Figure S1), peptides derived from pepsin digestion of apo-6-His-hMGL and inhibited enzyme (Figure S2), and deuterium uptake plots for all individual peptides derived from pepsin digestion of apo-6-His-hMGL and inhibited enzyme (Figure S3). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.De Petrocellis L, Di Marzo V. An introduction to the endocannabinoid system: from the early to the latest concepts. Best Pract. Res. Clin. Endocrinol. Metab. 2009;23:1–15. doi: 10.1016/j.beem.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Pertwee RG, Howlett AC, Abood ME, Alexander SP, Di Marzo V, Elphick MR, Greasley PJ, Hansen HS, Kunos G, Mackie K, Mechoulam R, Ross RA. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: beyond CB1 and CB2. Pharmacol. Rev. 2010;62:588–631. doi: 10.1124/pr.110.003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blankman JL, Simon GM, Cravatt BF. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem. Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ueda N, Tsuboi K, Uyama T, Ohnishi T. Biosynthesis and degradation of the endocannabinoid 2-arachidonoylglycerol. Biofactors. 2011;37:1–7. doi: 10.1002/biof.131. [DOI] [PubMed] [Google Scholar]
- 5.Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat. Rev. Drug Discovery. 2012;11:52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang JW, Niphakis MJ, Lum KM, Cognetta AB, Wang C, Matthews ML, Niessen S, Buczynski MW, Parsons LH, Cravatt BF. Highly selective inhibitors of monoacylglycerol lipase bearing a reactive group that is biosteric with endocannabinoid substrates. Chem. Biol. 2012;19:579–588. doi: 10.1016/j.chembiol.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulvihill MM, Nomura DK. Therapeutic potential of monoacylglycerol lipase inhibitors. Life Sci. 2013;92:492–497. doi: 10.1016/j.lfs.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feledziak M, Lambert DM, Marchand-Brynaert J, Muccioli GG. Inhibitors of the endocannabinoid-degrading enzymes, or how to increase endocannabinoid’s activity by preventing their hydrolysis. Recent Pat. CNS Drug Discovery. 2012;7:49–70. doi: 10.2174/157488912798842223. [DOI] [PubMed] [Google Scholar]
- 9.Miller LK, Devi LA. The highs and lows of cannabinoid receptor expression in disease: mechanisms and their therapeutic implications. Pharmacol. Rev. 2011;63:461–470. doi: 10.1124/pr.110.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pisani V, Madeo G, Tassone A, Sciammana G, Maccarrone M, Stanzione P, Pisani A. Homeostatic changes of the endocannabinoid system in Parkinson’s disease. Mov. Disord. 2011;26:216–222. doi: 10.1002/mds.23457. [DOI] [PubMed] [Google Scholar]
- 11.Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, Hoover HH, Cravatt BF. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem. Biol. 2011;18:846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MCG, Ward AM, Hahn YK, Lichtman AH, Conti B, Cravatt BF. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuro-inflammation. Science. 2011;334:809–813. doi: 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chanda PK, Gao Y, Mark L, Btesh J, Strassle BW, Lu P, Piesla MJ, Zhang MY, Bingham B, Uveges A, Kowal D, Garbe D, Kouranova EV, Ring RH, Bates B, Pangalos MN, Kennedy JD, Whiteside GT, Samad TA. Monoacylglycerol lipase activity is a critical modulator of the tone and integrity of the endocannabinoid system. Mol. Pharmacol. 2010;78:996–1003. doi: 10.1124/mol.110.068304. [DOI] [PubMed] [Google Scholar]
- 14.Schlosburg JE, Blankman JL, Long JZ, Nomura DK, Pan B, Kinsey SG, Nguyen SG, Ramesh D, Booker L, Burston JJ, Thomas EA, Selley DE, Sim-Selley LJ, Liu QS, Lichtman AH, Cravatt BF. Chronic monoacylglyerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat. Neurosci. 2010;13:1113–1119. doi: 10.1038/nn.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comelli F, Giagnoni G, Bettoni I, Colleoni M, Costa B. The inhibition of monoacylglycrol lipase by URB602 showed an anti-inflammatory and anti-nociceptive effect in a murine model of acute inflammation. Br. J. Pharmacol. 2007;152:787–794. doi: 10.1038/sj.bjp.0707425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hohmann AG. Inhibitors of monoacylglycerol lipase as novel analgesics. Br. J. Pharmacol. 2007;150:673–675. doi: 10.1038/sj.bjp.0707153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sciolino NR, Zhou W, Hohmann AG. Enhancement of endocannabinoid signaling with JZL184, an inhibitor of the 2-arachidonoylglycerol hydrolyzing enzyme monoacylglycerol lipase, produces anxiolytic effects under conditions of high environmental aversiveness in rats. Pharmacol. Res. 2011;64:226–234. doi: 10.1016/j.phrs.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naidoo V, Karanian DA, Vadival SK, Locklear JR, Wood JT, Nasr M, Quizon PMP, Graves EE, Shukla V, Makriyannis A, Bahr BA. Equipotent inhibition of fatty acid amide hydrolase and monoacylglycerol lipase–dual targets of the endocannabinoid system to protect against seizure pathology. Neurotherapeutics. 2012;9:801–813. doi: 10.1007/s13311-011-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson M, Contreras JA, Hellmann U, Tornqvist H, Holm C. cDNA Cloning, tissue distribution, and identification of the catalytic triad of monoacylglycerol lipase. J. Biol. Chem. 1997;272:27218–27223. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- 20.Karageorgos I, Tyukhtenko S, Zvonok N, Janero DR, Sallum C, Makriyannis A. Identification by nuclear magnetic resonance spectroscopy of an active-site hydrogen-bond network in human monoacylglycerol lipase (hMGL): implications for hMGL dynamics, pharmacological inhibition, and catalytic mechanism. Mol. BioSyst. 2010;6:1381–1388. doi: 10.1039/c004515b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karageorgos I, Zvonok N, Janero DR, Vemuri VK, Shukla V, Wales TE, Engen JR, Makriyannis A. Endocannabinoid enzyme engineering: soluble human thio-mono-acylglycerol lipase (sol-S-hMGL) ACS Chem. Neurosci. 2012;3:393–399. doi: 10.1021/cn3000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zvonok N, Pandarinathan L, Williams J, Johnston M, Karageorgos I, Janero DR, Krishnan SC, Makriyannis A. Covalent inhibitors of human monoacylglycrol lipase: ligand-assisted characterization of the catalytic site by mass spectrometry and mutational analysis. Chem. Biol. 2008;15:854–862. doi: 10.1016/j.chembiol.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavón FJ, Serrano AM, Selley DE, Parsons LH, Lichtman AH, Cravatt BF. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat. Chem. Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoornaert C. Triazolopyridine carboxamides derivatives, preparation thereof, and therapeutic uses thereof. 7868007B2. U.S. Patent. 2011
- 25.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Differential hydrogen/ deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev. Proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zvonok N, Williams J, Johnston M, Pandarinathan L, Janero DR, Krishnan SC, Makriyannis A. Full mass spectrometric characterization of human monoacylglycerol lipase generated by large-scale expression and single-step purification. J. Proteome Res. 2008;7:2158–2164. doi: 10.1021/pr700839z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiyanont K, Wales TE, Aste-Amezaga M, Aster JC, Engen JR, Blacklow SC. Evidence for increased exposure of the Notch1 metalloprotease cleavage site upon conversion to an activated conformation. Structure. 2011;19:546–554. doi: 10.1016/j.str.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcsisin SR, Narute PS, Emert-Sedlak LA, Kloczewiak M, Smithgall TE, Engen JR. On the solution conformation and dynamics of the HIV-1 viral infectivity factor. J. Mol. Biol. 2011;410:1008–1022. doi: 10.1016/j.jmb.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertrand T, Augé F, Houtmann J, Rak A, Vallée F, Mikol V, Berne PF, Michot N, Cheuret D, Hoornaert C, Mathieu M. Structural basis for human monoglyceride lipase inhibition. J. Mol. Biol. 2010;396:663–673. doi: 10.1016/j.jmb.2009.11.060. [DOI] [PubMed] [Google Scholar]
- 31.Labar G, Bauvois C, Borel F, Ferrer J-L, Wouters J, Lambert DM. Crystal structure of the human mono-acylglycerol lipase, a key actor in endocannabinoid signaling. ChemBioChem. 2010;11:218–227. doi: 10.1002/cbic.200900621. [DOI] [PubMed] [Google Scholar]
- 32.Doshi U, McGowan LC, Ladani ST, Hamelberg D. Resolving the complex role of enzyme conformational dynamics in catalytic function. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5699–5704. doi: 10.1073/pnas.1117060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat. Rev. Drug Discovery. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 34.Broglia R, Levy Y, Tiana G. HIV-1 protease folding and the design of drugs which do not create resistance. Curr. Opin. Struct. Biol. 2008;18:60–66. doi: 10.1016/j.sbi.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Busenlehner LS, Armstrong RN. Insights into enzyme structure and dynamics elucidated by amide H/D exchange mass spectrometry. Arch. Biochem. Biophys. 2005;433:34–46. doi: 10.1016/j.abb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Morgan CR, Engen JR. Investigating solution-phase protein structure and dynamics by hydrogen-exchange mass spectrometry. Curr. Protoc. Protein Sci. 2009;58:17.6.1–17.9.7. doi: 10.1002/0471140864.ps1706s58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith DL, Deng Y, Zhang Z. Probing the non-covalent structure of proteins by amide hydrogen exchange and mass spectrometry. J. Mass Spectrom. 1997;32:135–146. doi: 10.1002/(SICI)1096-9888(199702)32:2<135::AID-JMS486>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 38.Shen ML, Johnson KL, Mays DC, Lipsky JJ, Naylor S. Identification of the protein-drug adduct formed between aldehyde dehydrogenase and S-methyl-N,N-diethylthiocarbamoyl sulfoxide by on-line proteolytic digestion high performance liquid chromatography electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2000;14:918–923. doi: 10.1002/(SICI)1097-0231(20000530)14:10<918::AID-RCM966>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 39.Tomlinson AJ, Johnson KL, Lam-Holt J, Mays DC, Lipsky JJ, Naylor S. Inhibition of human mitochondrial aldehyde dehydrogenase by the disulfiram metabolite S-methyl-N,N-diethylthiocarbamoyl sulfoxide. Biochem. Pharmacol. 1997;54:1253–1260. doi: 10.1016/s0006-2952(97)00359-6. [DOI] [PubMed] [Google Scholar]
- 40.Carr PD, Ollis DL. α/β Hydrolase fold: an update. Protein Pept. Lett. 2009;16:1137–1148. doi: 10.2174/092986609789071298. [DOI] [PubMed] [Google Scholar]
- 41.Holmquist M. α/β-Hydrolase fold enzymes: structures, functions and mechanisms. Curr. Protein Pept. Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- 42.Kourist R, Jochens H, Bartsch S, Kuipers R, Padhi SK, Gall M, Böttcher D, Joosten H-J, Bornscheuer UT. The α/β-hydrolase fold 3DM database (ABHDB) as a tool for protein engineering. ChemBioChem. 2010;11:1635–1643. doi: 10.1002/cbic.201000213. [DOI] [PubMed] [Google Scholar]
- 43.Bowman AL, Makriyannis A. Refined homology model of monoacylglycerol lipase: toward a selective inhibitor. J. Comput.-Aided Mol. Des. 2009;23:799–806. doi: 10.1007/s10822-009-9289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rengachari S, Bezerra GA, Riegler-Berket L, Gruber CC, Sturm C, Taschler U, Boeszoermanyi A, Dreveny I, Zimmermann R, Gruber K, Oberer M. The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes. Biochim. Biophys. Acta. 2012;1821:1012–1021. doi: 10.1016/j.bbalip.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schalk-Hihi C, Schubert C, Alexander R, Bayoumy S, Clemente JC, Deckman I, Des Jarlais RL, Dzordzorme KC, Flores CM, Grasberger B, Kranz JK, Lewandowski F, Liu L, Ma H, Maguire D, Macielag MJ, McDonnell ME, Mezzasalma Haarlander T, Miller R, Milligan C, Reynolds C, Kuo LC. Crystal structure of a soluble form of human monoacylglycerol lipase in complex with an inhibitor at 1.35 Å resolution. Protein Sci. 2011;20:670–683. doi: 10.1002/pro.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acharya KR, Lloyd MD. The advantages and limitations of protein crystal structures. Trends Pharmacol. Sci. 2005;26:10–14. doi: 10.1016/j.tips.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 47.Nasr M, Hi X, Bowman AL, Johnson M, Zvonok N, Janero DR, Vemuri VK, Wales TE, Engen JR, Makriyannis A. Membrane phospholipid bilayer as a determinant of monoacylglycerol lipase kinetic profile and conformational repertoire. Protein Sci. 2013;22:774–787. doi: 10.1002/pro.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engen JR, Wales TE, Shi X. Encyclopedia of Analytical Chemistry. New York: Wiley; 2011. Hydrogen exchange mass spectrometry for conformational analysis of proteins. [Google Scholar]
- 49.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.