Abstract
RNA transcripts that do not code for proteins have been long known to lie at the heart of many biological processes, such as splicing and translation. Yet their full potential has only been appreciated recently and non-coding RNAs (ncRNAs) are now attracting increasing attention. Pioneering work in yeast and plant systems has revealed that non-coding RNAs can have a major influence on the deposition of histone and DNA modifications. This can introduce heritable variation into gene expression and, thus, be the basis of epigenetic phenomena. Mechanistically, such processes have been studied extensively in the fission yeast Schizosaccharomyces pombe, providing an important conceptual framework for possible modes of action of ncRNAs also in other organisms. In this review, we highlight mechanistic insights into chromatin-associated ncRNA activities gained from work with fission yeast, and we draw parallels to studies in other eukaryotes that indicate evolutionary conservation.
Keywords: non-coding RNA, chromatin, genome regulation
Introduction
Recent advances in technologies enabling the profiling of transcriptomes at unprecedented depth have revealed the production of RNA in eukaryotic cells from a proportion of their genomes much larger than anticipated (Guttman et al. 2009, 2011; Willingham et al. 2006; Cheng et al. 2005; Birney et al. 2007; Kapranov et al. 2007a; Wilhelm et al. 2008). Estimates from the ENCODE consortium suggest that more than three quarters of the human genome is transcriptionally active (Djebali et al. 2012; Dunham et al. 2012), with the number of transcripts detected by far exceeding the number of known mRNAs coding for proteins. Whereas such non-protein coding RNA (ncRNA) catalogs are increasing at a great pace, the extent of ncRNA involvement in regulatory circuits and the mechanisms by which they might function remain largely unexplored. Furthermore, questions remain about what portion of the non-coding transcriptome is functionally relevant or simply reflects transcriptional “noise” (Struhl 2007). In particular, the observations that ncRNAs from genomic regions thought to be transcriptionally silent or from intergenic or promoter regions are often subjected to rapid degradation (Keller et al. 2012; Lubas et al. 2011; Houseley et al. 2007; Wyers et al. 2005; Vasiljeva et al. 2008; LaCava et al. 2005; Preker et al. 2008; Buhler et al. 2007; Woolcock et al. 2010), and that most ncRNAs are repressed below one copy per cell (Marguerat et al. 2012) have led to the hypothesis that these transcript are a consequence of an imperfect transcription machinery that produces spurious RNAs. Indeed, estimates of the fraction of the vast amount of ncRNAs that might be physiologically relevant differ considerably (Kowalczyk et al. 2012). Thus, biologists should be urged to dissect the various putative modes of action of ncRNAs, which is much more laborious and challenging than cataloguing but is essential for a solid comprehension of the non-protein coding genome.
Non-coding RNAs are typically classified into small and long ncRNAs based on an artificial length threshold of about 200 nucleotides (nt) (Kapranov et al. 2007b); this value simply reflects the nucleic acid absorption cutoff of the matrices most commonly used to isolate RNA molecules. Small ncRNAs include the well-studied transfer (t)RNAs, small nucleolar (sno)RNAs, small nuclear (sn)RNAs and micro (mi)RNAs, which range in size from 20 nt for miRNAs up to approximately 150 nt for snRNAs (Ma et al. 2013). Long ncRNAs (>200 nt) have received less attention, but a few examples have revealed that they can exert crucial functions in both the cytoplasmic and nuclear compartments of a cell. Intriguingly, there is growing evidence that ncRNAs are actively involved in genome regulation in various eukaryotes. Work with yeast and plant systems has revealed ncRNAs that have a major influence on the deposition of histone or on DNA modification, which can introduce heritable variation of gene expression without altering the DNA sequence and, thus, lead to epigenetic phenomena (Hall et al. 2002; Heo and Sung 2010; Herr et al. 2005; Onodera et al. 2005; Verdel et al. 2004). In particular, the fission yeast Schizosaccharomyces pombe has served as a tremendously powerful model organism to investigate the links between ncRNAs and chromatin at a mechanistic level, establishing guiding paradigms for studying ncRNA-mediated genome regulation in other eukaryotes. In this review, we focus on mechanistic insights into chromatin-associated ncRNA activities that have been gained with S. pombe and draw parallels to studies in other organisms that indicate evolutionary conservation.
ncRNAs acting as guide molecules
Arguably, the best established attribute of ncRNAs is that they can guide associated partner proteins to other nucleic acid target molecules by complementary base-pairing. In particular, small ncRNAs guide enzymatic activities to targets, endowing specificity on pathways that, for example, cleave and ligate RNA, modify RNA or DNA, regulate telomere length, or modify chromatin. Extensively studied examples of RNA guides are snRNAs, snoRNAs and tRNAs, which are crucial to the specificity of pre-mRNA splicing, the 2′-O-ribosylation and pseudo-uridylation of ribosomal RNA, or the decoding of the open reading frame of an mRNA during protein synthesis, respectively (Hopper and Phizicky 2003; Matera et al. 2007). An important feature of snRNAs and snoRNAs is that they contain several sequence motifs and RNA secondary structures that act as binding sites for specific partner proteins in cis, in addition to a trans-acting guide element (Matera et al. 2007). Thus, snoRNAs and snRNAs act at the same time as scaffolds assembling specific ribonucleoprotein (RNP) complexes and as guide molecules. We envision that this principle also applies to many of the recently discovered long ncRNAs (Fig. 1).
Fig. 1.
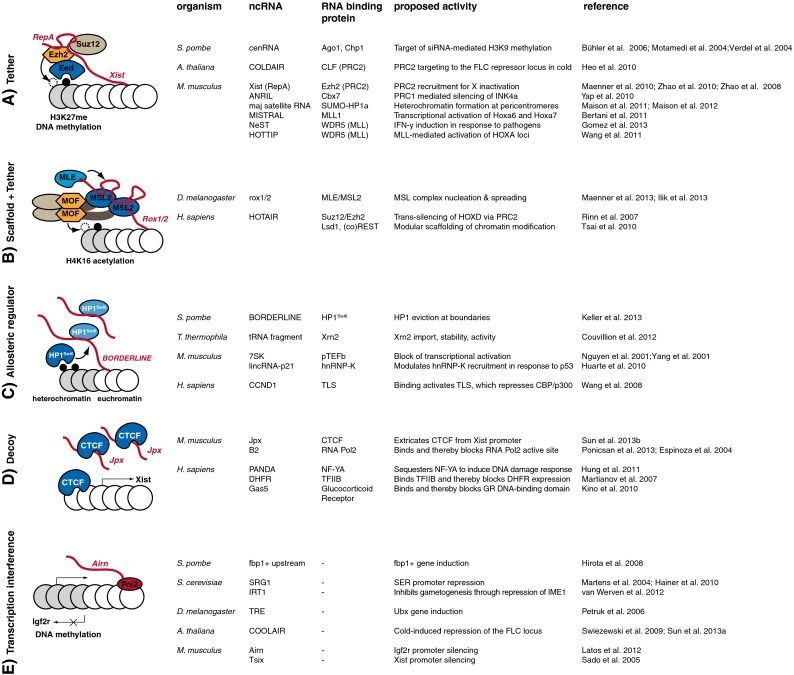
Various chromatin-associated activities of long ncRNAs. (A) ncRNA tethers link activities in cis via a nascent transcript. For example, the ncRNA Xist is transcribed from the X-chromosome and interacts via a conserved stem-loop sequence termed RepA with the PRC2 complex members Ezh2 and Suz12. This triggers H3K27 methylation, which is recognized by the H3K27 reader Eed. This leads to inactivation of the X chromosome. (B) In addition to acting as tethers, some long ncRNAs may also act as scaffolds. The ncRNA (e.g. rox1/2) not only serves as a nucleation site but is also an integral part of the chromatin-modifying activity (e.g. the MSL complex). In the case of the Drosophila MSL complex, this leads to H4K16 acetylation via the histone acetyltransferase MOF and transcriptional upregulation on the male X chromosome. (C) ncRNAs acting as allosteric regulators affect chromatin effectors by direct binding and tuning of their molecular properties. For example, RNA binding to the hinge region of HP1Swi6 induces conformational changes in the HP1Swi6 CD, which is incompatible with stable H3K9me3 association. This leads ultimately to HP1Swi6 eviction from heterochromatin. (D) Decoys mimicking a natural ligand of the molecule to be regulated compete for the binding site. For example, Jpx RNA contains a motif that effectively competes with CTCF DNA binding. This extricates CTCF from the Xist promoter and thereby activates Xist transcription, inducing X chromosome inactivation in female mice. (E) In the transcriptional interference model, the action of the transcribing RNA polymerase rather than the RNA product is functionally relevant. One example is transcriptional overlap of the Airn ncRNA, which is required for Igf2r promoter silencing. The table lists representative examples of each mode of action
In contrast, small ncRNAs that function in RNA interference (RNAi)-related pathways act solely as guide molecules (Hannon 2002). With a size range of 20–31 nt, depending on the type of small RNA and organism, they are simply too short to also encode scaffolding motifs. However, we note that such elements do exist in the precursors of the small RNAs and are important for their maturation (Yates et al. 2013).
RNAi, first recognized as a double-stranded RNA (dsRNA)-mediated process in Caenorhabditis elegans in 1998 (Fire et al. 1998), exists in various forms in a wide variety of eukaryotic organisms (Ghildiyal and Zamore 2009). A central feature of RNAi are the trans-acting Argonaute proteins that are guided to complementary targets by their bound small RNAs (Fig. 2) (Joshua-Tor and Hannon 2010; Meister 2013). Notably, Argonaute can be programmed with any small RNA guide and will target, in principle, any complementary sequence, may this be endogenous or exogenous (Brummelkamp et al. 2002; Elbashir et al. 2001). Therefore, RNAi has been widely exploited as an experimental technique and is considered to have great potential for therapeutics (Davidson and McCray 2011). The enormous impact on biotechnology and the still emerging physiological roles of small ncRNAs showcase the extraordinary power of ncRNAs in guiding protein complexes to their destination.
Fig. 2.
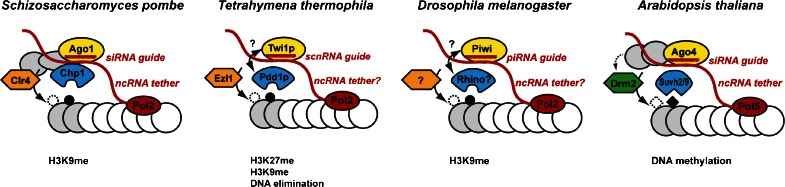
Small and long ncRNAs cooperate in the localization of effector complexes to chromatin. In S. pombe, siRNAs guide the RITS complex consisting of the Argonaute protein Ago1 (yellow), the chromodomain (CD) protein Chp1 (blue) and Tas3 (grey) to nascent transcripts originating from centromeric repeat sequences; this is followed by recruitment of the H3K9 methyltransferase Clr4. The CD of Chp1 recognizes H3K9-methylated histone tails and thereby stabilizes the complex on chromatin. The Chp1 CD also possesses intrinsic affinity for nucleic acids, which might further stabilize the complex on chromatin. In Tetrahymena thermophila, the Argonaute protein Twi1p is similarly involved in the recruitment of the H3K27me reader protein Pdd1p and the H3K27-methyltransferase Ezl1 via a 28-nt scnRNA guide. Whether this also involves nascent transcript interactions through a RITS-like complex remains to be shown. Ultimately, this process leads to DNA elimination. In Drosophila, Piwi interacts with a piRNA guide, which targets transposable elements (TEs) for transcriptional silencing via H3K9 methylation. Although direct evidence is still awaited, nascent TE transcripts are also thought to function as assembly platforms. The CD-protein Rhino has been implicated in piRNA generation. In Arabidopsis thaliana, Ago4-associated small RNAs guide the DNA methylation machinery to target sites. Ago4 can interact with nascent transcripts generated by RNA Polymerase 5 (Pol5) as well as with Pol5 itself
Small ncRNA-mediated guidance of effector complexes to chromatin
Whereas miRNAs are well-known guide molecules that act at a post-transcriptional level in the cytoplasm (Guo et al. 2010), small ncRNA guides are known to function in the nucleus in some organisms, specifying the targeting of protein effector complexes to their respective chromosomal loci. Classical examples are the targeting of histone or DNA methylation activities to chromatin, but the directing of other activities to chromatin may be equally possible.
RNA-mediated chromatin modification was first observed in plants (Wassenegger et al. 1994). Although the mechanism was not understood at the time, it is now known that small ncRNAs play a key role in RNA-directed DNA methylation (RdDM). Repetitive genomic sequences, including transposons and centromeric repeats, produce 24-nt siRNAs that target DNA methylation, silencing these regions and other loci homologous to the siRNAs (Matzke et al. 2009; Zhang and Zhu 2011). More recently, 21-nt small ncRNAs (diRNAs) produced from DNA double-stranded breaks (DSBs) have been proposed to guide Ago2 to damaged chromatin and mediate repair in Arabidopsis (Wei et al. 2012). Similarly, small ncRNAs (ddRNAs) have been linked to the DNA-damage response in humans, mouse and zebrafish (Francia et al. 2012; Wei et al. 2012). This is reminiscent of earlier studies in the filamentous fungus Neurospora crassa, where 20- to 21-nt ncRNAs (qiRNA) generated upon DNA damage were proposed to affect protein synthesis after DNA damage by inhibiting rRNA biogenesis (Lee et al. 2009). It is tempting to speculate that these small ncRNAs are actively involved in DNA repair also by guiding protein factors to the damaged site, an intriguing model that deserves further investigation in these organisms.
Small ncRNA guides in ciliated protozoa have also been directly linked to chromatin (Mochizuki 2011; Bracht et al. 2013). In Tetrahymena thermophila, an extreme example of RNAi-directed heterochromatin formation leads eventually to DNA elimination of transposon-related sequences from the newly developing somatic (macronuclear) genome (Kataoka and Mochizuki 2011). During sexual conjugation, the whole germline (micronuclear) genome is bidirectionally transcribed and processed in the nucleus to 28- to 29-nt “scan” RNAs (scnRNAs) (Malone et al. 2005; Mochizuki and Gorovsky 2005). These scnRNAs associate with an Argonaute protein and translocate to the parental macronucleus, where it is thought that base-pairing interactions between the scnRNAs and nascent non-coding transcripts from the parental macronuclear genome mediate scnRNA degradation. Therefore, this ‘scnRNA selection’ leaves behind only scnRNAs for which there are no homologous sequences in the parental macronuclear genome. The remaining scnRNAs, still bound to Argonaute, relocate to the newly developing macronucleus, where they target homologous sequences and mediate methylation of histone H3 lysine 9 and 27 (H3K9 and H3K27, respectively). Subsequent binding of chromodomain proteins marks these regions, which leads to excision by a PiggyBac transposase-like protein (Cheng et al. 2010). Therefore, this elegant ncRNA-mediated mechanism eliminates any sequences not present in the parental macronucleus (Fig. 2). Intriguingly, small ncRNAs in another ciliate, Oxytricha trifallax, also act in genome rearrangement but instead of eliminating they specify which sequences are retained (Bracht et al. 2013).
Furthermore, the more recently discovered piwi-interacting (pi)RNAs that control transposable elements in the germline of animals have been linked to chromatin regulatory processes. Mutations in piRNA pathway components led to defective DNA methylation in male mouse germ cells (Pillai and Chuma 2012; Aravin et al. 2006, 2008; Carmell et al. 2007; O’Donnell and Boeke 2007) and reduced H3K9 methylation at transposon loci in Drosophila ovarian somatic cells (Sienski et al. 2012; Rozhkov et al. 2013; Le Thomas et al. 2013). Finally, in C. elegans, artificially introduced or endogenous small ncRNAs can silence complementary, nuclear-retained RNAs or nuclear-localized polycistronic RNAs. This is accompanied by increased H3K9 methylation of the underlying chromosomal locus. These observations strongly argue that small ncRNAs can guide the H3K9 methylation machinery to chromatin also in worms (Burkhart et al. 2011; Burton et al. 2011; Guang et al. 2010; Lee et al. 2012; Bagijn et al. 2012)
Emerging mechanistic models for many of the above-mentioned examples are reminiscent of the basic concepts that arose from studies of RNAi-mediated heterochromatin formation in the fission yeast S. pombe, which is one of the most intensely studied examples of small ncRNA-guided chromatin modification. A decade ago, it was shown that all three major components of the RNAi machinery in S. pombe, namely Argonaute (Ago1), Dicer (Dcr1) and the RNA-dependent RNA polymerase (Rdp1), are essential for the formation of centromeric heterochromatin (Volpe et al. 2002; Provost et al. 2002) and small ncRNAs were identified that map to this supposedly transcriptionally inactive chromatin structure (Reinhart and Bartel 2002). Meanwhile, a number of protein complexes involved in this process have been identified (Buhler 2009; Lejeune and Allshire 2011; Reyes-Turcu and Grewal 2012; Moazed 2009; Castel and Martienssen 2013) and their intense study has provided solid evidence for a small ncRNA-guided model for chromatin modification.
In brief, the RNA-induced transcriptional silencing complex (RITS; consisting of Ago1, Chp1 and Tas3) (Verdel et al. 2004) accepts Dcr1-dependent single-stranded siRNAs that guide RITS to chromatin, where it recruits CLRC, a complex containing the sole S. pombe H3K9 methyltransferase Clr4 (Bayne et al. 2010). Similar to the situation in Tetrahymena, this RNAi-mediated histone methylation provides a binding site for HP1 homologues, such as Swi6 and Chp2, and can also stabilize binding of RITS via the chromodomain (CD)-containing Chp1 component. Interestingly, the Chp1 CD also possesses intrinsic nucleic acid-binding activity, a property that can further stabilize RITS association with chromatin (Ishida et al. 2012). RITS also helps recruit an Rdp1-containing complex, RDRC, which amplifies the process by generating more double-stranded RNA substrates for Dcr1 (Motamedi et al. 2004; Sugiyama et al. 2007) Thus, the RNAi machinery acts in a positive feedback loop on centromeric repeats, generating high levels of H3K9 methylation and rapid turnover of centromeric RNAs into siRNAs (Buhler and Moazed 2007).
Long ncRNAs act as binding sites for small ncRNA guides
A general concept for ncRNAs acting in association with chromatin that arose from studies of RNAi-mediated heterochromatin formation in S. pombe is that small ncRNA guides target the respective chromosomal regions through base-pairing interactions with long, chromatin-associated ncRNAs. Originally proposed by Shiv Grewal and Danesh Moazed (Grewal and Moazed 2003) this conception, also known as the “nascent transcript model”, has been corroborated by several lines of experimental evidence. Firstly, RITS and RDRC associate with centromeric heterochromatin and physically interact with non-coding centromeric RNAs (Motamedi et al. 2004; Noma et al. 2004; Woolcock et al. 2010; Verdel et al. 2004). Secondly, specific mutations in subunits of RNA polymerase II have been identified that do not generally affect transcription but lead to a loss of RNAi-dependent heterochromatin formation (Kato et al. 2005; Djupedal et al. 2005). Finally, direct proof-of-concept for the nascent transcript model was provided by the artificial tethering of RITS to the nascent transcript of a normally euchromatic gene. This was sufficient to trigger the formation of ectopic heterochromatin and activate the characteristic positive feedback loop that secured high levels of H3K9 methylation and efficient silencing of the newly formed heterochromatic locus (Buhler et al. 2006).
Although binding of the siRNA guide to single-stranded DNA cannot be formally ruled out, the experimental evidence for long, chromatin-associated ncRNAs acting as binding sites for small ncRNA guides is substantial. Strong support for this model also arose from studies of RdDM in Arabidopsis, which involves the action of two plant-specific RNA polymerases, RNA Pol IV and Pol V (Wierzbicki et al. 2008). Pol IV is thought to be responsible for the transcription of precursor RNAs that are processed by the RNA-dependent RNA polymerase (RdRP) RDR2 and DCL3 (DICER-LIKE 3), producing the 24-nt siRNAs that load onto AGO4 (Herr et al. 2005). Importantly, Pol V is thought to be the RNA polymerase dedicated to the production of transcripts that presumably act as scaffolds in the association of AGO4-siRNA complexes and subsequent chromatin modification (Wierzbicki et al. 2008, 2009) similar to the situation in S. pombe. Finally, scnRNAs in Tetrahymena and piRNAs in Drosophila and C. elegans are also thought to guide their associated Argonaute/Piwi proteins to the respective chromosomal loci via base-pairing with nascent transcripts (Luteijn and Ketting 2013).
Long ncRNAs acting as tethers: a recurrent theme
The nascent transcript model for small ncRNA-mediated chromatin regulation described above is reminiscent of proposals put forward to explain the role of long ncRNAs in X chromosomal dosage compensation in Drosophila and mouse. In flies, this process involves upregulated transcription on the male X chromosome via recruitment of the MSL complex, which deposits histone H4K16 acetylation (Conrad and Akhtar 2012). Following the discovery that the roX1 ncRNA specifically coats the X-chromosome, it was proposed more than a decade ago that ncRNAs change chromatin conformation by associating with MSL complexes, histone acetyltransferases, or chromatin components (Meller et al. 1997). Later, it was demonstrated that roX ncRNAs are an integral part of the MSL complex and thereby exert a scaffolding activity (Meller et al. 2000; Smith et al. 2000; Akhtar et al. 2000).
In vivo, the two functionally redundant ncRNAs roX1 and roX2 are encoded on the X chromosome itself. Only the roX1/2 double mutant results in male-specific lethality. This phenotype can be rescued by autosomal roX transgenes, which demonstrates that the roX1/2 lncRNAs can also function in trans (Meller and Rattner 2002). Autosomally expressed roX transgenes can trigger assembly and spreading of the MSL complex on the ectopic locus, but only if the X-linked roX sequences are mutated at the same time (Park et al. 2002). The roX RNAs, therefore, seem to compete for limiting MSL complexes and preferentially target the X chromosome, a property that may be provided by specific DNA sequence motifs (Alekseyenko et al. 2008). Recent iCLIP data indeed has confirmed that the MSL members MLE and MSL2 are involved in co-transcriptional incorporation of the roX ncRNAs into the MSL complex, which results in X-specific nucleation and spreading (Maenner et al. 2013; Park et al. 2002; Ilik et al. 2013). Taken together, the available data suggest that roX ncRNAs exert both scaffolding and cis-specific tethering activities.
In mammals, the mechanism of dosage compensation is fundamentally different in that it manifests in females, where one of the two X chromosomes is epigenetically silenced via H3K27me and DNA methylation. However, similar to the situation in Drosophila, a number of ncRNAs are involved in this process, with Xist being the most prominent example. When the Xist gene was identified about 20 years ago, it was noted that Xist RNA expression paradoxically occurs exclusively from the inactive X chromosome in females (Brown et al. 1991; Borsani et al. 1991; Brockdorff et al. 1991). While it was conceivable that Xist encodes for a protein (Borsani et al. 1991), Brown et al. hypothesized that “the XIST product is a cis-acting RNA molecule, perhaps involved structurally in the formation of the heterochromatic Barr body” (Brown et al. 1991). This hypothesis is supported by the fact that Xist contains no significant open reading frame, is not associated with the translational machinery, localizes predominantly to the nucleus (Brockdorff et al. 1992; Brown et al. 1992), and acts in a cis-specific manner (Penny et al. 1996). Brockdorff and colleagues proposed that Xist RNA molecules might (a) locally interact with chromatin, (b) form a complex with other nuclear factors, which might in turn interact with chromatin, or (c) change the subnuclear localization of the X chromosome, which eventually results in heterochromatinization. Alternatively, they suggested (d) that active transcription through the Xist locus, and not necessarily the Xist product, causes a change in chromatin structure (Brown et al. 1992; Brockdorff et al. 1992). Accordingly, autosomal expression of Xist is sufficient to mediate chromosome-wide changes and silence the transgenic chromosome (Lee et al. 1996; Lee and Jaenisch 1997; Wutz and Jaenisch 2000). Mapping experiments have demonstrated that the domains responsible for silencing lie within the 5′ repeat A (RepA). RepA folds into stem loops, which were proposed to act as binding sites for factors involved in heterochromatin formation (Wutz et al. 2002). These initial hypotheses gained further support recently when RepA was shown to bind to the PRC2 complex member Ezh2 (Zhao et al. 2008; Maenner et al. 2010; Kaneko et al. 2010). RepA might thereby recruit PRC2, which then triggers H3K27me and silencing of the X chromosome in cis.
Interestingly, apart from Xist, ncRNAs could play a more general role in PRC2 recruitment to chromatin. For example HOTAIR, a 2.2-kb ncRNA expressed at the boundary of two chromatin domains within the HOXC cluster, was shown to co-immunopurify with PRC2. Furthermore, knockdown of HOTAIR results in a global loss of H3K27me3 and increased RNA expression at the 40-kb remote HOXD locus. Thus, it is possible that HOTAIR recruits PRC2 to the HOXD cluster. In contrast to Xist, HOTAIR acts in trans (Rinn et al. 2007; Tsai et al. 2010). In general, the tethering or recruitment of chromatin-modifying complexes by ncRNAs has been attracting increasing attention. Therefore, it is not surprising that this also includes activating complexes such as the MLL/trithorax complex, which is recruited by the cis-acting ncRNA HOTTIP (Wang et al. 2011). We note that in many cases, very little is known about the kinetics and specificity of the proposed RNA–protein interactions, and we believe that generating specific mutations to disrupt the ncRNA–protein interaction will be essential to solve certain controversial issues regarding ncRNA-mediated recruitment of polycomb complexes (Brockdorff 2013; Schorderet and Duboule 2011). Equally valuable are tethering experiments (Buhler et al. 2006; Wang et al. 2011) that will help to define whether or not ncRNAs are sufficient to recruit/tether these complexes to chromatin, and will allow mechanistic dissection of the steps leading to chromatin modification.
ncRNAs acting as allosteric regulators
Whilst the list of ncRNAs that may function as guides or tethers is ever-expanding, additional paradigms of ncRNA activity are being uncovered. For example, recent work in our lab with S. pombe disclosed a rather unexpected activity of ncRNAs in regulating association of the heterochromatin protein HP1Swi6 with repressive chromatin. Results from a combination of in vivo and in vitro experiments strongly implied that RNA binding to HP1Swi6 prevents HP1Swi6 from stably associating with chromatin. In particular, we demonstrated that RNA binds directly to HP1Swi6, inducing a conformational change in its chromodomain that abolishes the affinity for methylated H3K9 (Buhler and Hiller 2012; Keller et al. 2012). Thus, HP1Swi6 binding to RNA is incompatible with stable heterochromatin association and ncRNA molecules can, therefore, evict HP1Swi6 from chromatin. This mode of action is functionally relevant for tight repression of heterochromatin and counteracting the spreading of heterochromatin into neighboring euchromatin (Keller et al. 2012, 2013).
Similarly, the ncRNA Jpx was proposed recently to act as an “evictor” of CTCF in mammals (Sun et al. 2013b). X-inactivation in mammals is triggered by transcription of the cis-acting long ncRNA Xist (Brockdorff et al. 1992; Brown et al. 1992). The zinc finger protein CTCF represses transcription of the Xist RNA. During the onset of dosage compensation, Jpx RNA is transcribed and binds to CTCF, thereby competing with CTCF binding to the Xist promoter DNA. As a result, Jpx RNA binding to CTCF results in eviction from the Xist promoter and, consequently, activates Xist transcription (Sun et al. 2013b). It remains to be tested whether Jpx RNA functions as an allosteric regulator, inducing a conformational change in CTCF. Alternatively, it is possible that Jpx simply binds the DNA-binding motif of CTCF and thus rather acts as a “decoy”. Similar decoy activities have been described for other mammalian ncRNAs that titrate transcription factors away from their DNA targets (Kino et al. 2010; Martianov et al. 2007).
The RNA-induced conformational change in the chromodomain of HP1Swi6 exemplifies the potential of ncRNAs to act as allosteric regulators of their partner proteins, a mode of action of ncRNAs that might turn out to be more prevalent than anticipated. Similar examples of such ncRNA-mediated protein regulation are the B2 or 7SK ncRNAs that are involved in transcriptional control. The murine 178-nt B2 lncRNA is induced in response to heat shock and globally represses transcription. Mechanistically, this is achieved by direct incorporation of the B2 ncRNA into the RNA polymerase II complex, which results in a block of RNA synthesis (Espinoza et al. 2004; Ponicsan et al. 2013). In metazoans, the 7SK ncRNA interacts with and thereby blocks pTEFb, a factor required in the release of proximally paused RNA polymerase II into productive elongation (Nguyen et al. 2001; Yang et al. 2001; Peterlin et al. 2011).
A further noteworthy example of ncRNAs that may fall into the same functional category are 3′ tRNA fragments in Tetrahymena that associate with the Argonaute protein Twi12p and are involved in rRNA processing via the exonuclease Xrn2. This is a particularly interesting case in the light of our above discussion of Argonaute-associated small ncRNAs functioning as guide molecules. Rather than acting as a guide, the tRNA fragment was proposed either to change the conformation of Xrn2 or to stabilize, catalytically activate, or import Xrn2 (Couvillion et al. 2012).
ncRNA-mediated epigenetic changes
A very intriguing aspect of ncRNAs acting on chromatin is, at least to us, the initiation of epigenetic alterations in some instances. For example, the RNAi machinery has been shown to be necessary for the assembly of heterochromatin at the silent mating type locus in S. pombe. However, once assembled, heterochromatin at the mating type locus is maintained independently of the RNAi pathway (Jia et al. 2004; Hall et al. 2002). Thus, small ncRNAs may constitute one of the signals inducing the formation of a different cellular state that can be maintained and perpetuated over many cell generations, even in the absence of the initial trigger (Ptashne 2007). Notably, the heterochromatic silencing of genes artificially inserted into the silent-mating type locus occurs in a stochastic way and requires a functional RNAi pathway. However, once the repressed state is acquired, it is stably transmitted to future generations through mitosis and meiosis (Noma et al. 2004; Sadaie et al. 2004; Nakayama et al. 2000).
Because ncRNAs would only be involved initially if they were indeed causing alterations in the epigenetic state of a cell, they may not be detected when simply profiling the transcriptome of a given cell. Fortunately, this is not the case for centromeric heterochromatin in S. pombe, for which RNAi is required to maintain heterochromatin and siRNAs are constantly produced, even though their abundance fluctuates during the cell cycle (Kloc et al. 2008; Chen et al. 2008; Buhler et al. 2008; Volpe et al. 2002). This requirement for RNAi in the maintenance of heterochromatin at centromeres is a property that has been crucial for elucidating the functional role of ncRNAs in heterochromatin formation. If ncRNAs act in other organisms like RNAi at the mating type locus of S. pombe, there may be many more ncRNA-mediated epigenetic changes to be discovered in the future. Along that line, small ncRNAs have been implicated in transgenerational inheritance of epigenetic traits in flies, plants, mice, and worms (Regulski et al. 2013; Kiani et al. 2013; Rassoulzadegan et al. 2006; Brennecke et al. 2008; Chandler 2010; Ashe et al. 2012; Bagijn et al. 2012; Daxinger and Whitelaw 2012; Shirayama et al. 2012) and it will be exciting to watch this field of research in the coming years.
Chromatin remodeling and transcriptional interference
As outlined above, there is ample evidence for ncRNAs being directly involved in genome regulation. However, all ncRNAs may not be functionally relevant, even if they appear to be highly abundant. Work on the S. pombe fbp1+ gene revealed that ncRNA transcription through the promoter region of the fbp1+ gene is important for efficient gene activation (Hirota et al. 2008). Notably, insertion of transcription termination sequences into the promoter abolished fbp1+ expression and this could not be rescued by expressing the ncRNA ectopically. In contrast, replacing the ncRNA sequence with unrelated sequences lacking terminators did not disturb fbp1+ activation. Thus, chromatin remodeling at the promoter region by the transcribing RNA polymerase seems to be important for fbp1+ activation. The ncRNA produced can be regarded as a non-functional byproduct in this case.
Similar ncRNA-transcription-dependent but ncRNA-independent regulatory processes have been described in other organisms. In Saccharomyces cerevisiae, transcription of the SRG1 ncRNA across the SER3 promoter is required for SER3 repression. It has been suggested that the mechanistic basis for this regulation is the interference of SRG1 transcription with the binding of activators to the SER3 promoter (Martens et al. 2004; Thebault et al. 2011). Similarly, gametogenesis in budding yeast is controlled by the transcription of two ncRNAs. Transcription of the ncRNA IRT1 interferes with expression of the master regulator of IME1 by inhibiting binding of transcription factors to the IME1 promoter (van Werven et al. 2012). In Drosophila, intergenic transcription of ncRNAs upstream of the Ultrabithorax (Ubx) gene is linked to Ubx expression. As in the case of SRG1, ncRNA transcription leads to Ubx repression. Finally, extensive truncation and promoter-swap experiments in mice have shown that silencing of the imprinted Ifg2r gene also results from transcriptional overlap between protein-coding and non-coding sequences, without a contribution by the long ncRNA Airn itself (Latos et al. 2012). In conclusion, these examples illustrate that the mere act of transcription and not necessarily the resultant ncRNA can be functionally relevant.
Perspectives
Much excitement has been triggered by the notion that eukaryotic cells transcribe a much greater proportion of their genomes than anticipated, and that these seemingly non-protein coding RNAs may add yet another layer of complexity to genome regulation. Clearly, the examples highlighted in this review and others demonstrate that some ncRNAs are functionally highly relevant. However, for most of the “non-coding transcriptome” of different organisms, the extent of ncRNA involvement in regulatory circuits and the mechanisms by which they might act remain largely unexplored. Thus, a future challenge will be to systematically probe ncRNAs for physiological relevance.
The power of yeast has been its genetic tractability, which has allowed specific manipulations of an ncRNA and its underlying genomic sequence in order to perform loss-of-function experiments. The ability to do so is of particular importance when the ncRNA under investigation acts in cis on chromatin. Given the recent advances in genome-editing technologies (Wang et al. 2013; Hockemeyer et al. 2011; Jinek et al. 2012), similar decisive experiments may sooner or later become more straightforward in mammalian systems. The insertion of premature transcription termination sites, mutation of the ncRNA sequence, or the inclusion of destabilizing elements such as ribozymes into the ncRNA sequence may provide more compelling evidence for the functional relevance of many ncRNAs that have been assigned a physiological role.
Most importantly, the properties and functions of ncRNAs, whether or not their activities are associated with chromatin, are likely to be heavily dependent on their interactions with specific proteins. Therefore, we must aim at developing strategies to determine the protein partners of ncRNAs, similar to a recent proteomic study in which close to 800 proteins were identified based on their ability to bind to polyadenylated transcripts (Baltz et al. 2012). Only when we know with which proteins a particular ncRNA interacts can we start to dissect its mechanism of action precisely.
Acknowledgments
We thank Patrick King for editorial comments on the manuscript. C.K. thanks Daniel Valsecchi for exellent assistance. Research in the laboratory of M.B. is supported by the Swiss National Science Foundation, the European Research Council, the Gebert Rüf Foundation, the EMBO Young Investigator Program, and the Novartis Research Foundation.
Abbreviations
- CD
Chromodomain
- CLRC
Clr4 methyltransferase complex
- CTCF
CCCTC-binding factor
- ddRNAs
DICER- and DROSHA-dependent small RNAs
- diRNAs
DNA double-stranded break-induced small RNAs
- DSB
DNA double-stranded break
- dsRNA
double-stranded RNA
- HOTAIR
HOX Antisense Intergenic RNA
- HP1
Heterochromatin protein 1
- iCLIP
Individual-nucleotide resolution UV cross-linking and immunoprecipitation
- lncRNA
Long non-coding RNA
- miRNAs
Micro RNAs
- MLL
Mixed lineage leukemia
- MSL
Male-specific lethal
- ncRNAs
Non-coding RNAs
- Piwi
P-element-induced wimpy testis
- piRNAs
Piwi-interacting RNAs
- PRC2
Polycomb repressive complex 2
- pTEFb
Positive transcription elongation factor b
- qiRNAs
QDE-2-interacting small RNAs
- RdDM
RNA-directed DNA methylation
- RDRC
RNA-directed RNA polymerase complex
- RdRP
RNA-dependent RNA polymerase
- RITS
RNA-induced transcriptional silencing complex
- RNAi
RNA interference
- RNP
Ribonucleoprotein
- scnRNAs
‘Scan’ RNAs
- snoRNAs
Small nucleolar RNAs
- snRNAs
Small nuclear RNAs
- tRNAs
Transfer RNAs
- Ubx
Ultrabithorax
References
- Akhtar A, Zink D, Becker PB. Chromodomains are protein–RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, Kuroda MI. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ & Miska EA (2012) Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337(6094):574–578. [DOI] [PMC free article] [PubMed]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, Allshire RC. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, et al. Identification and analysis of functional elements in 1 % of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, Willard HF, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- Bracht JR, Fang W, Goldman AD, Dolzhenko E, Stein EM, Landweber LF. Genomes on the edge: programmed genome instability in ciliates. Cell. 2013;152:406–416. doi: 10.1016/j.cell.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Buhler M. RNA turnover and chromatin-dependent gene silencing. Chromosoma. 2009;118:141–151. doi: 10.1007/s00412-008-0195-z. [DOI] [PubMed] [Google Scholar]
- Buhler M, Hiller S. Dynamic nature of heterochromatin highlighted by a HP1Swi6-dependent gene silencing mechanism. Cell Cycle. 2012;11:3907–3908. doi: 10.4161/cc.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 2011;7:e1002249. doi: 10.1371/journal.pgen.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL. Paramutation’s properties and puzzles. Science. 2010;330:628–629. doi: 10.1126/science.1191044. [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Vogt A, Mochizuki K, Yao MC. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell. 2010;21:1753–1762. doi: 10.1091/mbc.E09-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2012;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. A Tetrahymena Piwi bound to mature tRNA 3' fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell. 2012;48:509–520. doi: 10.1016/j.molcel.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson BL, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Kheradpour P, Lassmann T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Good PJ, Feingold EA, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Giddings MC, Gingeras TR, Guigo R, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Mikkelsen TS, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10(2):94–108 [DOI] [PMC free article] [PubMed]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2010;25:29–40. doi: 10.1101/gad.1975011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418(6894):244–251 [DOI] [PubMed]
- Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2010;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, Chang HY, Akhtar A. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell. 2013;51:156–173. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M, Shimojo H, Hayashi A, Kawaguchi R, Ohtani Y, Uegaki K, Nishimura Y, Nakayama J. Intrinsic nucleic acid-binding activity of Chp1 chromodomain is required for heterochromatic gene silencing. Mol Cell. 2012;47:228–241. doi: 10.1016/j.molcel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb Perspect Biol. 2010;3:a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Mochizuki K. Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism. Adv Exp Med Biol. 2011;722:156–173. doi: 10.1007/978-1-4614-0332-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Keller C, Adaixo R, Stunnenberg R, Woolcock KJ, Hiller S, Buhler M. HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol Cell. 2012;47:215–227. doi: 10.1016/j.molcel.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz HR, Buhler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nat Struct Mol Biol. 2013;20:994–1000. doi: 10.1038/nsmb.2619. [DOI] [PubMed] [Google Scholar]
- Kiani J, Grandjean V, Liebers R, Tuorto F, Ghanbarian H, Lyko F, Cuzin F, Rassoulzadegan M. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013;9:e1003498. doi: 10.1371/journal.pgen.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N & Chrousos GP (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3(107):ra8 [DOI] [PMC free article] [PubMed]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Higgs DR, Gingeras TR. Molecular biology: RNA discrimination. Nature. 2012;482:310–311. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, Aumayr K, Pasierbek P, Barlow DP. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Lee JT, Strauss WM, Dausman JA, Jaenisch R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Allshire RC. Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol. 2011;23:258–265. doi: 10.1016/j.ceb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10 [DOI] [PMC free article] [PubMed]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, Visvikis A, Branlant C. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Muller M, Frohlich J, Langer D, Becker PB. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol Cell. 2013;51:174–184. doi: 10.1016/j.molcel.2013.06.011. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, Almouzni G. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- Maison C, Romeo K, Bailly D, Dubarry M, Quivy JP, Almouzni G. The SUMO protease SENP7 is a critical component to ensure HP1 enrichment at pericentric heterochromatin. Nat Struct Mol Biol. 2012;19:458–460. doi: 10.1038/nsmb.2244. [DOI] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–457. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- Meller VH, Gordadze PR, Park Y, Chu X, Stuckenholz C, Kelley RL, Kuroda MI. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr Biol. 2000;10:136–143. doi: 10.1016/s0960-9822(00)00311-0. [DOI] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K. DNA rearrangements directed by non-coding RNAs in ciliates. Wiley Interdiscip Rev RNA. 2011;1:376–387. doi: 10.1002/wrna.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA. 2011;3:92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Chuma S. piRNAs and their involvement in male germline development in mice. Dev Growth Differ. 2012;54:78–92. doi: 10.1111/j.1440-169X.2011.01320.x. [DOI] [PubMed] [Google Scholar]
- Ponicsan SL, Houel S, Old WM, Ahn NG, Goodrich JA & Kugel JF (2013) The Non-Coding B2 RNA Binds to the DNA Cleft and Active-Site Region of RNA Polymerase II. J Mol Biol 425(19):3625–3638 [DOI] [PMC free article] [PubMed]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Provost P, Silverstein RA, Dishart D, Walfridsson J, Djupedal I, Kniola B, Wright A, Samuelsson B, Radmark O, Ekwall K. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc Natl Acad Sci U S A. 2002;99:16648–16653. doi: 10.1073/pnas.212633199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MT, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, Tingey S, Rafalski A, Hicks J, Ware D & Martienssen R (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. doi:10.1101/gr.153510.112 [DOI] [PMC free article] [PubMed]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22:156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC. The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013a;340:619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013b;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- Thebault P, Boutin G, Bhat W, Rufiange A, Martens J, Nourani A. Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II. Mol Cell Biol. 2011;31:1288–1300. doi: 10.1128/MCB.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell. 2008;29:313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Willingham AT, Dike S, Cheng J, Manak JR, Bell I, Cheung E, Drenkow J, Dumais E, Duttagupta R, Ganesh M, Ghosh S, Helt G, Nix D, Piccolboni A, Sementchenko V, Tammana H, Kapranov P, Gingeras TR. Transcriptional landscape of the human and fly genomes: nonlinear and multifunctional modular model of transcriptomes. Cold Spring Harb Symp Quant Biol. 2006;71:101–110. doi: 10.1101/sqb.2006.71.068. [DOI] [PubMed] [Google Scholar]
- Woolcock KJ, Gaidatzis D, Punga T, Buhler M. Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat Struct Mol Biol. 2010;18:94–99. doi: 10.1038/nsmb.1935. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14:142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]