Abstract
Background
The prognostic value of HIFs in colorectal cancer was evaluated in a large number of studies, but the conclusions were inconclusive. Meanwhile, clinicopathologic differences of HIF-1α and HIF-2α were rarely compared in recent studies.
Methodology
Identical search strategies were used to search relevant literatures in the PubMed and Web of Science databases. The prognostic significances and clinicopathological differences of HIFs in CRC were analyzed.
Principal Findings
A total of 23studies comprising 2984 CRC patients met the inclusion criteria. The results indicated that overexpressed HIFs were significantly associated with increase of mortality risk, including overall survival (OS) (HR 2.06 95%CI 1.55–2.74) and disease free survival (HR 2.84, 95%CI 1.87–4.31). Subgroup analysis revealed that both overexpressed HIF-1α and HIF-2α had correlations with worse prognosis. The pooled HRs were 2.01 (95% CI: 1.55–2.6) and 2.07(95% CI: 1.01–4.26). Further subgroup analysis on HIF-1α was performed by study location, number of patients, quality score and cut-off value. The results showed that HIF-1α overexpression was significantly associated with poor OS, particularly in Asian countries (HR 2.3, 95% CI: 1.74–3.01), while not in European or other countries. In addition, overexpression of HIF-1α was closely related with these clinicopathological features, including Dukes' stages (OR 0.39, 95% CI: 0.17–0.89), UICC stages (OR 0.42 95% CI: 0.3–0.59), depth of invasion (OR 0.71, 95% CI: 0.51–0.99), lymphnode status (OR 0.49, 95% CI: 0.32–0.73) and metastasis (OR 0.29, 95% CI: 0.11–0.81). While overexpression of HIF-2α was only associated with grade of differentiation (OR 0.48, 95% CI: 0.29–0.81).
Conclusions
This study showed that both HIF-1α and HIF-2α overexpression were associated with an unfavorable prognosis. HIF-1α overexpression seemed to be associated with worse prognosis in Asian countries. Additionally, HIF-1α and HIF-2α indicated distinct clinicopathologic features.
Introduction
Colorectal cancer is the third most common malignancy worldwide, and one of the leading causes of cancer-related deaths [1]. An increasing trend in the incidence of this carcinoma has been noticed in the Asian nations. Despite recent therapeutic advances, its 5-year survival rate is still pessimistic due to its recurrence and drug resistance [2]. Growing evidence suggests that hypoxia plays a pivotal role in disease progression and therapy resistance in most solid tumors, including colorectal cancer [3], [4]. Rapid oxygen consumption and aberrant tumor angiogenesis and blood flow result in a hypoxic tumor environment. Owing to the fundamental importance of oxygen for metabolism and survival, cells have evolved intricate response mechanisms to respond to hypoxia. The most important regulators mediating the primary transcriptional responses to hypoxic stress are hypoxia-inducible factors (HIFs). Given that hypoxia promotes tumor progression and therapy resistance, HIFs are expected to be useful biomarkers associated with progress disease and poor prognosis in CRC. Increased expression of HIFs has also been observed in a broad range of human cancer cell types, and has been associated with poor prognosis in many cases [5], but the prognostic value of HIFs for CRC patients is inconclusive.
HIFs are heterodimers composed of an inducible a-subunit (HIF-1α, HIF-2α or HIF-3α), and a constitutive HIF-1β subunit (also known as aryl hydrocarbon receptor nuclear translocator or ARNT), which together form the HIF-1, HIF-2 and HIF-3 transcriptional complexes, respectively. Of the three HIF family members, HIF-1 and HIF-2 are the most well-characterized. HIF-1α and HIF-2α are usually detected to measure tumor oxygen levels because the HIF-1β subunit is constitutive. HIF-1α and HIF-2α have 48% amino acid sequence identity and similar protein structures, but distinct target genes and mechanisms of regulation. HIF-1α preferentially induces glycolytic pathway, whereas HIF-2α regulates genes involved in tumor growth, cell cycle and maintaining stem cell pluripotency [6]. Thus, HIF1α and HIF2α can promote highly divergent, even opposing, outcomes, which results in distinct clinicopathologic features and prognosis. Multiple xenograft tumour models also support the hypothesis that HIF1α and HIF2α play different roles in tumor progression by regulating both shared and unique target genes [7]. However, clinicopathologic and prognostic differences of HIF1α and HIF2α in CRC were rarely compared in recent studies.
Therefore, we made a meta-analysis from eligible studies to investigate the relationship between HIF expression and prognosis of CRC patients. Meanwhile, we performed a subgroup analysis to assess the roles of HIF-1α and HIF-2α in clinicopathologic features and prognosis of CRC.
Materials and Methods
Identification and eligibility of relevant studies
We searched literature from PubMed, WanFang and Web of Science databases using the terms: “HIF”, “colorectal neoplasms”, “colorectal Cancer”, “colon cancer” “rectal cancer”, “prognosis” with all possible combinations. Bibliographies, review articles and other pertinent studies were searched manually for additional eligible studies.
The inclusion criteria for eligibility of a study in the meta-analysis were as follows: (1) evaluating HIF expression in the human CRC tissues; (2) assessing the relationships between HIFs expression with CRC clinicopathologic features or prognosis; (3) articles written in English or Chinese; (4) sufficient information provided to estimate hazard ratio (HR) about overall survival (OS) or disease free survival (DFS), or to estimate odds ratio (OR) about clinicopathologic features. In addition, letters, reviews, conference abstracts, and case reports were not in the scope of our analysis because of the limited data. Overlapping articles were also excluded from this meta-analysis, only the most recent or the most complete study was involved in the analysis.
Data extraction and management
Two investigators (Xin He and Wenjie Xia) reviewed each eligible study independently and extracted data from all the publications meeting the inclusion criteria. Controversial problems were arbitrated by the third investigator (Jinhong Xu). The following information was collected from each study: the first author's name, year of publication, country of origin, number of patients, gender of patients, HIF isoforms, source and dilution of antibody, cut-off value, tumor characteristics, condition of adjuvant therapy and survival data.
Methodological assessment
Newcastle–Ottawa quality assessment scale (NOS) was used to assess the quality of each study [8]. The score assessed eight items of methodology, categorized into three dimensions including selection, comparability, and outcome. A maximum of 1 score was awarded for each item with the exception of the item related to comparability that allowed the assignment of two scores. A total of 0 and 9 scores were respectively designated as lowest and highest quality, and the studies with 6 scores or more were graded as the high quality ones in the scale. The scores provided by two researchers were compared and a consensus value for each item was achieved.
Statistical methods
For the pooled analysis of the impact of HIF expression on survival outcome, HRs and its 95% CI were used. If these statistical variables were described in a literature, we pooled it directly; otherwise, they were calculated from available numerical data in the articles according to the methods described by Parmar [9]. In brief, if the trials offered the data such as log-rank test p values, number of total events. The number of aberrant HIF expression and number of preserved HIF expression were extracted to allow estimation of the HR and its 95% CI. If only Kaplan Meier graphs were published, Kaplan-Meier curves were read by Engauge Digitizer version4.1 (http://digitizer.sourceforge.net/). Time-to-event data from the Kaplan–Meier curves was extracted and HR and its 95% CI were calculated via SPSS16.0. Odds ratios (ORs) and their 95%CIs were combined to evaluate the association between HIF expression and clinicopathological factors, such as differentiation grade, Dukes' stages, depth of invasion, lymphnode status and metastasis. An observed HR>1 implies worse survival for the group with overexpressed/negative HIF expression. An observed OR<1 implies unfavorable parameters for the group with overexpressed/negative HIF expression. The impact of overexpressed/negative HIF expression on survival or clinicopathological factors was considered to be statistically significant if the 95%CI did not overlap with 1. Heterogeneity in between-study was assessed by Chi- square based Q statistical test [10]. And the I2 statistic to quantify the proportion of the total variation, which is due to inter-study heterogeneity rather than sampling error and is measured from 0% to 100% [11]. Higher values indicate a greater degree of heterogeneity. When the studies were found to be homogeneous(with P>0.10 for the Q test), the pooled ORs and HRs estimate of each study were calculated by the fixed-effects model (the Mantel-Haenszel method) [12]. Otherwise, we chose the random-effects model (the DerSimonian and Laird method) [13]. We assessed the possibility of publication bias by visually assessing a funnel plot for asymmetry and by quantitatively performing Egger's test. Publication bias was indicated when p value of Egger's test <0.05. The meta-analysis was performed using STATA version 12.0 software (Stata Corporation, Collage Station, Texas, USA). All the P values were for a two-side test and considered statistically significant when p<0.05.
Results
Description of studies
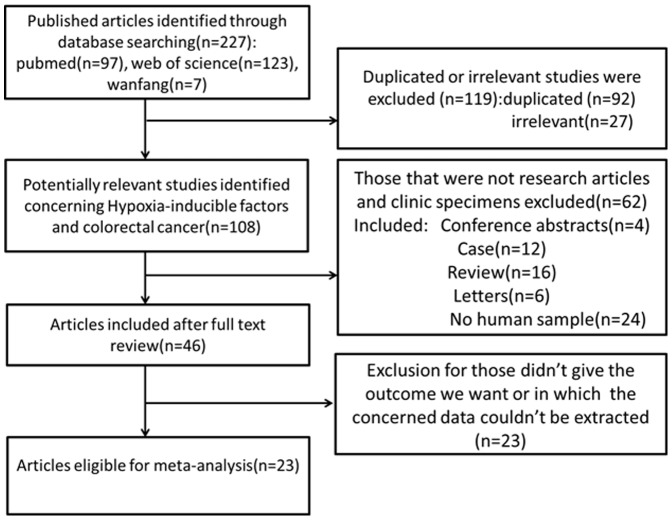
As shown in Figure 1 , 227 published records were identified from a search of the above databases using the search strategy as described above. After exclusion of the studies that were out of the scope of our systematic review, a total of 23 eligible studies were included in the final meta-analysis [4], [5], [14]–[32]. Of these 23 publications, 20 studies assessed the relationships between HIF-1 expression with CRC clinicopathologic features or prognosis, while 6 studies evaluated the association of HIF-2 expression and CRC pathological features or prognosis. The clinical features of these 23 included studies were summarized in Table 1 . These studies were published from 2003 to 2013, and total 2984 CRC patients were enrolled. Sample sizes ranged from 30 to 731 patients (mean 130). 14 of these studies enrolled less than 100 patients and 9 studies included more than 100 patients. 6 of these studies evaluated patients from China, 5 from Japan, 3 from England, others from America, Korea, Finland, Germany, Austrialia, Holand and Greece. 19 of these studies got 6 scores or more in methodological assessment, which meant they had high qualities.
Figure 1. Flow diagram of study selection procedure.
Table 1. Characteristics of studies included in the meta-analysis.
| First author | Year | Country | HIF isoforms | Number of total patient(positive) | Expression location | Method | Antibody source | Antibody dilution | Definition of HIF positive | HR estimation | Quality score |
| Xie | 2012 | China | HIF-1α | 93(46) | C and N | IHC | Santa Cruz | 1∶100 | Multiplying the intensitys core by expressions core≥median value | NA | 8 |
| Korkeila | 2011 | Finland | HIF-1α | 168(68) | N | IHC | BD | 1∶100 | Weak,moderate or strong staining | HR for DFS | 7 |
| Shioya | 2011 | Japan | HIF-1α | 50(21) | N | IHC | Neomarkers | 1∶20000 | 40% | HR for OS and survival curves for DFS | 8 |
| Havelund | 2011 | Denmark | HIF-1α | 86(39) | N | IHC | BD | 1∶75 | Summing the intensitys core and expressions core≥3 | Survival curves for OS | 7 |
| Saigusa | 2011 | Japan | HIF-1α | 52(NA) | NA | RT-PCR | NA | NA | 0.0212 for PFS and 0.1274 for OS | HR for OS and DFS | 7 |
| Mohammed | 2011 | Spain | HIF-2α | 154(NA) | NA | RT-PCR | NA | NA | NA | NA | 6 |
| Kwon | 2010 | Korea | HIF-1α | 311(196) | N | IHC | Novus | 1∶50 | 10% | HR for OS and DFS | 7 |
| Wu | 2010 | China | HIF-1α | 68(30) | C and N | IHC | Abcam | 1∶200 | Multiplying the intensitys core by expressions core≥5 | NA | 6 |
| Zheng | 2010 | China | HIF-1α | 62(39) | C and N | IHC | Boster | 1∶400 | Summing the intensitys core and expressions core≥5 | NA | 5 |
| Baba | 2010 | United States | HIF-1α | 731(142) | C | IHC | Santa Cruz | 1∶500 | Moderate staining>50% or Any strong staining | HR for OS | 7 |
| HIF-2α | 695(322) | C | IHC | Santa Cruz | 1∶250 | Weak to strong expression. | HR for OS | ||||
| Toiyama | 2010 | England | HIF-1α | 40(NA) | NA | RT-PCR | NA | NA | 0.3649 | HR for DFS | 5 |
| Gao | 2009 | China | HIF-1α | 71(39) | C and N | IHC | Zymed | 1∶50 | Any staining | HR for OS | 8 |
| Jubb | 2009 | Austrialia | HIF-2α | 155(60) | N | IHC | BD | NA | Any staining | HR for OS | 6 |
| Rajaganeshan | 2009 | England | HIF-1α | 55(25) | C or N | IHC | Abcam | 1∶100 | >10% in nuclear or distinct, Strong staining in cytoplasm | HR for DFS | 4 |
| Schmitz | 2009 | Germany | HIF-1α | 129(34) | N | IHC | Transduction Laboratories | 1∶10 | Any staining | NA | 8 |
| Rasheed | 2009 | England | HIF-1α | 90(48) | C and N | IHC | Novus | 1∶500 | NA | HR for DFS | 7 |
| HIF-2α | 90(58) | C and N | IHC | Novus | 1∶100 | NA | NA | ||||
| Cleven | 2007 | Holand | HIF-2α | 133(110) | N | IHC | BD | 1∶120 | 5% | HR for OS | 4 |
| Lu | 2006 | China | HIF-1α | 30(19) | N | IHC | NA | 1∶200 | 10% | Survival curves for OS | 7 |
| Theodoropoulos | 2006 | Greece | HIF-1α | 92(44) | N | IHC | Stress Gene | 1∶1200 | Moderate or strong staining | HR for OS and DFS | 7 |
| Yoshimura | 2004 | Japan | HIF-1α | 87(39) | N | IHC | Novus | 1∶1000 | 5% | Survival curves for OS | 6 |
| HIF-2α | 87(26) | N | IHC | Novus | 1∶1000 | 5% | Survival curves for OS | ||||
| Kuwai | 2003 | Japan | HIF-1α | 139(81) | C and N | IHC | Dako | 1∶1000 | 9.60% | NA | 6 |
| Shimomura | 2013 | Japan | HIF-1α | 64(20) | C | IHC | Novus | 1∶50 | Sum the intensity and percentage scores≥6 | Survival curves for OS | 7 |
| Yu | 2012 | China | HIF-1α | 124(67) | C | IHC | Abcam | 1∶100 | 10% | Survival curves for OS | 6 |
NA, not available; HR, hazard ratio; OS, overall survival; DFS, disease free survival; IHC, immunohistochemistry; C, cytoplasm; N, nucleus.
Impact of HIFs expression on overall survival and disease free survival of colorectal cancer
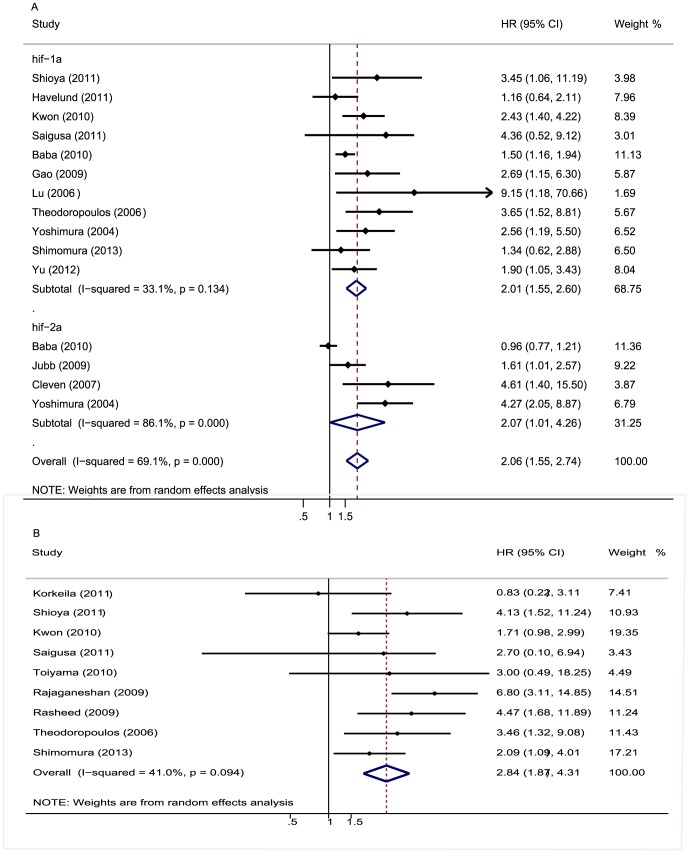
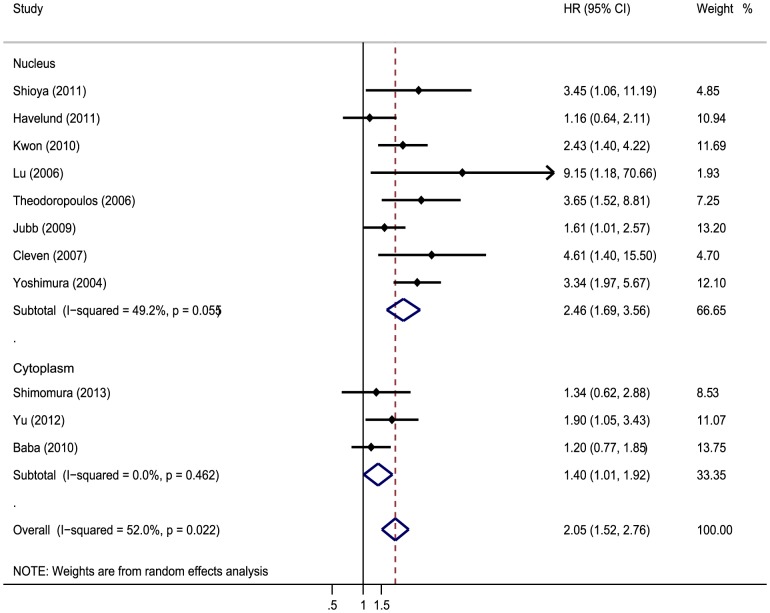
The meta-analysis was performed on 15 studies assessing the association of HIFs expression with OS. The pooled HR was 2.06 (95%CI 1.55–2.74; I2 69.1%) ( Figure 2A ). Nine studies evaluating the correlation of HIFs expression with DFS were all about HIF-1α. The pooled HR was 2.84 (95%CI 1.87–4.31; I2 41%) ( Figure 2B ). It suggested that overexpressed HIF was significantly associated with increase of mortality risk. In addition, sensitive analysis was performed. We removed one study at a time and evaluated the rest, pooled HR of HIFs overexpression on OS ranged from 1.98(95% CI: 1.5–2.61) to 2.28(95% CI: 1.74–2.98) ( Table 2 ), and combined HR of HIFs overexpression on DFS ranged from 2.34(95% CI: 1.68–3.26)to 3.22(95% CI: 2.08–4.99) ( Table 3 ). We also performed subgroup analysis about association of HIFs expression with OS by HIF isoforms, the results showed that both HIF-1α and HIF-2α were associated with worse prognosis. The pooled HR was 2.01 (95% CI: 1.55–2.6, I2 33.1%) and 2.07(95% CI: 1.01–4.26, I2 86.1%) respectively ( Figure 2A ). Subgroup analysis about association of different subcellular localization of HIFs expression with OS was performed, and the results showed that the correlation was not changed no matter where HIF located in (nucleus or cytoplasm). The pooled HR was 2.456 (95% CI: 1.694–3.561, I2 49.2%) and 2.049(95% CI: 1.519–2.764, I2 0%), respectively ( Figure 3 ).
Figure 2. Forrest plot of Hazard ratio (HR) for the association of different HIF isoforms expression with overall survival (OS) and disease free survival (DFS).
A. HRs with corresponding 95% CIs of the HIFs expression with OS. B. HRs with corresponding 95% CIs of the HIFs expression with DFS. HR>1 implied worse survival for the group with increased HIFs/negative expression and overexpressed HIFs was significantly with the worse prognosis of CRC patients.
Table 2. HRs (95% CI) of sensitivity analysis for HIFs overexpression on OS.
| Study omitted | Estimated HR | low value of 95%CI | High value of 95%CI |
| Korkeila (2011) | 3.08576 | 2.080869 | 4.575933 |
| Shioya (2011) | 2.716886 | 1.716758 | 4.299657 |
| Kwon (2010) | 3.219644 | 2.075922 | 4.993494 |
| Saigusa (2011) | 2.852396 | 1.829702 | 4.446713 |
| Toiyama (2010) | 2.839389 | 1.815004 | 4.441936 |
| Rajaganeshan (2009) | 2.338718 | 1.678648 | 3.258336 |
| Rasheed (2009) | 2.684669 | 1.706306 | 4.224007 |
| Theodoropoulos (2006) | 2.776092 | 1.734063 | 4.444295 |
| Shimomura (2013) | 3.028906 | 1.852393 | 4.95266 |
| Combined | 2.8408349 | 1.8734429 | 4.3077604 |
Table 3. HRs (95% CI) of sensitivity analysis for HIFs overexpression on DFS.
| Study omitted | Estimated HR | low value of 95%CI | High value of 95%CI |
| Shioya (2011) | 2.0867274 | 1.567879 | 2.7772751 |
| Havelund (2011) | 2.2600963 | 1.7024611 | 3.0003829 |
| Kwon (2010) | 2.1164002 | 1.5588678 | 2.8733358 |
| Saigusa (2011) | 2.0795095 | 1.5696353 | 2.7550092 |
| Baba (2010) | 2.2764547 | 1.7367126 | 2.9839401 |
| Gao (2009) | 2.1033344 | 1.5671818 | 2.8229115 |
| Lu (2006) | 2.0664124 | 1.5748442 | 2.7114177 |
| Theodoropoulos (2006) | 2.0455844 | 1.541568 | 2.7143891 |
| Yoshimura (2004) | 1.9817994 | 1.5027167 | 2.6136189 |
| Shimomura (2013) | 2.2203045 | 1.6552455 | 2.97826 |
| Yu (2012) | 2.1814032 | 1.602671 | 2.9691179 |
| Jubb (2009) | 2.2353566 | 1.6377747 | 3.0509808 |
| Cleven (2007) | 2.0521758 | 1.554372 | 2.7094064 |
| Combined | 2.127789 | 1.6131618 | 2.8065913 |
Figure 3. Forrest plot of Hazard ratio (HR) for the association of HIF in different subcellular localization with overall survival (OS).
Moreover, further subgroup analysis on HIF-1α was performed by study location, number of patients, antibody dilution, cut-off value. Subgroup analysis indicated a significant relation between HIF-1α overexpression and OS was exhibited in Asian countries (HR 2.3, 95% CI: 1.74–3.01, I2 0%). Other factors comprising number of patients, antibody dilution and cut-off value did not alter the significant OS of overexpressed HIF-1α ( Table 4 ).
Table 4. Stratified analysis of pooled hazard ratios for colorectal cancer patients with overexpressed HIF-1α.
| Heterogeneity | |||||||
| Stratified analysis | Number of studies | Number of patients | Pooled HR(95%CI) | P value | I2(%) | P value | Model used |
| Study location | |||||||
| Asia | 8 | 789 | 2.3(1.74–3.01) | 0.000 | 0 | 0.598 | FEM |
| Europe | 2 | 178 | 1.96(0.64–6.03) | 0.239 | 77.7 | 0.034 | REM |
| Nubmer of patients | |||||||
| >100 | 3 | 1166 | 1.67(1.34–2.07) | 0.000 | 23.3 | 0.272 | FEM |
| <100 | 8 | 532 | 2.11(1.54–2.88) | 0.000 | 35.7 | 0.144 | FEM |
| Cut off value | |||||||
| Percentage | 5 | 602 | 2.41(1.72–3.38) | 0.000 | 0 | 0.621 | FEM |
| Staining | 2 | 163 | 3.12(1.69–5.75) | 0.000 | 0 | 0.624 | FEM |
| Percentage+staining | 3 | 881 | 1.43(1.14–1.79) | 0.002 | 0 | 0.727 | FEM |
| Dilution | |||||||
| ≤1∶500 | 6 | 686 | 1.85(1.39–2.46) | 0.000 | 29.1 | 0.217 | REM |
| >1∶500 | 4 | 960 | 1.73(1.37–2.17) | 0.000 | 52.2 | 0.099 | FEM |
REM, random-effectsmodel; FEM, fixed-effectsmodel; HR, hazard ratio; CI, confidenceinterval.
Correlation of HIFs expression with clinicopathological parameters
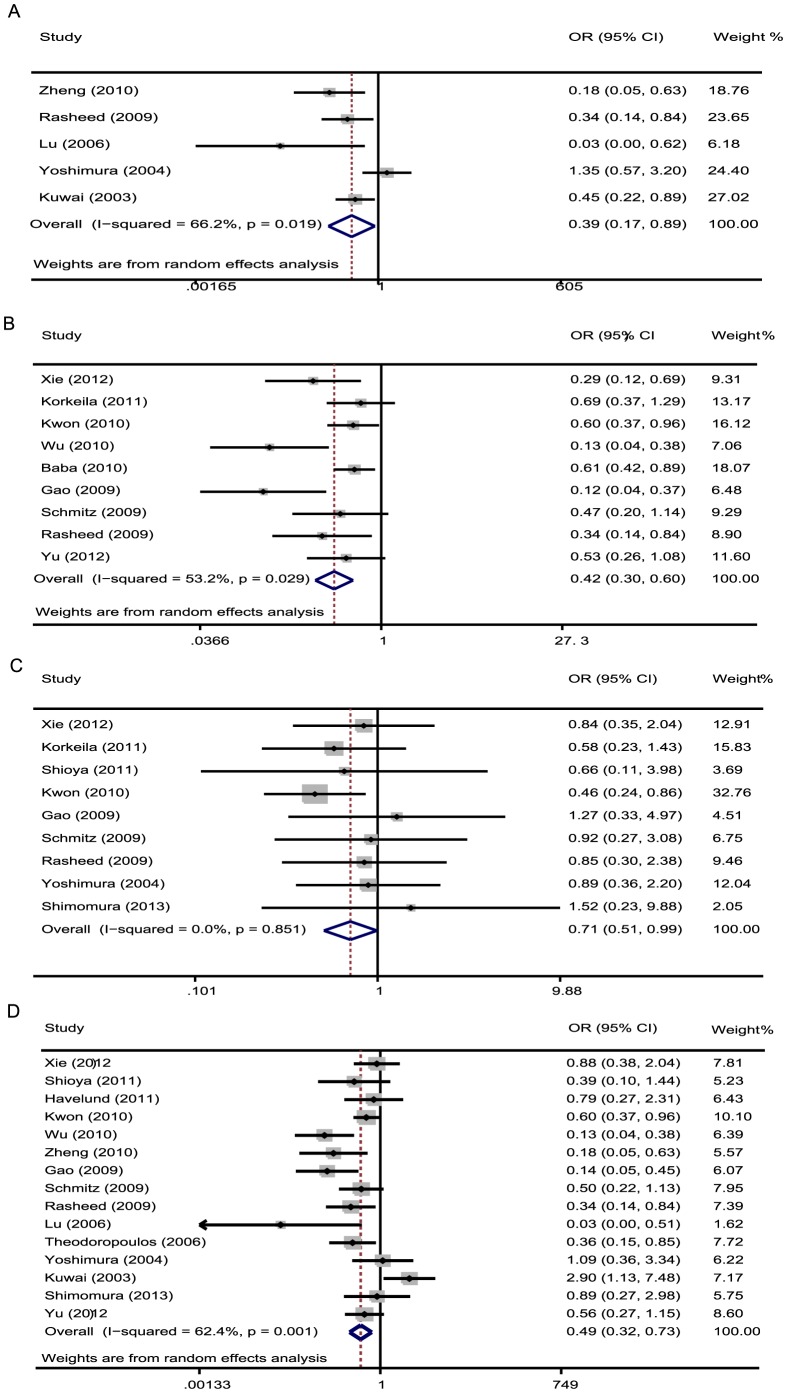
The meta-analysis was also assessed the correlation between HIF-1α expression and clinicopathological characteristics of CRC. As shown in Table 5 , overexpression of HIF-1α was significantly associated with Dukes' stages (OR 0.39, 95% CI: 0.17–0.89), UICC stages (OR 0.42 95% CI: 0.3–0.59), depth of invasion (OR 0.71, 95% CI: 0.51–0.99), lymphnode status (OR 0.49, 95% CI: 0.32–0.73) and metastasis (OR 0.29, 95% CI: 0.11–0.81). Furthermore, there was no significant association between HIF-1α expression with grade of differentiation. The pooled OR was 0.97 (95% CI: 0.67–1.39). ( Figure 4 ).
Table 5. HIF-1α and HIF-2α expression and clinicopathological features for colorectal cancer.
| Heterogeneity | |||||||
| Clinicopathological features | Nuber of studies | Nuber of patients | Pooled OR (95%CI) | P value | I2(%) | P value | Model used |
| HIF-1α | |||||||
| Differentiation grade | 15 | 2226 | 0.97 (0.67–1.39) | 0.864 | 39.2 | 0.06 | REM |
| Dukes' stages | 5 | 408 | 0.39(0.17–0.89) | 0.025 | 66.2 | 0.019 | REM |
| Lymphnode status | 15 | 1490 | 0.49(0.32–0.73) | 0.001 | 62.4 | 0.001 | REM |
| Metastasis | 5 | 480 | 0.29(0.11–0.81) | 0.018 | 52.8 | 0.076 | REM |
| UICC stage | 9 | 1733 | 0.42(0.3–0.59) | 0.000 | 53.2 | 0.029 | REM |
| Depth of invasion | 9 | 1016 | 0.71(0.51–0.99) | 0.045 | 0.00 | 0.851 | FEM |
| HIF-2α | |||||||
| Differentiation grade | 2 | 782 | 0.484(0.289–0.812) | 0.006 | 58 | 0.123 | FEM |
| Dukes' stages | 2 | 177 | 0.9(0.197–4.168) | 0.9 | 82 | 0.019 | REM |
| Lymphnode status | 3 | 329 | 0.95(0.418–2.16) | 0.904 | 63.7 | 0.064 | REM |
| Depth of invasion | 2 | 177 | 0.379(0.038–3.798) | 0.409 | 83.4 | 0.014 | REM |
REM, random-effects model; FEM, fixed-effects model; OR, odds ratio; CI, confidence interval.
Figure 4. Forrest plot of odds ratios (ORs) for the association of HIF-1α expression with clinicopathological features.
A. ORs with corresponding 95% CIs of the HIF-1α expression with Dukes' stages. OR<1 suggested that unfavorable parameters for the group with increased HIF-1α expression/negative. Overexpressed HIF-1α was associated with advanced Dukes' stage of CRC. B. ORs with corresponding 95% CIs of the HIF-1α overexpression with UICC stage. OR<1 suggested that unfavorable parameters for the group with increased HIF-1α expression/negative and HIF-1α overexpression was associated with advanced stage of CRC. C. ORs with corresponding 95% CIs of the HIF-1α overexpression with depth of invasion. D. ORs with corresponding 95% CIs of the HIF-1α overexpression with lymphnode metastasis.
In addition, we evaluated the correlation between HIF-2α overexpression with clinicopathological characteristics of CRC. The result showed that overexpression of HIF-2α was significantly associated with grade of differentiation (OR 0.48, 95% CI: 0.29–0.81). There was no significant association between HIF-2α expression with Dukes' stages, depth of invasion and lymphnode status. The pooled OR was 0.91(95% CI: 0.20–4.17), 0.38 (95% CI: 0.04–3.80), and 0.95 (95% CI: 0.428–2.16), respectively ( Table 5 ).
Publication bias
Egger's test indicated that there was no evidence of significant publication bias after assessing the funnel plot (Figure S1–S3) for the studies included in our meta-analysis.
Discussion
Hypoxia has been recognized as a common feature of solid tumors and a negative prognostic factor for response to treatment and survival of cancer patients. In 1993, Höckel reported that cervix cancer patients with hypoxic tumors (median pO2<10 mmHg) had a significantly lower overall and recurrence-free survival [33]. Since then, hypoxia has been found to indicate a highly aggressive disease phenotype associated with poor prognosis in many cancers, including brain, breast, prostate, pancreas, cervix, bladder and ovary [34]–[37]. HIFs are the best characterized markers mediating transcriptional responses to hypoxic stress and expected to be unfavorable prognostic indicators. Hypoxia and consequently HIF activation is regarded as an important stimulus of CRC angiogenesis. HIF binds to the HRE in the VEGF promoter region, leading to up- regulation of VEGF transcription and the formation of new blood vessels [38]. Surprisingly, both HIF-1 and HIF-2 can function as tumor suppressors in certain cancers [39]. Many studies were also performed to assess the prognostic value of HIF for CRC patients, but the conclusions were also inconclusive. On the other hand, HIF-1 and HIF-2 have distinct target genes, but few studies compared the clinicopathologic and prognostic differences between HIF-1 and HIF-2.
This meta-analysis aimed to examine the association between HIFs expression and the prognosis of CRC patients, and assess the roles of HIF-1α and HIF-2α in clinicopathologic features. Our analysis combined the outcomes of 23 studies comprising 2984 CRC patients, indicating that overexpressed HIF was significantly associated with increase of mortality risk, including OS (2.06 95%CI 1.55–2.74; Z = 4.95; P = 0.000) and DFS (2.84,, 95%CI 1.87–4.31; Z = 4.92; P = 0.000). Additionally, the results of sensitivity analysis showed that the association was not changed after removing any study. Subgroup analysis revealed that both overexpressed HIF-1α and HIF-2α were associated with worse prognosis in CRC. On the basis of different HIF isoforms, further subgroup analysis was performed by study location, number of patients, antibody dilution, cut-off value. HIF-1α overexpression was significantly associated with poor OS in Asian countries (HR 2.3, 95% CI: 1.74–3.01, Z = 5.76, P = 0.000), while not in European or other countries. It indicated that HIF-1α overexpression seemed to be associated with disease progress and unfavorable prognosis in Asian CRC patients. Other factors did not alter the significant OS of overexpressed HIF-1α. In addition, significant correlations were observed between HIF-1α overexpression with clinicopathological features including Dukes' stages, UICC stages, depth of invasion, lymphnode status and metastasis. Our results concurred with previous study that HIF-1α expression had a significant inverse correlation in T1 and T2 CRC. On the other hand, overexpression of HIF-2α was significantly associated with grade of differentiation. Thus, HIF-1 and HIF-2 indicate distinct clinicopathologic features.
In this meta-analysis, we had dealt with highly significant heterogeneity among the 23 studies. Although we used random effects models to analyze the data, it did not identify the source of heterogeneity. Thus, we performed stratified analysis according to study location, number of patients, cut-off value. When the analysis on OS was performed without consideration of other factors, heterogeneity was detected (I2 69.1% P = 0.000). While when the studies included were classified into three groups according to evaluation criterion (percentage, staining and percentage plus staining), the heterogeneity was not detected (I2 0% P = 0.621, I2 0% P = 0.624, I2 0% P = 0.727). Therefore, the heterogeneity in this study could be explained by the evaluation standards. Meanwhile, there were some limitations in this meta-analysis. First, the study included in our meta-analysis was restricted only to articles published in English or Chinese, which probably provided additional bias. Second, HRs calculated from data or extracted from survival curves might be less reliable than direct analysis of variance. Third, the sample size in European studies was not big enough so that the difference of HIF-1expression on survival was not significant.
In summary, we showed that both overexpressed HIF-1α and HIF-2α were significantly associated with worse prognosis in CRC. Subgroup analysis indicated that HIF-1α overexpression was associated with progress disease and unfavorable prognosis in Asian CRC patients. Significant correlations were observed between HIF-1α overexpression with Dukes' stages, UICC stages, depth of invasion, lymphnode status and metastasis, but there was no significant association between overexpressed HIF-1α with grade of differentiation. While overexpressed HIF-2α was only associated with differentiation. Large, well-designed prospective studies are required to investigate the precise prognostic significance and clinicopathologic differences of HIFs expression.
Supporting Information
Egger's publication bias plot showed no publication bias for studies regarding overexpressed HIF-1α and overall survival (OS) in the meta-analysis: the relationship between the effect size of individual studies (HR, vertical axis) and the precision of the study estimate (standard error, horizontal axis).
(TIF)
Egger's publication bias plot showed no publication bias for studies regarding overexpressed HIF-1α and disease free survival (DFS) in the meta-analysis.
(TIF)
Egger's publication bias plot showed no publication bias for studies regarding overexpressed HIF-2α and overall survival (OS) in the meta-analysis.
(TIF)
PRISMA Checklist.
(DOC)
Funding Statement
This work was supported by the National Natural Scientific Foundation of China (NO. 91019005 and 81272672), the Zhejiang Provincial Program for the Cultivation of High-level Innovative Health Talents (HJ), the Key Discipline of Zhejiang Province Health Bureau and Traditional Chinese Medicine Administration (HJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA: A Cancer Journal for Clinicians 63: 11–30. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics, 2010. CA: A Cancer Journal for Clinicians 60: 277–300. [DOI] [PubMed] [Google Scholar]
- 3. Griffiths EA, Pritchard SA, Valentine HR, Whitchelo N, Bishop PW, et al. (2007) Hypoxia-inducible factor-1alpha expression in the gastric carcinogenesis sequence and its prognostic role in gastric and gastro-oesophageal adenocarcinomas. British Journal of Cancer 96: 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba Y, Nosho K, Shima K, Irahara N, Chan AT, et al. (2010) HIF1A overexpression is associated with poor prognosis in a cohort of 731 colorectal cancers. American Journal of Pathology 176: 2292–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshimura H, Dhar DK, Kohno H, Kubota H, Fujii T, et al. (2004) Prognostic impact of hypoxia-inducible factors 1alpha and 2alpha in colorectal cancer patients: correlation with tumor angiogenesis and cyclooxygenase-2 expression. Clinical Cancer Research 10: 8554–8560. [DOI] [PubMed] [Google Scholar]
- 6. Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC (2003) Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Molecular and Cellular Biology 23: 9361–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mazumdar J, Hickey MM, Pant DK, Durham AC, Sweet-Cordero A, et al. (2010) HIF-2alpha deletion promotes Kras-driven lung tumor development. Proceedings of the National Academy of Sciences of the United States of America 107: 14182–14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology 25: 603–605. [DOI] [PubMed] [Google Scholar]
- 9. Parmar MK, Torri V, Stewart L (1998) Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in Medicine 17: 2815–2834. [DOI] [PubMed] [Google Scholar]
- 10. Handoll HH (2006) Systematic reviews on rehabilitation interventions. Archives of Physical Medicine and Rehabilitation 87: 875. [DOI] [PubMed] [Google Scholar]
- 11. Ioannidis JP, Patsopoulos NA, Evangelou E (2007) Uncertainty in heterogeneity estimates in meta-analyses. BMJ 335: 914–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute 22: 719–748. [PubMed] [Google Scholar]
- 13. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie G, Zheng L, Ou J, Huang H, He J, et al. (2012) Low expression of prolyl hydroxylase 2 is associated with tumor grade and poor prognosis in patients with colorectal cancer. Exp Biol Med (Maywood) 237: 860–866. [DOI] [PubMed] [Google Scholar]
- 15. Korkeila EA, Sundstrom J, Pyrhonen S, Syrjanen K (2011) Carbonic anhydrase IX, hypoxia-inducible factor-1alpha, ezrin and glucose transporter-1 as predictors of disease outcome in rectal cancer: multivariate Cox survival models following data reduction by principal component analysis of the clinicopathological predictors. Anticancer Research 31: 4529–4535. [PubMed] [Google Scholar]
- 16. Shioya M, Takahashi T, Ishikawa H, Sakurai H, Ebara T, et al. (2011) Expression of hypoxia-inducible factor 1alpha predicts clinical outcome after preoperative hyperthermo-chemoradiotherapy for locally advanced rectal cancer. J Radiat Res 52: 821–827. [DOI] [PubMed] [Google Scholar]
- 17. Havelund BM, Sorensen FB, Lindebjerg J, Spindler KL, Jakobsen A (2011) Pretreatment HIF-1alpha and GLUT-1 expressions do not correlate with outcome after preoperative chemoradiotherapy in rectal cancer. Anticancer Research 31: 1559–1565. [PubMed] [Google Scholar]
- 18. Kwon HC, Kim SH, Oh SY, Lee S, Kwon KA, et al. (2010) Clinicopathological significance of p53, hypoxia-inducible factor 1alpha, and vascular endothelial growth factor expression in colorectal cancer. Anticancer Research 30: 4163–4168. [PubMed] [Google Scholar]
- 19. Saigusa S, Tanaka K, Toiyama Y, Yokoe T, Okugawa Y, et al. (2011) Clinical significance of CD133 and hypoxia inducible factor-1alpha gene expression in rectal cancer after preoperative chemoradiotherapy. Clinical Oncology (Royal College of Radiologists) 23: 323–332. [DOI] [PubMed] [Google Scholar]
- 20. Wu Y, Jin M, Xu H, Shimin Z, He S, et al. (2010) Clinicopathologic significance of HIF-1alpha, CXCR4, and VEGF expression in colon cancer. Clin Dev Immunol 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng J, Sun X, Wang W, Lu S (2010) Hypoxia-inducible factor-1alpha modulates the down-regulation of the homeodomain protein CDX2 in colorectal cancer. Oncology Reports 24: 97–104. [PubMed] [Google Scholar]
- 22. Toiyama Y, Inoue Y, Saigusa S, Okugawa Y, Yokoe T, et al. (2010) Gene expression profiles of epidermal growth factor receptor, vascular endothelial growth factor and hypoxia-inducible factor-1 with special reference to local responsiveness to neoadjuvant chemoradiotherapy and disease recurrence after rectal cancer surgery. Clinical Oncology (Royal College of Radiologists) 22: 272–280. [DOI] [PubMed] [Google Scholar]
- 23. Cao D, Hou M, Guan YS, Jiang M, Yang Y, et al. (2009) Expression of HIF-1alpha and VEGF in colorectal cancer: association with clinical outcomes and prognostic implications. BMC Cancer 9: 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jubb AM, Turley H, Moeller HC, Steers G, Han C, et al. (2009) Expression of delta-like ligand 4 (Dll4) and markers of hypoxia in colon cancer. British Journal of Cancer 101: 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rajaganeshan R, Prasad R, Guillou PJ, Scott N, Poston G, et al. (2009) Expression patterns of hypoxic markers at the invasive margin of colorectal cancers and liver metastases. European Journal of Surgical Oncology 35: 1286–1294. [DOI] [PubMed] [Google Scholar]
- 26. Schmitz KJ, Muller CI, Reis H, Alakus H, Winde G, et al. (2009) Combined analysis of hypoxia-inducible factor 1 alpha and metallothionein indicates an aggressive subtype of colorectal carcinoma. International Journal of Colorectal Disease 24: 1287–1296. [DOI] [PubMed] [Google Scholar]
- 27. Rasheed S, Harris AL, Tekkis PP, Turley H, Silver A, et al. (2009) Hypoxia-inducible factor-1alpha and -2alpha are expressed in most rectal cancers but only hypoxia-inducible factor-1alpha is associated with prognosis. British Journal of Cancer 100: 1666–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cleven AH, van Engeland M, Wouters BG, de Bruine AP (2007) Stromal expression of hypoxia regulated proteins is an adverse prognostic factor in colorectal carcinomas. Cell Oncol 29: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu XG, Xing CG, Feng YZ, Chen J, Deng C (2006) Clinical significance of immunohistochemical expression of hypoxia-inducible factor-1alpha as a prognostic marker in rectal adenocarcinoma. Clin Colorectal Cancer 5: 350–353. [DOI] [PubMed] [Google Scholar]
- 30. Theodoropoulos GE, Lazaris AC, Theodoropoulos VE, Papatheodosiou K, Gazouli M, et al. (2006) Hypoxia, angiogenesis and apoptosis markers in locally advanced rectal cancer. International Journal of Colorectal Disease 21: 248–257. [DOI] [PubMed] [Google Scholar]
- 31. Kuwai T, Kitadai Y, Tanaka S, Onogawa S, Matsutani N, et al. (2003) Expression of hypoxia-inducible factor-1alpha is associated with tumor vascularization in human colorectal carcinoma. International Journal of Cancer 105: 176–181. [DOI] [PubMed] [Google Scholar]
- 32. Mohammed N, Rodriguez M, Garcia V, Garcia JM, Dominguez G, et al. (2011) EPAS1 mRNA in plasma from colorectal cancer patients is associated with poor outcome in advanced stages. Oncol Lett 2: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hockel M, Knoop C, Schlenger K, Vorndran B, Baussmann E, et al. (1993) Intratumoral pO2 predicts survival in advanced cancer of the uterine cervix. Radiotherapy and Oncology 26: 45–50. [DOI] [PubMed] [Google Scholar]
- 34. Birner P, Gatterbauer B, Oberhuber G, Schindl M, Rossler K, et al. (2001) Expression of hypoxia-inducible factor-1 alpha in oligodendrogliomas: its impact on prognosis and on neoangiogenesis. Cancer 92: 165–171. [DOI] [PubMed] [Google Scholar]
- 35. Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, et al. (2003) Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer 97: 1573–1581. [DOI] [PubMed] [Google Scholar]
- 36. Shibaji T, Nagao M, Ikeda N, Kanehiro H, Hisanaga M, et al. (2003) Prognostic significance of HIF-1 alpha overexpression in human pancreatic cancer. Anticancer Research 23: 4721–4727. [PubMed] [Google Scholar]
- 37. Theodoropoulos VE, Lazaris A, Sofras F, Gerzelis I, Tsoukala V, et al. (2004) Hypoxia-inducible factor 1 alpha expression correlates with angiogenesis and unfavorable prognosis in bladder cancer. European Urology 46: 200–208. [DOI] [PubMed] [Google Scholar]
- 38. Sutter CH, Laughner E, Semenza GL (2000) Hypoxia-inducible factor 1alpha protein expression is controlled by oxygen-regulated ubiquitination that is disrupted by deletions and missense mutations. Proc Natl Acad Sci U S A 97: 4748–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lidgren A, Hedberg Y, Grankvist K, Rasmuson T, Vasko J, et al. (2005) The expression of hypoxia-inducible factor 1alpha is a favorable independent prognostic factor in renal cell carcinoma. Clinical Cancer Research 11: 1129–1135. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Egger's publication bias plot showed no publication bias for studies regarding overexpressed HIF-1α and overall survival (OS) in the meta-analysis: the relationship between the effect size of individual studies (HR, vertical axis) and the precision of the study estimate (standard error, horizontal axis).
(TIF)
Egger's publication bias plot showed no publication bias for studies regarding overexpressed HIF-1α and disease free survival (DFS) in the meta-analysis.
(TIF)
Egger's publication bias plot showed no publication bias for studies regarding overexpressed HIF-2α and overall survival (OS) in the meta-analysis.
(TIF)
PRISMA Checklist.
(DOC)