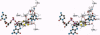Abstract
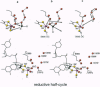
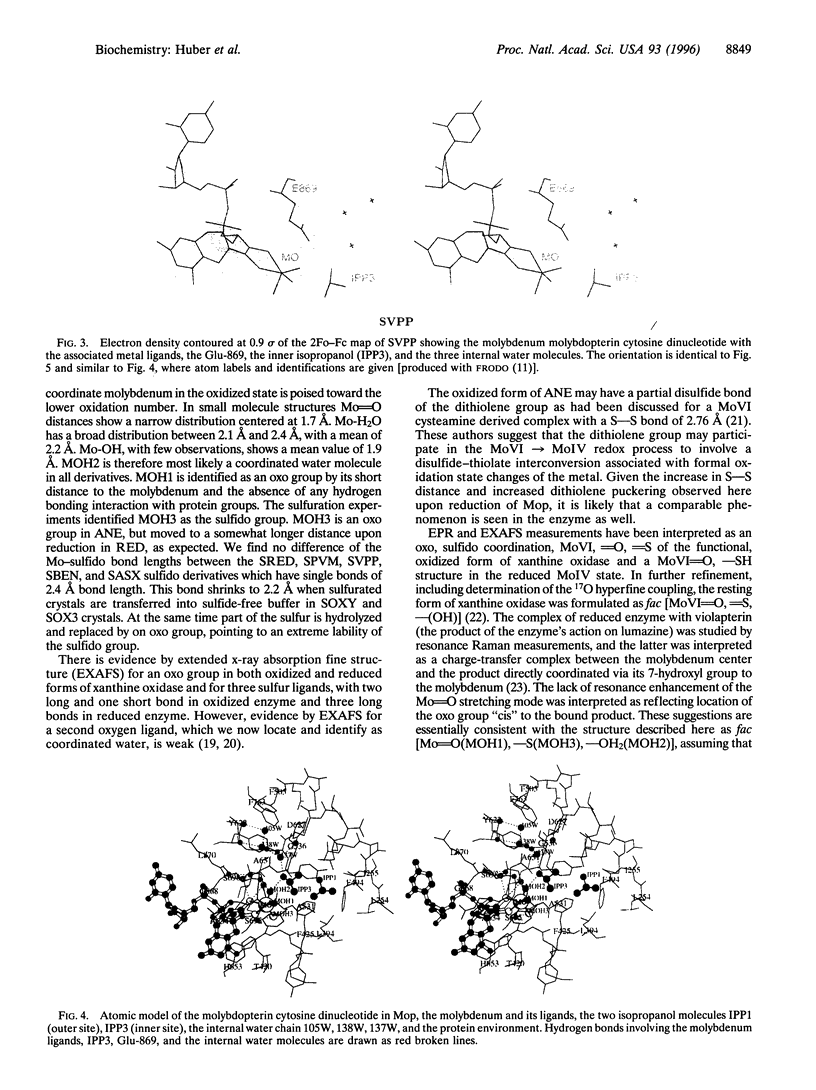
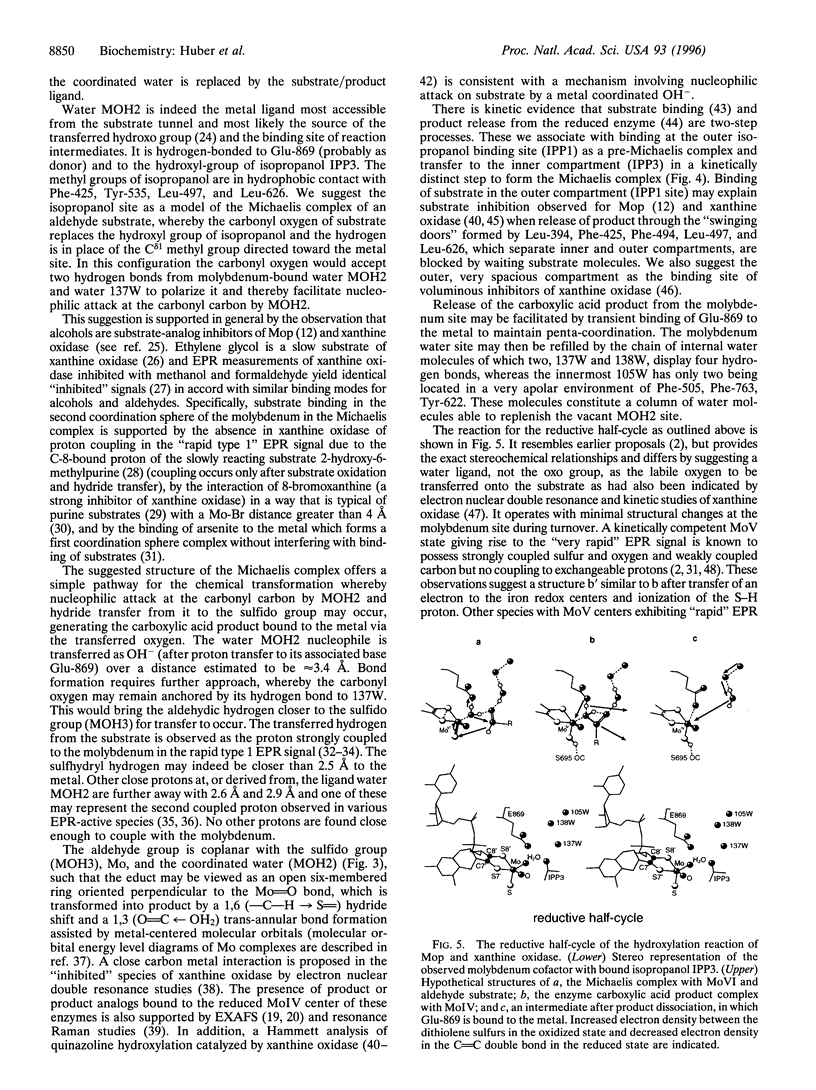
The crystal structure of the xanthine oxidase-related molybdenum-iron protein aldehyde oxido-reductase from the sulfate reducing anaerobic Gram-negative bacterium Desulfovibrio gigas (Mop) was analyzed in its desulfo-, sulfo-, oxidized, reduced, and alcohol-bound forms at 1.8-A resolution. In the sulfo-form the molybdenum molybdopterin cytosine dinucleotide cofactor has a dithiolene-bound fac-[Mo, = O, = S, ---(OH2)] substructure. Bound inhibitory isopropanol in the inner compartment of the substrate binding tunnel is a model for the Michaelis complex of the reaction with aldehydes (H-C = O,-R). The reaction is proposed to proceed by transfer of the molybdenum-bound water molecule as OH- after proton transfer to Glu-869 to the carbonyl carbon of the substrate in concert with hydride transfer to the sulfido group to generate [MoIV, = O, -SH, ---(O-C = O, -R)). Dissociation of the carboxylic acid product may be facilitated by transient binding of Glu-869 to the molybdenum. The metal-bound water is replenished from a chain of internal water molecules. A second alcohol binding site in the spacious outer compartment may cause the strong substrate inhibition observed. This compartment is the putative binding site of large inhibitors of xanthine oxidase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barata B. A., LeGall J., Moura J. J. Aldehyde oxidoreductase activity in Desulfovibrio gigas: in vitro reconstitution of an electron-transfer chain from aldehydes to the production of molecular hydrogen. Biochemistry. 1993 Nov 2;32(43):11559–11568. doi: 10.1021/bi00094a012. [DOI] [PubMed] [Google Scholar]
- Barata B. A., Liang J., Moura I., Legall J., Moura J. J., Huynh B. H. Mössbauer study of the native, reduced and substrate-reacted Desulfovibrio gigas aldehyde oxido-reductase. Eur J Biochem. 1992 Mar 1;204(2):773–778. doi: 10.1111/j.1432-1033.1992.tb16693.x. [DOI] [PubMed] [Google Scholar]
- Bray R. C., George G. N. Electron-paramagnetic-resonance studies using pre-steady-state kinetics and substitution with stable isotopes on the mechanism of action of molybdoenzymes. Biochem Soc Trans. 1985 Jun;13(3):560–567. doi: 10.1042/bst0130560. [DOI] [PubMed] [Google Scholar]
- Bray R. C., Gutteridge S. Numbers and exchangeability with water of oxygen-17 atoms coupled to molybdenum (V) in different reduced forms of xanthine oxidase. Biochemistry. 1982 Nov 9;21(23):5992–5999. doi: 10.1021/bi00266a041. [DOI] [PubMed] [Google Scholar]
- Bray R. C. The inorganic biochemistry of molybdoenzymes. Q Rev Biophys. 1988 Aug;21(3):299–329. doi: 10.1017/s0033583500004479. [DOI] [PubMed] [Google Scholar]
- D'Ardenne S. C., Edmondson D. E. Kinetic isotope effect studies on milk xanthine oxidase and on chicken liver xanthine dehydrogenase. Biochemistry. 1990 Sep 25;29(38):9046–9052. doi: 10.1021/bi00490a023. [DOI] [PubMed] [Google Scholar]
- Davis M. D., Olson J. S., Palmer G. The reaction of xanthine oxidase with lumazine. Characterization of the reductive half-reaction. J Biol Chem. 1984 Mar 25;259(6):3526–3533. [PubMed] [Google Scholar]
- George G. N., Bray R. C. Reaction of arsenite ions with the molybdenum center of milk xanthine oxidase. Biochemistry. 1983 Mar 1;22(5):1013–1021. doi: 10.1021/bi00274a003. [DOI] [PubMed] [Google Scholar]
- Grams F., Reinemer P., Powers J. C., Kleine T., Pieper M., Tschesche H., Huber R., Bode W. X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design. Eur J Biochem. 1995 Mar 15;228(3):830–841. doi: 10.1111/j.1432-1033.1995.tb20329.x. [DOI] [PubMed] [Google Scholar]
- Guasch A., Coll M., Avilés F. X., Huber R. Three-dimensional structure of porcine pancreatic procarboxypeptidase A. A comparison of the A and B zymogens and their determinants for inhibition and activation. J Mol Biol. 1992 Mar 5;224(1):141–157. doi: 10.1016/0022-2836(92)90581-4. [DOI] [PubMed] [Google Scholar]
- Gutteridge S., Tanner S. J., Bray R. C. Comparison of the molybdenum centres of native and desulpho xanthine oxidase. The nature of the cyanide-labile sulphur atom and the nature of the proton-accepting group. Biochem J. 1978 Dec 1;175(3):887–897. doi: 10.1042/bj1750887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge S., Tanner S. J., Bray R. C. The molybdenum centre of native xanthine oxidase. Evidence for proton transfer from substrates to the centre and for existence of an anion-binding site. Biochem J. 1978 Dec 1;175(3):869–878. doi: 10.1042/bj1750869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOFSTEE B. H. On the mechanism of inhibition of xanthine oxidase by the substrate xanthine. J Biol Chem. 1955 Sep;216(1):235–244. [PubMed] [Google Scholar]
- Hille R., Kim J. H., Hemann C. Reductive half-reaction of xanthine oxidase: mechanistic role of the species giving rise to the "rapid type 1" molybdenum(V) electron paramagnetic resonance signal. Biochemistry. 1993 Apr 20;32(15):3973–3980. doi: 10.1021/bi00066a018. [DOI] [PubMed] [Google Scholar]
- Hille R., Kim J. H., Hemann C. Reductive half-reaction of xanthine oxidase: mechanistic role of the species giving rise to the "rapid type 1" molybdenum(V) electron paramagnetic resonance signal. Biochemistry. 1993 Apr 20;32(15):3973–3980. doi: 10.1021/bi00066a018. [DOI] [PubMed] [Google Scholar]
- Hille R., Sprecher H. On the mechanism of action of xanthine oxidase. Evidence in support of an oxo transfer mechanism in the molybdenum-containing hydroxylases. J Biol Chem. 1987 Aug 15;262(23):10914–10917. [PubMed] [Google Scholar]
- Hille R., Stewart R. C., Fee J. A., Massey V. The interaction of arsenite with xanthine oxidase. J Biol Chem. 1983 Apr 25;258(8):4849–4856. [PubMed] [Google Scholar]
- Hille R., Stewart R. C. The inhibition of xanthine oxidase by 8-bromoxanthine. J Biol Chem. 1984 Feb 10;259(3):1570–1576. [PubMed] [Google Scholar]
- Hille R., Stewart R. C. The inhibition of xanthine oxidase by 8-bromoxanthine. J Biol Chem. 1984 Feb 10;259(3):1570–1576. [PubMed] [Google Scholar]
- Howes B. D., Bray R. C., Richards R. L., Turner N. A., Bennett B., Lowe D. J. Evidence favoring molybdenum-carbon bond formation in xanthine oxidase action: 17Q- and 13C-ENDOR and kinetic studies. Biochemistry. 1996 Feb 6;35(5):1432–1443. doi: 10.1021/bi9520500. [DOI] [PubMed] [Google Scholar]
- Howes B. D., Pinhal N. M., Turner N. A., Bray R. C., Anger G., Ehrenberg A., Raynor J. B., Lowe D. J. Proton electron-nuclear double-resonance spectra of molybdenum(V) in different reduced forms of xanthine oxidase. Biochemistry. 1990 Jul 3;29(26):6120–6127. doi: 10.1021/bi00478a002. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Hille R. Reductive half-reaction of xanthine oxidase with xanthine. Observation of a spectral intermediate attributable to the molybdenum center in the reaction of enzyme with xanthine. J Biol Chem. 1993 Jan 5;268(1):44–51. [PubMed] [Google Scholar]
- Malthouse J. P., Bray R. C. The nature of the sulphur atom liberated from xanthine oxidase by cyanide. Evidence from e.p.r. spectroscopy after 35S substitution. Biochem J. 1980 Oct 1;191(1):265–267. doi: 10.1042/bj1910265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey V., Edmondson D. On the mechanism of inactivation of xanthine oxidase by cyanide. J Biol Chem. 1970 Dec 25;245(24):6595–6598. [PubMed] [Google Scholar]
- Morpeth F. F., Bray R. C. Inhibition of xanthine oxidase by various aldehydes. Biochemistry. 1984 Mar 13;23(6):1332–1338. doi: 10.1021/bi00301a047. [DOI] [PubMed] [Google Scholar]
- Morpeth F. F. Studies on the specificity toward aldehyde substrates and steady-state kinetics of xanthine oxidase. Biochim Biophys Acta. 1983 May 18;744(3):328–334. doi: 10.1016/0167-4838(83)90207-8. [DOI] [PubMed] [Google Scholar]
- Nar H., Huber R., Auerbach G., Fischer M., Hösl C., Ritz H., Bracher A., Meining W., Eberhardt S., Bacher A. Active site topology and reaction mechanism of GTP cyclohydrolase I. Proc Natl Acad Sci U S A. 1995 Dec 19;92(26):12120–12125. doi: 10.1073/pnas.92.26.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertling W. A., Hille R. Resonance-enhanced Raman scattering from the molybdenum center of xanthine oxidase. J Biol Chem. 1990 Oct 15;265(29):17446–17450. [PubMed] [Google Scholar]
- Oertling W. A., Hille R. Resonance-enhanced Raman scattering from the molybdenum center of xanthine oxidase. J Biol Chem. 1990 Oct 15;265(29):17446–17450. [PubMed] [Google Scholar]
- Okamoto K., Nishino T. Mechanism of inhibition of xanthine oxidase with a new tight binding inhibitor. J Biol Chem. 1995 Apr 7;270(14):7816–7821. doi: 10.1074/jbc.270.14.7816. [DOI] [PubMed] [Google Scholar]
- Phillips M. A., Fletterick R., Rutter W. J. Arginine 127 stabilizes the transition state in carboxypeptidase. J Biol Chem. 1990 Nov 25;265(33):20692–20698. [PubMed] [Google Scholar]
- Pick F. M., McGartoll M. A., Bray R. C. Reaction of formaldehyde and of methanol with xanthine oxidase. Eur J Biochem. 1971 Jan 1;18(1):65–72. doi: 10.1111/j.1432-1033.1971.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Romão M. J., Archer M., Moura I., Moura J. J., LeGall J., Engh R., Schneider M., Hof P., Huber R. Crystal structure of the xanthine oxidase-related aldehyde oxido-reductase from D. gigas. Science. 1995 Nov 17;270(5239):1170–1176. doi: 10.1126/science.270.5239.1170. [DOI] [PubMed] [Google Scholar]
- Romão M. J., Barata B. A., Archer M., Lobeck K., Moura I., Carrondo M. A., LeGall J., Lottspeich F., Huber R., Moura J. J. Subunit composition, crystallization and preliminary crystallographic studies of the Desulfovibrio gigas aldehyde oxidoreductase containing molybdenum and [2Fe-2S] centers. Eur J Biochem. 1993 Aug 1;215(3):729–732. doi: 10.1111/j.1432-1033.1993.tb18085.x. [DOI] [PubMed] [Google Scholar]
- Skibo E. B., Gilchrist J. H., Lee C. H. Electronic probes of the mechanism of substrate oxidation by buttermilk xanthine oxidase: role of the active-site nucleophile in oxidation. Biochemistry. 1987 Jun 2;26(11):3032–3037. doi: 10.1021/bi00385a013. [DOI] [PubMed] [Google Scholar]
- Tanner S. J., Bray R. C. Ethylene glycol as a very slow substrate for xanthine oxidase [proceedings]. Biochem Soc Trans. 1978;6(6):1331–1333. doi: 10.1042/bst0061331. [DOI] [PubMed] [Google Scholar]
- Thoenes U., Flores O. L., Neves A., Devreese B., Van Beeumen J. J., Huber R., Romão M. J., LeGall J., Moura J. J., Rodrigues-Pousada C. Molecular cloning and sequence analysis of the gene of the molybdenum-containing aldehyde oxido-reductase of Desulfovibrio gigas. The deduced amino acid sequence shows similarity to xanthine dehydrogenase. Eur J Biochem. 1994 Mar 15;220(3):901–910. doi: 10.1111/j.1432-1033.1994.tb18693.x. [DOI] [PubMed] [Google Scholar]
- Turner N. A., Bray R. C., Diakun G. P. Information from e.x.a.f.s. spectroscopy on the structures of different forms of molybdenum in xanthine oxidase and the catalytic mechanism of the enzyme. Biochem J. 1989 Jun 1;260(2):563–571. doi: 10.1042/bj2600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N., Barata B., Bray R. C., Deistung J., Le Gall J., Moura J. J. The molybdenum iron-sulphur protein from Desulfovibrio gigas as a form of aldehyde oxidase. Biochem J. 1987 May 1;243(3):755–761. doi: 10.1042/bj2430755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl R. C., Rajagopalan K. V. Evidence for the inorganic nature of the cyanolyzable sulfur of molybdenum hydroxylases. J Biol Chem. 1982 Feb 10;257(3):1354–1359. [PubMed] [Google Scholar]