Abstract
We investigated how asparagine mutagenesis of conserved aspartic acids in helix two (D2.50) and three (D3.32) of M1 – M4 muscarinic receptors alters the irreversible binding of acetylcholine mustard and BR384 (4-[(2-bromoethyl)methyl-amino]-2-butynyl N-(3-chlorophenyl)carbamate), a nitrogen mustard derivative of McN-A-343 ([4-[[N-(3-chlorophenyl)carbamoyl]oxy]-2-butynyl] trimethylammonium chloride). The D2.50N mutation moderately increased the affinity of the aziridinium ions of acetylcholine mustard and BR384 for M2 – M4 receptors and had little effect on the rate constant for receptor alkylation. The D3.32N mutation greatly reduced the rate constant for receptor alkylation by acetylcholine mustard, but not by BR384, although the affinity of BR384 was reduced. The combination of both mutations (D2.50N/D3.32N) substantially reduced the rate constant for receptor alkylation by BR384 relative to wild type and mutant D2.50N and D3.32N receptors. The change in binding affinity caused by the mutations suggests that the D2.50N mutation alters the interaction of acetylcholine mustard with D3.32 of M1 and M3 receptors, but not that of the M4 receptor. BR384 exhibited the converse relationship. The simplest explanation is that acetylcholine mustard and BR384 alkylate at least two residues on M1 – M4 receptors and that the D2.50N mutation alters the rate of alkylation of D3.32 relative to another residue, perhaps D2.50 itself.
Irreversible ligands are often useful in studies on drug-receptor interactions. One approach for synthesizing an irreversible ligand is to incorporate a nitrogen mustard group into its structure (1). This strategy has yielded several anticancer agents that react covalently with DNA to form guanine adducts (2, 3). Muscarinic receptor ligands seem ideally suited for this modification because the reactive aziridinium ion derived from a nitrogen mustard group resembles the quaternary ammonium group of acetylcholine. Indeed, the nitrogen mustard analog of acetylcholine, acetylcholine mustard (AChM), has been shown to bind irreversibly with a highly conserved aspartic acid residue (D3.32, nomenclature scheme of Ballesteros and Weinstein (4)) in helix three of muscarinic receptors (5). Thus, 2-haloethylamine muscarinic ligands may have the capacity to react covalently with the receptor at D3.32.
We have used acetylcholine mustard (AChM) (Figure 1) as a probe to investigate whether a test ligand interacts competitively or allosterically with the orthosteric site alkylated by AChM (6, 7). The method involves first incubating the muscarinic receptor with AChM and different concentrations of the test ligand. After stopping the reaction and washing the receptor preparation, residual unalkylated receptors are estimated using a suitable radioligand, like [3H]N-methylscopolamine ([3H]NMS). Competitive and allosteric modulators exhibit a difference in how they affect receptor alkylation. This approach has advantages over equilibrium and kinetic binding methods for analyzing allosteric interactions (6, 7).
Figure 1.
Structures of acetylcholine, McN-A-343 and their nitrogen mustard derivatives and transformation products in aqueous solution at neutral pH.
We have shown that mutation of aspartic acid 3.32 to asparagine in M1 and M2 muscarinic receptors causes a large decrease in the rate constant for receptor alkylation by AChM, which is consistent with the postulate that AChM alkylates D3.32 (8). This mutation does not completely prevent receptor alkylation, however, particularly at high concentrations of AChM. We also found that the D3.32N greatly inhibited the alkylation of M1 and M2 receptors by a nitrogen mustard analog (BR384; 4-[(2-bromoethyl) methyl-amino]-2-butynyl N-(3-chlorophenyl)carbamate) of the functionally selective muscarinic agonist, McN-A-343 ([4-[[N-(3-chlorophenyl)carbamoyl]oxy]-2-butynyl]tri-methylammonium chloride) (Figure 1). The inhibition was due primarily to a reduction in affinity, however, and not to a decreased rate constant for alkylation, suggesting perhaps that BR384 does not alkylate the D3.32 residue of M1 and M2 receptors. Nonetheless, the orthosteric muscarinic antagonist, NMS, competitively inhibited alkylation of wild type M1, wild type M2 and the D103N mutant of the M2 receptor by BR384, whereas the known allosteric modulator, gallamine, allosterically prevented alkylation (6, 8, 9). Thus, BR384 probably alkylates another residue within the orthosteric-binding pocket of the M2 receptor.
McN-A-343 is an important muscarinic agonist because it exhibits a preference for activating M1 and M4 muscarinic receptors relative to the other subtypes (10, 11). Intravenous administration of McN-A-343 elicits a pressor response due to activation of M1 muscarinic receptors in sympathetic ganglia (12, 13). In contrast, nonselective muscarinic agonists activate the M3 muscarinic receptor on peripheral blood vessels causing vasodilatation and a reduction in blood pressure (14, 15). The aziridinium ion of BR384 behaves like McN-A-343 with regard to its pressor response in vivo and to its lack of contractile action on the guinea pig ileum in vitro (16). The latter response is mediated through the M3 muscarinic receptor (17). Understanding the mode interaction of BR384 with muscarinic receptors may shed light on the development of novel selective orthosteric ligands.
In this study, we have investigated how the D3.32N mutation (Figure 2) affects the alkylation of M3 and M4 receptors by AChM and BR384. We have also investigated how mutation of a highly conserved residue in helix 2 (D2.50, Figure 2) affects M1 – M4 receptor alkylation, by itself and in combination with D3.32N. Our results are consistent with the postulate that AChM and BR384 alkylate D3.32 in addition to another residue within the binding pockets of M1 – M4 muscarinic receptors.
Figure 2.
Crystal structures of the helices and extracellular loops of human M2 and rat M3 muscarinic receptors bound with 3-quinuclidinyl-benzilate and tiotropium, respectively (26, 27). The secondary structure (helices 1 – 6) and backbone (helix 7) are shown. Aspartic acids 2.50 (M2 D69 and M3 D113 (corresponds to human M3 D114)) and 3.32 (M2 D103 and M3 D147 (corresponds to human M3 D148)) are indicated on helices two and three (PDB ID: 3uon and 4daj for M2 and M3 receptors, respectively).
Experimental Procedures
Materials
Reagents were obtained from the following sources: Dulbecco’s Modified Eagle Medium with high glucose plus L-glutamine, Luria-Bertani broth, trypsin-EDTA, and penicillin-streptomycin (Invitrogen, Carlsbad, CA); Fetal calf serum (HyClone Laboratories Inc., South Logan, UT); G418 (InvivoGen, San Diego, CA); NMS, atropine, acetylcholine perchlorate, HEPES, EDTA, scopolamine and Na2S2O3 (Sigma-Aldrich, Inc., St. Louis, MO); salts for phosphate buffer and binding buffer, HCl and NaOH (Thermo Fisher Scientific, Waltham, MA); Zyppy Plasmid Miniprep Kit (Zymo Research, Irvine, CA); NucleoBond Xtra Midi Plus (Clontech Laboratories Inc., Mountain View, CA); GeneJammer (Agilent Technologies, Cedar Creek, TX) and Oligonucleotide primers (Integrated DNA Technologies, Inc., San Diego, CA). AChM and McN-A-343 were synthesized as described previously (7). BR384 was synthesized by the method of Ringdahl et al. (16).
Both AChM and BR384 were first cyclized to their reactive aziridinium ions as described previously (7, 9) before being used in the assays described below.
Site-directed mutagenesis
The human M1, M2, M3 and M4 muscarinic receptor cDNAs, cloned into a modified expression vector (pCD-hM1, pCD-hM2, pCD-hM3, pCD-hM4), were obtained from Dr. Tom Bonner at the National Institute of Mental Health (Bethesda, MD). Mutations were introduced into pCD-hM1, pCD-hM2, pCD-hM3, and pCD-hM4 using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies) and mutagenesis primers. Sequences of mutant receptors were verified by Laragen, Inc. (Culver City, CA). The mutant plasmids were purified using Zyppy Plasmid Miniprep Kit or NucleoBond Xtra Midi Plus kit following the manufacturers’ protocols.
Cell culture and transfection
Chinese hamster ovary (CHO) cells stably expressing the human M1, M2, M3 and M4 muscarinic receptors were obtained from Acadia Pharmaceuticals (San Diego, CA) and cultured as described previously (7). Human embryonic kidney (HEK) 293 cells were cultured as described previously (8) and transfected with plasmids encoding mutated muscarinic receptors using GeneJammer following the manufacturer's protocols. After transfection, the cells were incubated for 48 h and harvested for assays.
Preparation of cellular homogenates
CHO or HEK 293 cells expressing muscarinic receptors were grown to confluence in 100-mm dishes (Corning Life Sciences, Acton, MA) and scraped into binding buffer (20 mM sodium-HEPES, pH 7.4, 100 mM NaCl, and 10 mM EDTA) using a Teflon spatula. The mixture was centrifuged at low speed (1247g, 10 min) and the supernatant discarded. The pellet was suspended in binding buffer using a Polytron homogenizer (Kinematica, Littau-Lucerne, Switzerland; setting #4, 10 sec). Homogenates of cells expressing M1 D71N/D105N, M2 D69N/D103N, M3 D114N/D148N, or M4 D78N/D112N receptors were centrifuged once more at high speed (39,400g, 10 min, 4°C) and suspended in fresh binding buffer.
Homogenate was prepared at varying concentrations depending on the assay and receptor construct so that the receptor concentration in the final binding assay would only result in minimal depletion of the free concentration of [3H]NMS (see below).
Treatment of cellular homogenate with cyclized AChM and BR384
The covalent interactions of AChM and BR384 with muscarinic receptors were investigated by first incubating homogenates of cells expressing muscarinic receptors with the aziridinium ions of AChM and BR384, and then measuring residual muscarinic receptors using the radioligand [3H]NMS as described previously (7).
Cellular homogenate (200 µl) was incubated at 37°C in a shaking water bath, and an aliquot (50 µl) of cyclized AChM, BR384 or binding buffer (control) was added to yield a final volume of 0.25 ml. The reaction was allowed to proceed for specific times as described under “Results.” An aliquot (0.75 ml) of stopping solution (see below) was added at the end of the incubation, and the mixture was incubated another 20 min to allow inactivation of the aziridinium ions of AChM or BR384.
The reaction tubes were centrifuged (25,000g, 15 min, 4°C), and the pellets were suspended in fresh buffer to remove the transformation products of AChM and BR384. If the stopping solution contained scopolamine (see below), the centrifugation step was repeated two more times. Ultimately, the final pellets were suspended in a volume of 1 ml of binding buffer. Triplicate measurements of [3H]NMS binding were made on each homogenate as described below.
The reactions were stopped in two ways depending on the goal of the experiment. In the first method, the stopping solution contained sodium thiosulfate (1.33 mM) in binding buffer. Thiosulfate forms a covalent adduct with the remaining aziridinium ion derived from AChM and BR384, and this first order process is complete in about 15 min. During this time, additional alkylation of the receptor can occur. In the second method, the stopping solution contained both scopolamine (10 µM) and sodium thiosulfate (1.33 mM). The scopolamine immediately stops the reaction and the thiosulfate slowly inactivates the aziridinium ion. The kinetic constants of AChM and BR384 (see equations 4 and 6) were estimated only from reactions that were stopped using the latter method.
We previously showed that our stopping procedure with scopolamine immediately prevents receptor alkylation by BR384 (100 µM) and that the associated washing step is adequate to remove residual scopolamine (8). When the concentration of BR384 was increased to 300 µM, however, 28% of wild type M1 and M2 muscarinic receptors were alkylated after adding the stopping solution. In contrast, 3 mM AChM only caused 1% inhibition of [3H]NMS binding in the presence of the stopping solution. Thus, measurements of [3H]NMS binding after treatment with 300 µM BR384 were multiplied by a factor (1.39; i.e., 1/(1 – 0.28)) to correct for this continued alkylation after addition of scopolamine and thiosulfate.
[3H]NMS binding assays
The residual amount of free muscarinic receptors in cellular homogenates treated with AChM and BR384 was estimated using a binding assay with the muscarinic antagonist radioligand, [3H]NMS (specific activity, 82 Ci/mmol; PerkinElmer Life and Analytical Sciences, Waltham, MA).
For those experiments involving M1 D71N/D105N, M2 D69N/D103N, M3 D114N/D148N, or M4 D78N/D112N receptors, a centrifugation assay was used to measure [3H]NMS binding (8) because these mutants exhibited low affinity for [3H]NMS (pKD, 7.7 – 8.0). We were concerned that [3H]NMS-receptor complexes might dissociate during the washing phase of the filtration assay that was used for the other receptors (see below). An aliquot (0.3 ml) of cellular homogenate was incubated in a microcentrifuge tube (G-tube; Thermo Fisher Scientific) for 30 min at 37°C in a final volume of 0.5 ml containing binding buffer and [3H]NMS (3.0 nM). The equilibration period was stopped by centrifugation (30,000g, 20 min, 4°C). The supernatant was aspirated and the residual pellet washed twice with 0.6 ml of ice-cold binding buffer. An aliquot (0.2 ml) of 1 M NaOH was added to dissolve the pellet. Following an overnight incubation, the solubilized material was acidified with 1 M HCl (0.25 ml) and transferred to a scintillation vial (Research Products International Corp., Mount Prospect, IL). Following addition of scintillation cocktail (Budget-Solve; Research Products International Corp), radioactivity was measured using a liquid scintillation counter (LS 6500; Beckman Coulter, Fullerton, CA). Nonspecific binding was defined as the residual binding in the presence of 10 µM atropine. All measurements were done in triplicate.
For the experiments involving the other receptor constructs, a filtration assay was used to measure [3H]NMS binding (7) in homogenates previously treated with AChM or BR384. An aliquot (0.3 ml) of cellular homogenate was incubated for 30 min at 37°C in a final volume of 1 ml containing binding buffer and 1.0 nM [3H]NMS. The equilibration was stopped by rapid filtration over glass fiber filters (Whatman GFB) using a cell harvester (Brandel Inc., Gaithersburg, MD). The filters were washed three times with ice-cold 0.9% saline (approximately three ml per wash). The filters were placed in scintillation vials, and radioactivity was measured using liquid scintillation spectroscopy as described above.
Ligand/[3H]NMS competition experiments
The competitive inhibition of [3H]NMS binding to muscarinic receptors by acetylcholine, McN-A-343 and NMS was measured in cellular homogenates using the centrifugation or filtration assay, depending on the receptor construct (double mutant or all others, respectively). The assay was done as described above except that fresh cellular homogenate was used, and the assay included various concentrations of the nonlabeled competitors.
Analysis of data
The IC50 values were estimated by nonlinear regression analysis of the ligand/[3H]NMS competition curves using Prism 6.0 (GraphPad Software Inc., San Diego, CA) and the following equation:
| (1) |
In this equation, P and B denote the specific binding of [3H]NMS in the absence and presence of nonlabeled inhibitor, respectively, IC50, the concentration of inhibitor causing half-maximal inhibition of specific binding, and n, the Hill coefficient.
The concentration of homogenate was such that less than 2% of the total concentration of [3H]NMS was depleted at the IC50 point of the competition curve. Equilibrium dissociation constants (Ki, units of molarity) were estimated from the IC50 values (concentration of competitor causing half-maximal displacement of specific binding) using the standard competitive inhibition relationship (18).
| (2) |
in which, [[3H]NMS] denotes the free concentration of [3H]NMS at the IC50 point of the competition curve, and KNMS, the dissociation constant of [3H]NMS (units of molarity, M). The latter was estimated by rearrangement of equation 2 for the case where the competitor and radioligand are the same:
| (3) |
In the analysis of the NMS/[3H]NMS competition curves, the Hill coefficient in equation 1 was constrained to one. The concentration of [3H]NMS was established with accuracy by first making a concentrated stock solution and calculating its concentration from the amount of radioactivity associated with a small aliquot of it. This solution was diluted to a working stock solution that was used in the competition experiment, and its concentration was subsequently determined for each experiment as just described.
For a given experiment, the competitive inhibition of [3H]NMS binding by acetylcholine, McN-A-343 and nonlabeled NMS was measured simultaneously so that individual values of log Ki or log KNMS could be estimated for each nonlabeled ligand. The text, figures and tables report the mean and SEM of these estimates.
The basis for the estimation of the dissociation constant (K1) and rate constant for alkylation (k1) of AChM and BR384 is described by Suga et al. (7). Our analysis rests on the assumption that the rate constants describing the reversible interaction of the aziridinium ion with the receptor are much faster than that of the alkylation step (k1) (see scheme 1).
Scheme 1.

Quasi-equilibrium model for the interaction of the aziridinium ions of AChM and BR384 with the muscarinic receptor. The dissociation constant, K1 (units of M), describes the reversible interaction of the aziridinium ion (A) with the receptor (R) to yield the reversible receptor complex (AR). The rate constant, k1 (units of inverse time, min−1), describes the instantaneous rate of receptor alkylation (k1AR) to yield the irreversible receptor complex (A-R).
The data needed for this kinetic analysis comes from an experiment in which aliquots of a given receptor preparation are incubated with different concentrations of the nitrogen mustard (AChM or BR384) for single or multiple incubation times. The amount of unalkylated receptors is estimated subsequently by measuring the binding of [3H]NMS at a single concentration (1 or 3 nM).
The kinetic analysis for AChM involves fitting the following regression equation to the measurements of [3H]NMS binding:
| (4) |
In this equation, Yt denotes [3H]NMS binding after incubation with the irreversible ligand, Y0, the estimate of [3H]NMS binding in the absence of irreversible ligand, k1, the rate constant for alkylation, t, the time of incubation, O, receptor occupancy by the aziridinium ion and b, the fraction of receptors that can bind [3H]NMS but are resistant to receptor alkylation. The variable O is given by:
| (5) |
in which k1, denotes the dissociation constant of the aziridinium ion for the receptor (molar units), and X, the concentration of the aziridinium ion.
A different equation was used to analyze data obtained with BR384 because the concentration of the aziridinium ion decays substantially during the incubation. The basis of this equation is described by Ehlert and Jenden (19):
| (6) |
In this equation, τ denotes the macroscopic time constant for the decay in the concentration of the aziridinium ion from its peak concentration. The parameter, τ, was constrained to a constant (0.07 min−1) based on the values of the microscopic constants for formation and decay of the aziridinium ion estimated by Ringdahl et al. (16).
For all of the kinetic experiments, the homogenate concentration during the subsequent binding assay was adjusted so that the maximal depletion of the free concentration of [3H]NMS was less than 2.5%.
For each kinetic experiment, individual estimates of log k1 and k1 were made, and the text and tables report the mean and SEM of these values. We noted that the error in the estimate of k1 tended to be proportional to the measurement such that a log transformation yielded a more uniform variance. Consequently, all statistical analyses were done using the log k1 values. The tables in the Supporting Information also include the mean ± SEM values for log k1, and Figure 7 illustrates these values as well.
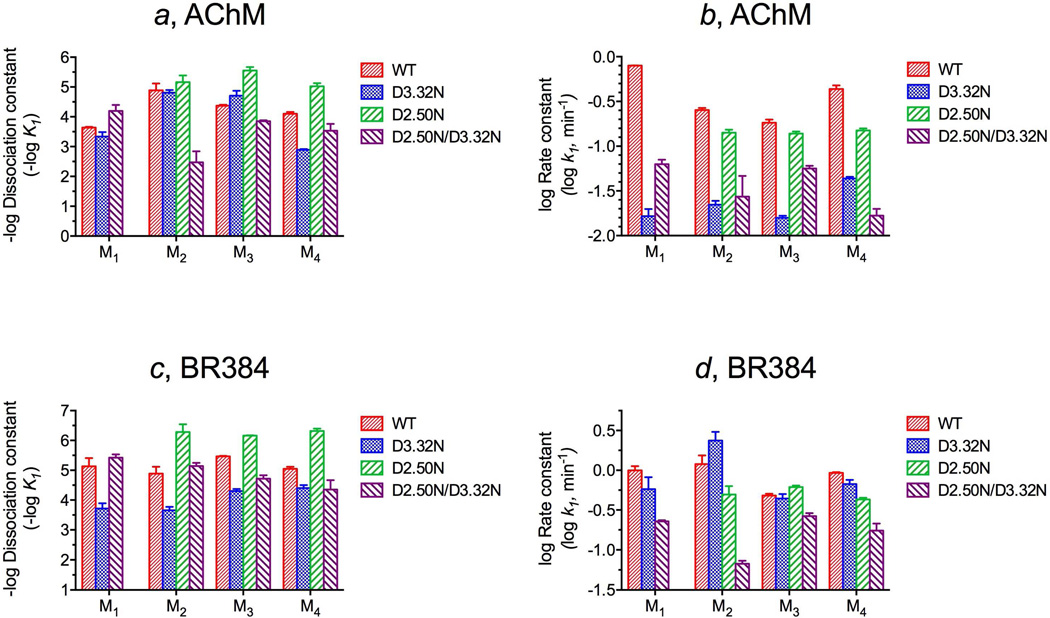
Figure 7.
Estimates of the dissociation constants (a and c) and rate constants for alkylation (b and d) for AChM (a and b) and BR384 (c and d) at wild type and mutant M1 – M4 muscarinic receptors. The parameters were estimated from the data in Figures 3 and 6. The parameter estimates for wild type and D3.32N mutants of M1 and M2 receptors are from our prior studies (6–9). Analysis of variance showed that for both AChM and BR384, there were significant differences among the parameter estimates for the different mutants at each receptor subtype (Supporting Information, Tables S1 and S3). A summary of post hoc comparisons of the parameter estimates is given in Tables S2 and S4 of the Supporting Information, and the numerical values of the parameters are listed in Tables S1 and S3 of the Supporting Information.
The mutation-induced change in the log k1 values of the nitrogen mustards (AChM and BR384) or the log Ki values of the nonlabeled competitors (NMS, acetylcholine and McN-A-343) (Δ log Kmutant) was estimated using the following equation:
| (7) |
in which the subscripts mutant and WT denote whether the dissociation constant (K) is associated with a mutant or wild type receptor, respectively. The variance of Δ log Kmutant is equal to the sum of the variances of the individual log Kmutant and log KWT estimates.
The significance of differences in the estimates of a given parameter among wild type and mutants of a given receptor subtype were determined by one-way analysis of variance with post hoc comparisons (Holm-Sidak’s multiple comparisons test) using Prism 6.0.
Results
Kinetics of alkylation of the M4 muscarinic receptor by AChM and BR384
We measured the kinetics of alkylation of the M4 muscarinic receptor at various concentrations of AChM and BR384 (Figure 3). Homogenates of CHO cells expressing the wild type M4 receptor were incubated with different concentrations of cyclized AChM or BR384 for different times, and the reaction was subsequently stopped with scopolamine and thiosulfate. The homogenates were washed, and the specific binding of [3H]NMS was measured to estimate residual unalkylated receptors. Figure 3a shows that following treatment with AChM, there is a concentration-dependent loss of [3H]NMS binding and that the magnitude of the loss increases with an increase in time. This behavior suggests that the aziridinium ion binds rapidly to the M4 receptor to form a reversible complex that converts to a covalent complex at a slower rate (Scheme 1). To test this model, we fitted equation 4 to the data to obtain an estimate of the affinity constant of the aziridinium ion of AChM for the M4 receptor (log K1 = −4.10 ± .058), its rate constant for alkylation (k1 = 0.44 ± .038 min−1), and the estimate of the proportion of receptors that can bind [3H]NMS but not be alkylated by AChM (b = 17 ± 2.0%). Regression analysis yielded a good fit of equation 4 to the data.
Figure 3.
Alkylation of the wild type M4 muscarinic receptor by AChM and BR384. Homogenates of CHO cells expressing the human M4 muscarinic receptor were incubated with various concentrations of AChM (a) or BR384 (b) for different times, the reactions were stopped with scopolamine and thiosulfate, the homogenates washed, and the binding of [3H]NMS measured at a single concentration of 1.0 nM. The theoretical curves represent the global fit of equations 4 (a) and 6 (b) to the data with the estimates of b, k1 and k1 shared. Mean values ± SEM of three experiments are shown. In a, the incubation times were 2 (○), 4 (●), 8 (△) and 15 (▲) min. In b, the incubation times were 1 (○), 2 (●), 4 (△) and 8 (▲) min.
Similar behavior was observed in experiments with BR384 as shown in Figure 3b. Regression analysis of these data was done using equation 6 to account for the decay in the aziridinium ion of BR384, which is less stable than that of AChM. This analysis yielded estimates of the dissociation constant of aziridinium ion of BR384 for the M4 receptor (log K1 = −5.05 ± 0.072) and its rate constant for alkylation (k1 = 0.93 ± .020 min−1). These estimates of K1 and k1 are approximately ten- and two-fold greater, respectively, than those of AChM. The estimate of the proportion of receptors that can bind [3H]NMS but not be alkylated by BR384 was 7.7 ± 1.8%.
Effects of various concentrations of AChM and BR384 on wild type and D2.50N and D3.32N mutants of M2 and M4 muscarinic receptors
The conclusion that a component of the M4 muscarinic receptor population can bind [3H]NMS but not be alkylated by AChM or BR384 (i.e., b values of 17 and 7.7%, respectively; see equations 4 and 6) seems puzzling because both [3H]NMS and the aziridinium ions of AChM and BR384 should have access to the same pool of receptors in cellular homogenates. For example, the charged aziridinium ions might be unable to penetrate ER vesicles containing a high density of wild type or mutant receptor in cellular homogenates, but so would the quaternary ligand [3H]NMS, particularly at the low concentration used in the binding assay. In addition, when solutions of AChM and BR384 are cyclized to yield their maximal concentrations of aziridinium ion, there is still a modest amount of the parent mustard in solution (2% and 30% of starting concentrations of AChM and BR384, respectively). This species of weak base is expected to penetrate lipid barriers and alkylate any receptors inaccessible to [3H]NMS following cyclization within the putative lipid compartment. Perhaps there is cleavage of the covalent AChM- and BR384-receptor bonds or unalkylated receptors trapped within a membrane compartment may be exposed over time with repetitive washing and trituration of homogenate. The rate of recovery of [3H]NMS binding over four hours is negligible in cerebral cortical homogenates having about 85% of their muscarinic receptors alkylated with BR384 indicating that the binding of BR384 to cerebral muscarinic receptors is nearly irreversible over four hours (Supporting Information, Figure S1).
Because of this complication, we investigated a more rapid experiment employing a single incubation time and washing step. In these experiments, various concentrations of the irreversible agonists were incubated with M2 and M4 muscarinic receptors for 15 (AChM) and 4 min (BR384), and the reaction was stopped with thiosulfate only and washed once as described under “Experimental Procedures” (Figure 4).
Figure 4.
The interaction of AChM and BR384 with wild type and mutant M2 (a and b), and M4 (c and d) muscarinic receptors. Homogenates of cells expressing muscarinic receptors were incubated with different concentrations of AChM (a and c) or BR384 (b and d) for 15 (AChM, a and c) or 4 (BR384, b and d) min. The reactions were stopped with thiosulfate, the homogenates washed, and residual unalkylated muscarinic receptors were estimated by measuring [3H]NMS binding at a concentration of 3 nM (D2.50N/D3.32N) or 1 nM (wild type, D2.50N and D3.32N). The different receptors and their mutants are indicated as: wild type, ○; D2.50N, △; D3.32N, ● and D2.50N/D3.32N, ▲. Mean values ± SEM from three - four experiments are shown.
With regard to wild type and the D2.50N mutants of M2 and M4 receptors, the data suggest that all of the [3H]NMS binding sites are capable of being alkylated by AChM (Figure 4a and c) and BR384 (Figure 4b and d). The same also applies to the effect of BR384 on the D3.32N mutants (Figure 4b and d). Presumably, AChM alkylates the D3.32N and D2.50N/D3.32N mutants too slowly for near complete alkylation to occur during the incubation period (Figure 4a and c). The same is true with regard to the alkylation of the D2.50N/D3.32N mutant by BR384 (Figure 4b and d). The data also show that the D2.50N mutation increases receptor alkylation, whereas the D3.32N mutation has the opposite effect (Figure 4a, b, c and d). In contrast, the D2.50N mutation clearly inhibited receptor alkylation when added in combination with the D3.32N mutation with regard to AChM at the M2 receptor (Figure 4a) and to BR384 at both M2 and M4 receptors (Figure 4b and d).
To obtain further evidence for the mutually exclusive binding of [3H]NMS and the aziridinium ions of AChM and BR384, we measured the competitive displacement of [3H]NMS binding to the slowly alkylated D2.50N/D3.32N mutants of M2 and M4 receptors under conditions where the rate of alkylation of the receptor is negligible or greatly reduced (60 min at 0°C). Under this condition, the highest concentration AChM tested (3 mM) caused 14 ± 0.5% and 20 ± 2.2% alkylation of the double mutants of M2 and M4 receptors (Figure 5a and b). The corresponding values for BR384 (0.3 mM) were 48 ± 4.2% and 44 ± 0.6%, respectively. In the competitive displacement assay, the highest concentration of AChM tested (3 mM) caused 73 ± 1.7% and 74 ± 0.4% inhibition of [3H]NMS binding in the double mutants of M2 and M4 receptors, with no evidence of a non-zero plateau in the inhibition curve (Figure 5c and d). The corresponding values for BR384 (0.3 mM) were 97 ± 1.3% and 88 ± 0.7% respectively. All of the competition curves are consistent with a model of competitive inhibition (i.e., 100% displacement of specific binding). The affinity was too low, however, for near complete displacement of [3H]NMS binding at the highest concentrations of AChM and BR384 used, except for BR384 at M2 D69N/D103N. The log molar IC50 values of AChM at the D2.50N/D3.32N mutants of M2 and M4 receptors were −2.80 ± 0.32 and −2.86 ± 0.32 (N = 3 each), respectively. The corresponding values for BR384 were −4.54 ± 0.32 and −4.16 ± 0.04 (N = 3 each). Thus, the data are consistent with the postulate that the aziridinium ions of AChM and BR3884 have access to all of the sites labeled by [3H]NMS in the D2.50N/D3.32N mutants of M2 and M4 receptors.
Figure 5.
Effects of pretreatment (a and b) or coincubation (c and d) with AChM (○) and BR384 (●) (60 min at 0°C) on the specific binding of [3H]NMS to M2 D69N/D103N (a and c) and M4 D78N/D112N receptors (b and d). a, Various concentrations of AChM or BR384 were incubated with homogenates of HEK 293 cells expressing M2 D69N/D103N receptors for 60 min at 0°C. The reaction was stopped immediately with scopolamine and thiosulfate and washed. Residual binding was measured with [3H]NMS (3 nM). b, Same as panel a except that the receptor preparation was M4 D78N/D112N. c, The specific binding of [3H]NMS (3 nM) to M2 D69N/D103N receptors was measured in the presence of various concentrations of AChM or BR384. The incubation lasted 60 min at 0°C. d, Same as panel c except that the receptor preparation was M4 D78N/D112N. The data represent mean values ± SEM of three experiments, each done in triplicate. The theoretical curve represents the least-squares fit of equation 1 to the data in panels c and d.
Single time-point kinetic assay for estimation of the affinities and rate constants of AChM and BR384
Data like those shown in Figure 4 are sufficient for estimating the dissociation (K1) and rate (k1) constants of the irreversible ligand provided that the covalent reaction is stopped quickly and the fraction of [3H]NMS binding sites insensitive to the alkylating agent is known.
For example, we analyzed each inhibition curve in Figure 3 by regression analysis using equations 4 (AChM, panel a) and 6 (BR384, panel b) with the value of the unreactive sites (b) constrained to that obtained in the global analysis in Figure 3 (17% for AChM; 7.7% for BR384). When applied to the data in panel a, this analysis yielded independent estimates of the log dissociation constant of AChM for the 2-, 4-, 8- and 15-min time points (−4.04 ± 0.11, −4.01 ± 0.025, −4.25 ± 0.10 and −4.20 ± 0.052). The corresponding estimates of the rate constant were 0.45 ± 0.030, 0.51 ± 0.020, 0.38 ± 0.090 and 0.36 ± 0.02 min−1, respectively. Analysis of the data in panel b yielded estimates of the log dissociation constants (−4.91 ± 0.11, −5.05 ± 0.044, −5.10 ± 0.045 and −5.17 ± 0.043) and rate constants (0.99 ± 0.045, 1.05 ± 0.10, 0.75 ± 0.06 and 0.74 ± 0.086 min−1) of BR384 at incubation times of 1, 2, 4 and 8 min, respectively. The average estimates of log K1 (AChM, −4.13; BR384, −5.06) and k1 (AChM, 0.42 min−1; BR384, 0.82 min−1) are nearly the same as those described above in connection with Figure 3.
For a given curve, it is possible to obtain good fit of equations 4 (AChM) and 6 (BR384) as long as b is constrained to a value less than the plateau level of the curve. Over this domain, the estimate of k1 is correlated with that of b and K1 such that the ratio k1/K1 is constant. This relationship occurs because incomplete receptor alkylation at receptor saturating concentrations of AChM or BR384 could be attributed to a significant fraction of receptors resistant to alkylation (significant b value) or a rate constant for alkylation of insufficient magnitude for complete alkylation during the incubation period.
Kinetics of the Interaction of AChM and BR384 with wild type and D2.50N and D3.32N mutants of M1 – M4 muscarinic receptors
We applied the single time-point assay just described to investigate the kinetics of alkylation of M1 – M4 muscarinic receptors by AChM and BR384 (Figure 6). In these experiments, we stopped the alkylation step quickly with scopolamine and thiosulfate and washed cellular homogenates three times before measuring residual muscarinic receptors. Our rationale was that by reducing the number of data points, the assay for a given receptor could be completed more quickly, and hence, the appearance of new unalkylated receptors (i.e., significant b value) would be greatly reduced or eliminated. The results obtained with the more rapidly alkylated receptors (wild type and D2.50N mutants) suggest a b value of zero, given the length of the incubation of receptor with AChM (15 min) and BR384 (6 min).
Figure 6.
The interaction of AChM and BR384 with wild type and mutant M1 (a and b), M2 (c and d), M3 (e and f) and M4 (g and h) muscarinic receptors. Homogenates of cells expressing muscarinic receptors were incubated with different concentrations of AChM (a, c, e and g) or BR384 (b, d, f and h) for 15 (AChM) or 6 (BR384) min. The reactions were stopped with thiosulfate and scopolamine, the homogenates washed, and residual unalkylated muscarinic receptors were estimated by measuring [3H]NMS binding at a single concentration (1 or 3 nM). Mean values ± SEM from three experiments are shown. The different receptors and their mutants are indicated as: wild type, ○; D2.50N, △; D3.32N, ● and D2.50N/D3.32N, ▲.
Figure 6 shows our data on M1 (a and b), M2 (c and d), M3 (e and f) and M4 (g and h) receptors with regard to wild type M3, the D2.50N mutant of M2 – M4 receptors, the D3.32N mutant of M3 and M4 receptors and the D2.50N/D3.32N mutant of M1 – M4 receptors. Data on the wild type M4 receptor are only shown in Figure 4, and data for the wild type and D3.32N mutants of M1 and M2 receptors are also not shown in Figure 6, but have been published previously (6–9). The data in Figure 6 were analyzed as described above with the parameter b in equations 4 (AChM) and 6 (BR384) constrained to 0. The results of this analysis are illustrated in Figure 7, which shows the estimates of -log K1 (a and c) and k1 (b and d) for AChM and BR384. Also shown are the parameter estimates for the wild type M4 receptor from the data in Figure 3 and those for wild type and D3.32N mutants of M1 and M2 receptors from our prior studies (6–9). We did not investigate the D2.50N mutant of the M1 receptor.
With regard to AChM (Figure 7a and b), the D3.32N mutation (M1 D105N, M2 D103N, M3 D148N and M4 D112N) reduced the value of the rate constant for alkylation to only one-fiftieth to one-tenth that of the corresponding wild type receptor and had little effect on the affinity of AChM for M1 – M3 receptors. In the case of the M4 receptor, the affinity for AChM was reduced to one-tenth that of wild type.
In contrast, the D2.50N mutation had smaller effects on the rate constant for receptor alkylation by AChM (one-third to three-fourths that of wild type), but increased affinity 8- to 17-fold relative to wild type M2 – M4 receptors. The combination of both mutations (D2.50N/D3.32N) reduced the rate constant (one-thirtieth to one-third that of wild type) and affinities (one five-hundredth to one-thirtieth that of wild type) of AChM for M2 – M4 receptors. The rate constants for alkylation of double mutant of M1 and M3 receptors was greater than those of the corresponding D3.32N mutants, whereas the rate constant for alkylation of M4 D78N/D112N was smaller than that of M4 D112N.
These mutations had qualitatively similar effects on the interaction of BR384 with muscarinic receptors (Figure 7c and d). The D3.32N mutation had little or no inhibitory effect on the rate constant for alkylation, but reduced affinity to only about one-twentieth (M1 – M3) and one-fourth (M4) that of wild type. In contrast, the D2.50N mutation increased affinity about 30-, 5-and 30-fold at M2 – M4 receptors while having a modest effect on the rate constant for alkylation of the M3 receptor. This mutation also reduced the rate constants for alkylation of M2 and M4 receptors to values about one-half that of wild type. The combination of both mutations (D2.50N/D3.32N) reduced the rate constant for alkylation to values about one-twentieth (M2), one-fourth (M1), one-third (M4) and one-half (M3) that of wild type. The associated changes in affinity represented both increases of two-fold (M1 and M2) and decreases to about one-fifth of wild type (M3 and M4).
The numerical values for the estimates of log K1 and k1 for AChM and BR384 are listed in Tables S1 and S3 of the Supporting Information. Tables S2 and S4 of the Supporting Information summarize post hoc comparisons of the former data, respectively. Analysis of variance showed that the mutations had significant effects on the estimates of log K1 and log k1 for AChM and BR384 at each receptor subtype (Supporting Information, Tables S1 and S3). Post hoc comparisons showed a significant effect of each mutation on the log k1 value for AChM at each receptor subtype. For BR384, all of the mutations had a significant effect on log k1 except the D105N mutation of the M1 receptor, the D103N mutation of the M2 receptor, the D114N and D148N mutations of the M3 receptor, and the D112N mutation of the M4 receptor. With regard to the log K1 estimates for AChM, all of the mutations had significant effects except the D105N mutation of the M1 receptor and the D103N and D69N mutations of the M2 receptor. Finally, with regard to the log K1 estimates for BR384, the mutations had significant effects except for the D71N/D105N and D69N/D103N mutants of M1 and M2 receptors, respectively. Tables S2 and S4 of the Supporting information also list the results of all other possible comparisons among a given receptor subtype.
Because the parameter estimates depend on the value to which b is constrained in equations 4 and 6, we searched parameter space to identify the maximum value of b that would still yield a non-significant increase in the residual sum of squares. This maximum estimate of b was lowest for alkylation of M3 (0.067) and M4 (0.085) receptors by BR384. These low values suggest that constraining b to 0 during regression analysis is reasonable. Constraining b to 0.065, for example, had little or no effect on parameter estimates for M3 and M4 receptor constructs when expressed relative to that of another receptor construct examined in the same analysis (e.g., wild type or D2.50N).
The estimate of the log ratio of k1 to K1 is more accurate than either single parameter and independent of the value of b over the range of 0 to a low value (e.g., 10%). The value of log k1/K1 represents the combined effect of the mutation on affinity and the rate constant for alkylation. Figure 8 shows a plot of the estimates of the log k1/K1 values of AChM and BR384 for wild type M1, M2, M3 and M4 receptors and the mutants thereof. The numerical values of these estimates are listed in Tables S1 and S3 of the Supporting Information. One-way analysis of variance showed a highly significant effect of the mutations on each receptor subtype. Post hoc comparisons showed that the estimate of log k1/K1 was significantly different (P < 0.05) for every comparison among the various constructs of a given receptor subtype, except for M1 wild type vs M1 D2.50N/D3.32N (AChM, P = 0.062; BR384, P = 0.26) and comparisons between of D3.32N and D2.50N/D3.32N with regard to AChM at M3 (P = 0.051) and M4 (P = 0.25) receptors and BR384 at the M2 (P = 0.69) receptor.
Figure 8.
A combined measure of the affinity and reactivity of AChM and BR384 with M1 – M4 wild type receptors and their D2.50N, D3.32N and D2.50N/D3.32N mutants. The histograms show the value of the log ratio of the rate constant for alkylation (k1) and of the affinity constant (K1) of the aziridinium ion of AChM and BR384. The value of the log of this ratio (log k1/K1) is given for wild type and mutant M1 (a), M2 (b), M3 (c) and M4 (d) muscarinic receptors. Mean values ± SEM from four experiments are shown. A summary of the numerical values is listed in Tables S1 and S3 of the Supporting Information.
To quantify the effect of single and double point mutations on the binding affinities of AChM and BR384, we calculated the corresponding change in the log K1 value relative to wild type (Δ log K1). These are listed in Table 1 for the single (D2.50N and D3.32N) and double (D2.50N/D3.32N) point mutations of M2 – M4 receptors. Also listed is the sum of the Δ log K1 values for the two single mutants (D2.50N + D3.32N). At M2 and M3 receptors, AChM exhibited a significant increase in the Δ log K1 value of the double mutant compared to the sum of the Δ log K1 values of the single mutants, indicating an interaction between the mutations. At the M4 receptor, the sum of the Δ log K1 values of AChM for the single mutants was approximately equal to the Δ log K1 value of the double mutant. In contrast, there was no evidence of an interaction between the mutations with regard to the binding affinity of BR384 for M2 and M3 receptors, but there was for the M4 receptor.
Table 1.
Changes in the log K1 of AChM and BR384 associated with single and double point mutations and the sum of the two single mutations (e.g., M2 D103N + D69N).
| AChM (Δ log K1) |
BR384 (Δ log K1) |
|
|---|---|---|
| M2 D103N | −0.36 ± 0.16 | 1.23 ± 0.26 |
| M2 D69N | −0.71 ± 0.26 | −1.39 ± 0.35 |
| M2 D103N + M2 D69N | −1.07 ± 0.31 | −0.17 ± 0.43 |
| M2 D69N/D103N | 1.98 ± 0.40 a | −0.25 ± 0.25 |
| M3 D148N | −0.34 ± 0.16 | 1.17 ± 0.07 |
| M3 D114N | −1.18 ± 0.13 | −0.69 ± 0.02 |
| M3 D148N + M3 D114N | −1.51 ± 0.20 | 0.48 ± 0.08 |
| M3 D114N/D148N | 0.52 ± 0.04b | 0.75 ± 0.12 |
| M4 D112N | 1.21 ± 0.06 | 0.65 ± 0.13 |
| M4 D78N | −0.92 ± 0.12 | −1.27 ± 0.11 |
| M4 D1 12N + M4 D78N | 0.29 ± 0.14 | −0.62 ± 0.17 |
| M4 D78N/D112N | 0.57 ± 0.24 | 0.69 ± 0.32 c |
Significantly different from D2.50N + D3.32N, P < 10−4.
Significantly different from D2.50N + D3.32N, P < 10−6.
Significantly different from D2.50N + D3.32N, P < 0.01.
Interaction of acetylcholine, McN-A-343 and NMS with wild type and D2.50N and D3.32N mutants of M1 – M4 muscarinic receptors
We initially expected that the dissociation constants of acetylcholine and McN-A-343 for the different receptor mutants might be similar to the aziridinium ions of AChM and BR384 because of the close structural similarity of the compounds (Figure 1). To explore this question, we measured the competitive inhibition of [3H]NMS binding by acetylcholine and McN-A-343 as well as nonlabeled NMS. For each competition curve, the IC50 value was estimated and corrected for the competitive effect of [3H]NMS to yield the Ki value of the competitor. These estimates are illustrated in Figure 9, and the numerical values are listed in Table S5 of the Supporting Information together with the Hill slopes of the competition curves for acetylcholine and McN-A-343. The Hill coefficients of the competition curves for McN-A-343 were approximately equal to one at each receptor mutant, indicating that the Ki value is a good estimate of the dissociation constant (K1) of the compound. With regard to acetylcholine, its competition curves at M2 wild type, M2 D103N and M3 D114N/D148N receptors had Hill slopes that were substantially less than one (0.69, 0.60 and 0.56, respectively), indicating behavior consistent with at least two types of binding sites. A likely explanation in the case of the M2 receptor is that a fraction of the receptor population interacts with Gi/o, resulting in higher observed affinity. Thus, in most, but not all cases, the -log Ki values of acetylcholine adequately represent the -log dissociation constant for the receptor (-log K1).
Figure 9.
Negative log dissociation constants of NMS (a), acetylcholine (b) and McN-A-343 (c) for wild type and mutant M1, M2, M3 and M4 muscarinic receptors. The competitive inhibition of the binding of [3H]NMS to the different wild type and mutant receptors was measured, and the data were analyzed to estimate the dissociation constant of each ligand. Mean values ± SEM from three experiments are shown. Analysis of variance showed that for each ligand, there were significant differences among the estimates of pKi for the different mutants at each receptor subtype (Supporting Information, Table S5). A summary of post hoc comparisons of the pKi estimates is given in Tables S6 (NMS) and S7 (acetylcholine and McN-A-343) of the Supporting Information, and a list of the numerical values of the parameter estimates is given in Table S5 of the Supporting Information.
In most instances, the effects of the mutations on ligand affinity are qualitatively similar to those observed with the corresponding aziridinium ions (compare Figures 7 and 9). Two striking exceptions are the effects of the D3.32N mutation on M2 and M3 receptors. This mutation reduced the affinities of acetylcholine to values only one-fiftieth and one-thirtieth those of the wild type receptors, respectively, but had little effect on the aziridinium ion of AChM. This mutation clearly altered how the aziridinium ion interacted with M2 and M3 receptors, however, because its rate constant for alkylation was substantially reduced. A more moderate difference was noted in the effect of the D2.50N/D3.32N mutation. This mutation always greatly reduced the affinities of acetylcholine and McN-A-343, but caused less of a decrease in the affinities of their analogous aziridinium ions for M3 and M4 receptors and an increase in affinity for M1 and M2 receptors.
Analysis of variance showed that all of the mutations had significant effects on the affinities of NMS, acetylcholine and McN-A-343 for M1 – M4 receptors (Supporting Information, footnotes to Table S5). Post hoc comparisons showed that all of the mutations had significant effects on the affinity of NMS, acetylcholine and McN-A-343 for M1 – M4 receptors relative to wild type except for the affinities of acetylcholine and NMS for the M2 D69N mutant. A summary of these and all other post hoc comparisons is given in Tables S6 (NMS) and S7 (acetylcholine and McN-A-343) of the Supporting Information.
We also calculated the Δ log Ki values for the effects of the single and double point mutations on the binding of acetylcholine, McN-A-343 and NMS, and these values are listed in Table 2. With the exception of NMS at the M3 receptor, the increase in the Δ log Ki value associated with the double mutation (D2.50N/D3.32N) was always significantly greater than the sum of the Δ log Ki values of the two single mutations (D2.50N + D3.32N).
Table 2.
Changes in the log Ki of acetylcholine, McN-A-343 and NMS associated with single and double point mutations and the sum of the two single mutations (e.g., M2 D103N + D69N).
| Acetylcholine (Δ log Ki) |
McN-A-343 (Δ log Ki) |
NMS (Δ log Ki) |
|
|---|---|---|---|
| M2 D103N | 2.71 ± 0.15 | 0.46 ± 0.040 | 1.28 ± 0.11 |
| M2 D69N | −0.11 ± 0.13 | −0.57 ± 0.12 | −0.21 ± 0.19 |
| M2 D103N + M2 D69N | 2.60 ± 0.20 | −0.12 ± 0.12 | 1.07 ± 0.21 |
| M2 D69N/D103N | 3.81 ± 0.089 c | 1.04 ± 0.16 d | 1.72 ± 0.16 a |
| M3 D148N | 2.36 ± 0.22 | 1.19 ± 0.075 | 1.36 ± 0.096 |
| M3 D114N | −1.09 ± 0.22 | −0.91 ± 0.086 | −0.33 ± 0.095 |
| M3 D148N + M3 D114N | 1.27 ± 0.31 | 0.28 ± 0.11 | 1.03 ± 0.14 |
| M3 D114N/D148N | 2.29 ± 0.090 a | 1.27 ± 0.073 d | 1.28 ± 0.092 |
| M4 D112N | 2.75 ± 0.13 | 1.22 ± 0.095 | 1.59 ± 0.093 |
| M4 D78N | −0.96 ± 0.20 | −1.11 ± 0.18 | −0.43 ± 0.19 |
| M4 D1 12N + M4 D78N | 1.80 ± 0.23 | 0.11 ± 0.21 | 1.15 ± 0.21 |
| M4 D78N/D112N | 2.71 ± 0.13 b | 1.37 ± 0.096 c | 1.92 ± 0.10 d |
Significantly different from D2.50N + D3.32N, P < 0.05.
Significantly different from D2.50N + D3.32N, P < 10−2.
Significantly different from D2.50N + D3.32N, P < 10−3.
Significantly different from D2.50N + D3.32N, P < 10−5.
Discussion
Our method of estimating the dissociation (K1) and rate (k1) constants is based on the assumption that AChM and BR384 reduce the binding capacity of [3H]NMS for a given population of receptors without affecting its affinity for the unalkylated receptors. We begin by explaining the evidence for this assumption and then describe how our conclusions would be modified if our assumption is incorrect.
Three types of experimental evidence can be used to demonstrate that prior treatment of muscarinic receptors with AChM or BR384 causes a reduction in the binding capacity of [3H]NMS: 1) direct demonstration that AChM or BR384 treatment reduces the binding capacity of [3H]NMS without affecting affinity for the residual receptors, 2) competitive and allosteric protection from the inhibitory effects of AChM and BR384 by known orthosteric and allosteric ligands, respectively, and 3) complete inhibition of [3H]NMS binding by AChM or BR384.
With regard to the first type of evidence, we have previously shown that prior treatment of M1 and M2 receptors with AChM and BR384 causes a reduction in the binding capacity of [3H]NMS without affecting affinity for the residual receptors (6, 7, 9). It has also been shown that prior treatment of rodent forebrain or cerebral cortex with BR384 causes a reduction in the binding capacity of [3H]quinuclidinyl benzilate and [3H]NMS without affecting for the residual receptors affinity (16). These brain regions are abundant in M1, M2 and M4 muscarinic receptors (20–24).
We have also shown that orthosteric ligands, like acetylcholine, NMS and McN-A-343, competitively protect M1, M2 and M2 D103N receptors from alkylation by AChM and BR384, whereas gallamine allosterically inhibits alkylation (6–9) (Type 2 evidence). Similarly, NMS and atropine competitively protect cerebral cortical muscarinic receptors from alkylation by BR384 (6, 16) (Type 2 evidence).
In this report, we show that treatment with AChM (15 min) or BR384 (4 min) causes a near complete inhibition of [3H]NMS binding to wild type and D2.50N mutants of M3 and M4 receptors as well as BR384 treatment of the corresponding D3.32N mutants (Figure 4). Given the brief time of incubation and the concentration of the alkylating agents, the data are consistent with the postulate that populations of the former M3 and M4 receptor are potentially sensitive to complete inactivation by AChM and BR384 (Type 3 evidence). If the irreversible ligands acted at an allosteric site to alter the affinity of [3H]NMS, then the requisite negative cooperativity would have to be very great to cause near complete inhibition of [3H]NMS binding and our method of estimating K1 and k1 would be appropriate, nonetheless.
A fourth type of evidence that can be marshaled to support the postulated reduction in binding capacity is a consistency of the alkylation process with Scheme 1, provided that orthosteric ligands competitively prevent alkylation and complete alkylation is possible under the appropriate conditions (lengthy incubation and receptor saturating concentration of irreversible ligand). We have previously shown that alkylation of M1 and M2 muscarinic receptors is consistent with Scheme 1 (6, 7, 9), and in this report, we show the same for the M4 receptor.
We found, however, that a good fit of the data in Figure 3 to Scheme 1 requires the assumption that a fraction of the sites that bind [3H]NMS do not react covalently with the irreversible ligands. In the case of our prior work on intact cells, recycling of intracellular receptors to the plasma membrane after AChM and BR384 treatment can explain the presence of a fraction of receptors that appear resistant to alkylation. Even in cellular homogenates, there is evidence that the tertiary amine ligand, [3H]quinuclidinyl benzilate, has access to muscarinic receptors in membrane compartments that [3H]NMS does not (22). Nonetheless, we would expect that the aziridinium ions of AChM and BR384 would have access to all of the sites labeled by [3H]NMS, and we described such evidence for M2 and M4 receptors under “Results”. As described under “Results”, we suggest that redistribution of receptors during the washing process might account for the small proportion of [3H]NMS sites in Figure 3 that exhibit a so-called resistance to alkylation.
To summarize, our evidence suggests that AChM and BR384 cause a reduction in the binding capacity of [3H]NMS at M1 – M4 receptors (Type 1 and 2 evidence), the D2.50 mutants of M3 and M4 receptors (Type 3 evidence), and the D3.32N mutant of the M2 receptor (Type 2 evidence). We also have strong evidence that the D3.32N mutants of M3 and M4 receptors undergo a reduction in binding capacity following treatment with BR384 (Type 3 evidence). Because we still observe a small to moderate inhibitory effect of AChM and BR384 on [3H]NMS binding in the D2.50N/D3.32N mutants, then a residue or residues other than D2.50 and D3.32 must be involved in the covalent binding. If the inhibitory effect of AChM and BR384 on [3H]NMS binding in the D2.50N/D3.32N mutants is due, in part, to a change in affinity of [3H]NMS, then this would imply alkylation of an allosteric site. Such an effect would not invalidate our conclusions regarding the participation of D3.32 or D2.50 in receptor alkylation of the wild type receptor (see below). It would also imply that the rate of alkylation of the orthosteric site in the double mutant might very well be essentially zero.
One might expect little difference in the reversible binding properties of acetylcholine and McN-A-343 and their respective aziridinium ions derived from AChM and BR384 because the latter atomic structures differ by only two hydrogens (see Figure 1). With regard to acetylcholine and the aziridinium ion of AChM, both had similar affinities for wild type M1 and M3 receptors, whereas acetylcholine had about 15- and five-fold higher affinity at wild type M2 and M4 receptors, respectively. In the case of the aziridinium ion of BR384, it had four- to seven-fold higher affinity than McN-A-343 at wild type M1 – M4 receptors after taking into account that the aziridinium ion only accounts for 54% of the initial amount of BR384 (16). It seems likely that the differences in affinity can be attributed to the smaller bond angles (~60°) in the aziridinium rings of cyclized AChM and BR384 compared to the tetrahedral structure of the trimethylammonium head groups of acetylcholine and McN-A-343 (bond angles, ~110°). In contrast there is little difference in the affinity of the aziridinium ion of BM123 and its corresponding stable analog (oxotremorine-M) for rat cerebral cortical muscarinic receptors (19).
Our competitive binding data indicate that McN-A343 has similar affinities for M1 through M4 muscarinic receptors (Figure 9 and Supporting Information, Table S5). These data are consistent with the idea that the functional selectivity of McN-A-343 for M1 and M4 muscarinic receptors (10) is based on its ability to activate these receptors subtypes selectively. It has been demonstrated that relative to carbachol, McN-A-343 exhibits higher affinity for the active state of M1 and M4 receptors relative to those of M2 and M3 (11). The combination of equivalent observed affinity for muscarinic receptor subtypes and selectivity for the active states of M1 and M4 receptors implies that McN-A-343 has higher efficacy for M1 and M4 receptors.
Using a peptide mapping strategy Spalding et al. (5) and Curtis et al. (25) showed that the aziridinium ions of AChM and the muscarinic antagonist, benzilylcholine mustard, bind covalently with D3.32 in the M1 muscarinic receptor. In the crystal structures of the M2 and M3 muscarinic receptors bound with the antagonists, 3-quinuclidinyl benzilate and tiotropium, respectively, the basic amino group of each antagonist is coordinated with D3.32 (26, 27). Given the large reduction in the alkylation rate constant of AChM caused by the D3.32N mutation of M1 – M4 receptors, it seems likely that AChM primarily alkylates D3.32N. A slower alkylation process occurs at high concentrations of AChM, which presumably represents an interaction with another residue.
In the crystal structures of M2 and M3 receptors, D2.50 is located two helical turns beneath D3.32, far from the orthosteric-binding site (see Figure 2) (26, 27). This residue is highly conserved among GPCRs, and its mutation to alanine causes a large reduction in receptor expression and ligand binding affinity to M1 muscarinic receptors (28). D2.50 is thought to maintain receptor structure by undergoing hydrogen bonding with adjacent asparagine residues in helix 1 (N1.50) and helix 7 (N7.49) (26–28). In the crystal structure of the inactive state of the human A2A adenosine receptor, a sodium ion is coordinated by D2.50, S3.39 and three water molecules within central cluster of 10 ordered water molecules (29). S3.39 is conserved across all muscarinic subtypes (human M2, S110 and rat M3, S154), and sodium is known to reduce agonist affinity and increase antagonist affinity at the M2 muscarinic receptor (30). These results are consistent with the stabilization of the inactive structure of the A2A adenosine receptor by sodium.
The hydrogen donating and accepting functions of D2.50 should be maintained with asparagine. Accordingly, we observed little or no loss in the binding affinities of the ligands for the D2.50N mutants, and usually, a moderate increase in affinity was observed. In a study on the human M2 muscarinic receptor, Vogel et al. (31) observed a large reduction in ligand affinity with the D2.50N mutation when binding was measured in hypotonic Na/Hepes (20 mM) containing MgCl2 (10 mM). These investigators also observed a large decrease in receptor signaling by M2 D2.50N.
If two residues are located within the binding pocket of a receptor, then an additive contribution to the Gibbs free energy of binding is expected. If a residue is located far from the binding pocket, then any effect on affinity can be attributed to a change in the conformation of the receptor. This change could alter how a given residue in the binding pocket interacts with the ligand, and thus, how mutations of such a residue alter ligand affinity. We expected, therefore, that the D2.50N mutation would alter how the D3.32N mutation affected the interaction of AChM and BR384 with the orthosteric binding site of muscarinic receptors.
The change in the Gibbs free energy of binding is proportional to the log dissociation constant. Consequently, we estimated the Δ log K1 values of AChM and BR384 associated with the different mutations. The D2.50N and D3.32N mutations caused small and large increases in the affinity, respectively, of AChM for M2 and M3 receptors, whereas the double mutation, D2.50N/D3.32N caused a large reduction in affinity, indicating a strong synergistic effect of the mutations on the binding affinity of AChM. In contrast, the mutations had near additive effects on the binding affinity of AChM for the M4 receptor.
The data with BR384 exhibited the opposite pattern. That is, the mutations had additive effects on the binding affinity of BR384 at M2 and M3 receptors, but synergistic effects at the M4 receptor.
With regard to the reversible ligands (i.e., acetylcholine, McN-A-343 and NMS), the mutations always had large synergistic inhibitory effects on affinity except in the case of NMS at the M3 receptor. With the exception of the latter result, the data are consistent with the postulate that the D2.50N mutation acts at a distance to modify how these ligands interact with the orthosteric binding pocket of the receptor. These results illustrate differences in how acetylcholine and the aziridinium ion of AChM interact with the M4 receptor and how McN-A-343 and the aziridinium ion of BR384 interact with M2 and M3 receptors.
The Hill slopes of the competitive binding curve for acetylcholine (Supporting Information, Table S5) were substantially less than one for M2 wild type (0.69), M2 D103N (0.65) and M3 D114N/D148N (0.56) receptors. This behavior most likely represents the contribution of at least two receptor populations exhibiting a difference in affinity for acetylcholine because of differences in coupling with G proteins. In these instances, we interpret the log Ki as a weighted average value of the Ki values of the different receptor populations, and hence, the Δ log Ki as being proportional to the weighted average change in the Gibbs free energy of binding for the different populations.
The rate constant for alkylation of wild type M1 – M4 muscarinic receptors by AChM was greatly reduced to values equal to or less than one-tenth that of wild type by the introduction of the D3.32N mutation, suggesting that this residue participates in the covalent reaction with AChM. In contrast, the D2.50N mutation only reduced the k1 value of AChM for alkylation of the M3 receptor to four-fifths that of wild type and that for alkylation of M2 and M4 receptors to values three-fifths and one-third those of the corresponding wild type receptor, respectively. In the double mutants of M1 and M3 receptors, however, the D2.50N mutation increased the k1 value of AChM three-fold relative to that observed in the D3.32 mutant. This result suggests that in the M1 D71N/D105N and M3 D114N/D148N mutants, the D2.50N mutation alters the conformation of the double mutant to increase the rate of alkylation of a residue other than D3.32. There was no significant difference between the k1 values of AChM at M2 D103N and M2 D69N/D103N receptors. With regard to the M4 receptor, the inhibitory effects of the two mutations on the k1 value of AChM were additive.
To summarize, the D3.32N mutation caused a large reduction (≥ 90%) in the rate constant for alkylation of the M1 – M4 subtypes, consistent with the idea that AChM alkylates the free carboxyl group of D3.32 in the wild type receptor. This mutation does not completely prevent receptor alkylation, however, and the D2.50N mutation appears to increase receptor alkylation of another residue by AChM in the D2.50N/D3.32N mutants of M1 and M3 receptors. Perhaps the D2.50N mutation reduces alkylation of the same residue in the D2.50N/D3.32N mutant of the M4 receptor.
In the case of BR384, the D3.32N mutation caused no significant reduction in the rate constant for receptor alkylation. While this might suggest that BR384 does not alkylate D3.32, and hence, the orthosteric site of M1 – M4 receptors, we found that irreversible alkylation of M1 and M2 receptors was competitively antagonized by NMS and allosterically inhibited by the allosteric modulator, gallamine (6, 9). We observed the same for the M2 D3.32N mutant (8), suggesting that BR384 can alkylate a residue in the orthosteric binding pocket of M1 and M2 receptors other than D3.32. Although the D3.32N mutation lacked an inhibitory effect on the rate constant for alkylation, it did reduce the affinity of the aziridinium ion to about one-twentieth that of wild type M1 - M3 receptors and to about one-fourth that of the wild type M4 receptor. The large effect on M1 – M3 receptors suggests an important role of D3.32 in the reversible binding of BR384 to these receptors.
At M1, M2 and M4 receptors, BR384 alkylated the double mutant (D2.50N/D3.32N) at a much slower rate than wild type, and the rate constant for alkylation of the D2.50N/D3.32N mutant was substantially and significantly smaller than those of wild type, D2.50N, and D3.32N. With regard to the binding affinity of BR384, the effects of the D2.50N and D3.32N mutations were additive at M2 and M3 receptors and synergistic at the M4. None of the mutations had large effects on the rate constant for alkylation of the M3 receptor by BR384 although the effect of the D2.50N/D3.32N mutation (reduced k1 to about three-fifths of wild type) was significant.
One interpretation of the data at M1, M2 and M4 receptors is that the remote D2.50N mutation alters the orientation of the aziridinium ion of BR384 with D3.32 to increase its alkylation of this residue while interfering with its alkylation of other nucleophiles. This hypothesis would explain the lack of substantial effects of the single mutations on the rate constant for alkylation of M1, M2 and M4 receptors despite the large inhibitory effects of the double mutation on the values of k1 (77%, 95% and 80% inhibition, respectively).
Another more speculative possibility is based on the crystal structures of the M2 and M3 muscarinic receptors, which show that the aqueous binding pocket extends down to a level one helical turn below D2.50 (26, 27). Perhaps BR384 is capable of alkylating either D3.32 or D2.50 in the wild type M1, M2 and M4 receptors and that it is necessary to mutate both residues to cause a substantial reduction in the rate constant for alkylation. This interpretation is consistent with the additive contribution of D2.50 and D3.32 to the binding affinity of the aziridinium ion at the M2 receptor, although the latter observation does not prove that BR384 interacts with either residue. Perhaps the additive effects of the mutations on the affinity of AChM for the M4 receptor might be explained by its ability to alkylate single receptors on either D2.50 or D3.32.
To sum up, BR384 probably alkylates more than one residue on M1 – M4 muscarinic receptors. Prior experiments on M1 and M2 receptors mentioned above indicate that BR384 alkylates the orthosteric binding pocket. The large decrease in the rate constant for alkylation in the D2.50N/D3.32N mutant of M1, M2 and M4 receptors suggests that D3.32 is alkylated by BR384. The D2.50N mutation may reduce the rate of alkylation of a non-D3.32 residue, or perhaps, it may prevent alkylation of D2.50. Although it seems unlikely that more than one residue is alkylated in the orthosteric binding pocket of a given receptor, within a population of receptors, different receptors may be alkylated on different residues.
Moderate to high concentrations of McN-A-343 and other orthosteric ligands inhibit the dissociation of [3H]NMS from the M2 muscarinic receptor suggesting that McN-A-343 interacts with an allosteric site (32, 33). Other kinetic and mutagenesis studies implicate a peripheral docking site on muscarinic receptors to which orthosteric ligands bind before shuttling to the primary activation site (34, 35). Recent modeling studies based on the crystal structure of the M2 receptor are consistent with the prior suggestion by Hulme and coworkers that W157 in the M1 receptor (W155 in the M2) is part of a docking site (26). It has been argued that it is difficult to explain the allosteric effect of gallamine on M2 muscarinic receptor-mediated inhibition of adenylate cyclase assuming a single binding site for orthosteric ligands, but not if allosteric modulation of a docking site is considered (36). Thus, the inhibitory effect of McN-A-343 on the kinetics of [3H]NMS binding might also be attributed to occupancy of a docking site.
A provocative study investigating hemi-ligands of McN-A-343 is consistent with the postulate that McN-A-343 interacts simultaneously with allosteric and orthosteric sites (37) causing competition between McN-A-343 and orthosteric ligands.
Given the structural resemblance of BR384 and McN-A-343, the studies mentioned in the prior two paragraphs suggest that BR384 might also alkylate the allosteric site of the muscarinic receptor. Our studies do not rule out this possibility. With regard to the irreversible inhibitory effect of BR384 on [3H]NMS binding, however, our prior results on M1 and M2 muscarinic receptors indicate that this effect is attributed to alkylation of the orthosteric site (6–9).
Supplementary Material
Acknowledgments
Supported by National Institutes of Health Grant GM 69829.
Abbreviations
- AChM
acetylcholine mustard
- BR384
4-[(2-bromoethyl)methyl-amino]-2-butynyl N-(3-chlorophenyl)carbamate
- CHO
Chinese hamster ovary
- HEK
Human embryonic kidney
- McN-A-343
[4-[[N-(3-chlorophenyl)carbamoyl]oxy]-2-butynyl] trimethylammonium chloride
- NMS
N-methylscopolamine
Footnotes
Supporting Information
Irreversible binding of BR384 to homogenates of rat cerebral cortex depicted in Figure S1, estimates of the dissociation constants (K1) and alkylation rate constants (k1) for the interaction of AChM (Table S1) and BR384 (Table S3) with M1 – M4 muscarinic receptors and their D3.32N, D2.50N and D2.50N/D3.32N mutants, summary of post hoc comparisons among the former estimates for AChM (Table S2) and BR384 (Table S4), estimates of the dissociation constants of NMS, acetylcholine and McN-A-343 for M1 – M4 muscarinic receptors and their D3.32N, D2.50N and D2.50N/D3.32N mutants (Table S5), and summary of post hoc comparisons among the former estimates for NMS (Table S6) and acetylcholine and McN-A-343 (Table S7). This information may be accessed free of charge at http://pubs.acs.org.
References
- 1.Fewtrell C, Rang HP. Distribution of bound 3 H-benzilylcholine mustard in subcellular fractions. Br J Pharmacol. 1971;43:417p–418p. [PMC free article] [PubMed] [Google Scholar]
- 2.Turner PR, Denny WA, Ferguson LR. Role of DNA minor groove alkylation and DNA cross-linking in the cytotoxicity of polybenzamide mustards. Anticancer Drug Des. 2000;15:245–253. [PubMed] [Google Scholar]
- 3.Hemminki K. Binding of metabolites of cyclophosphamide to DNA in a rat liver microsomal system and in vivo in mice. Cancer Res. 1985;45:4237–4243. [PubMed] [Google Scholar]
- 4.Ballesteros JA, Weinstein H. Integrated methods for modeling G-protein-coupled receptors: implications of the high-resolution structure of rhodopsin for structure-function analysis of rhodopsin-like receptors. Methods Neuroscience. 1995;25:366–428. [Google Scholar]
- 5.Spalding TA, Birdsall NJ, Curtis CA, Hulme EC. Acetylcholine mustard labels the binding site aspartate in muscarinic acetylcholine receptors. J Biol Chem. 1994;269:4092–4097. [PubMed] [Google Scholar]
- 6.Figueroa KW, Suga H, Ehlert FJ. Investigating the interaction of McN-A-343 with the M1 muscarinic receptor using its nitrogen mustard derivative and acetylcholine mustard. Br J Pharmacol. 2010;160:1534–1549. doi: 10.1111/j.1476-5381.2010.00810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suga H, Figueroa KW, Ehlert FJ. Use of acetylcholine mustard to study allosteric interactions at the M(2) muscarinic receptor. J Pharmacol Exp Ther. 2008;327:518–528. doi: 10.1124/jpet.108.141234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suga H, Sawyer GW, Ehlert FJ. Mutagenesis of nucleophilic residues near the orthosteric binding pocket of M1 and M2 muscarinic receptors: effect on the binding of nitrogen mustard analogs of acetylcholine and McN-A-343. Mol Pharmacol. 2010;78:745–755. doi: 10.1124/mol.110.065367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suga H, Ehlert FJ. Investigating the interaction of McN-A-343 with the M2 muscarinic receptor using its nitrogen mustard derivative. Biochem Pharmacol. 2010;79:1025–1035. doi: 10.1016/j.bcp.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazareno S, Farries T, Birdsall NJ. Pharmacological characterization of guanine nucleotide exchange reactions in membranes from CHO cells stably transfected with human muscarinic receptors m1-m4. Life Sci. 1993;52:449–456. doi: 10.1016/0024-3205(93)90301-i. [DOI] [PubMed] [Google Scholar]
- 11.Figueroa KW, Griffin MT, Ehlert FJ. Selectivity of Agonists for the Active State of M1 - M4 Muscarinic Receptor Subtypes. J Pharmacol Exp Ther. 2008;328:331–342. doi: 10.1124/jpet.108.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer R, Giachetti A. Muscarinic receptor subtypes: M1 and M2 biochemical and functional characterization. Life Sci. 1982;31:2991–2998. doi: 10.1016/0024-3205(82)90066-2. [DOI] [PubMed] [Google Scholar]
- 13.Roszkowski AP. An unusual type of sympathetic ganglionic stimulant. Journal of Pharmacology and Experimental Therapeutics. 1961;132:156–170. [PubMed] [Google Scholar]
- 14.Beny JL, Nguyen MN, Marino M, Matsui M. Muscarinic receptor knockout mice confirm involvement of M3 receptor in endothelium-dependent vasodilatation in mouse arteries. J Cardiovasc Pharmacol. 2008;51:505–512. doi: 10.1097/FJC.0b013e31816d5f2f. [DOI] [PubMed] [Google Scholar]
- 15.Cho AK, Haslett WL, Jenden DJ. The peripheral actions of oxotremorine, a metabolite of tremorine. The Journal of pharmacology and experimental therapeutics. 1962;138:249–257. [PubMed] [Google Scholar]
- 16.Ringdahl B, Mellin C, Ehlert FJ, Roch M, Rice KM, Jenden DJ. Tertiary 2-haloethylamine derivatives of the muscarinic agent McN-A-343, [4-[[N-(3-chlorophenyl)carbamoyl]oxy]-2-butynyl]trimethylammonium chloride. J Med Chem. 1990;33:281–286. doi: 10.1021/jm00163a046. [DOI] [PubMed] [Google Scholar]
- 17.Lambrecht G, Feifel R, Moser U, Wagner-Roder M, Choo LK, Camus J, Tastenoy M, Waelbroeck M, Strohmann C, Tacke R, Rodrigues de Miranda JF, Christophe J, Mutschler E. Pharmacology of hexahydro-difenidol, hexahydro-sila-difenidol and related selective muscarinic antagonists. Trends in Pharmacological Sciences Supplement. 1989:60–64. [PubMed] [Google Scholar]
- 18.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 19.Ehlert FJ, Jenden DJ. The binding of a 2-chloroethylamine derivative of oxotremorine (BM 123) to muscarinic receptors in the rat cerebral cortex. Mol Pharmacol. 1985;28:107–119. [PubMed] [Google Scholar]
- 20.Bertrand D, Bertrand S, Cassar S, Gubbins E, Li J, Gopalakrishnan M. Positive allosteric modulation of the alpha7 nicotinic acetylcholine receptor: ligand interactions with distinct binding sites and evidence for a prominent role of the M2-M3 segment. Mol Pharmacol. 2008;74:1407–1416. doi: 10.1124/mol.107.042820. [DOI] [PubMed] [Google Scholar]
- 21.Buckley NJ, Bonner TI, Brann MR. Localization of a family of muscarinic receptor mRNAs in rat brain. Journal of Neuroscience. 1988;8:4646–4652. doi: 10.1523/JNEUROSCI.08-12-04646.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehlert FJ, Tran LP. Regional distribution of M1, M2 and non-M1, non-M2 subtypes of muscarinic binding sites in rat brain. Journal of Pharmacology and Experimental Therapeutics. 1990;255:1148–1157. [PubMed] [Google Scholar]
- 23.Daval SB, Valant C, Bonnet D, Kellenberger E, Hibert M, Galzi JL, Ilien B. Fluorescent derivatives of AC-42 to probe bitopic orthosteric/allosteric binding mechanisms on muscarinic M1 receptors. J Med Chem. 2012;55:2125–2143. doi: 10.1021/jm201348t. [DOI] [PubMed] [Google Scholar]
- 24.Valant C, Felder CC, Sexton PM, Christopoulos A. Probe dependence in the allosteric modulation of a G protein-coupled receptor: implications for detection and validation of allosteric ligand effects. Mol Pharmacol. 2012;81:41–52. doi: 10.1124/mol.111.074872. [DOI] [PubMed] [Google Scholar]
- 25.Curtis CA, Wheatley M, Bansal S, Birdsall NJ, Eveleigh P, Pedder EK, Poyner D, Hulme EC. Propylbenzilylcholine mustard labels an acidic residue in transmembrane helix 3 of the muscarinic receptor. J Biol Chem. 1989;264:489–495. [PubMed] [Google Scholar]
- 26.Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bee MS, Hulme EC. Functional analysis of transmembrane domain 2 of the M1 muscarinic acetylcholine receptor. J Biol Chem. 2007;282:32471–32479. doi: 10.1074/jbc.M703909200. [DOI] [PubMed] [Google Scholar]
- 29.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, AP IJ, Cherezov V, Stevens RC. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberger LB, Yamamura HI, Roeske WR. Cardiac muscarinic cholinergic receptor binding is regulated by Na+ and guanyl nucleotides. J Biol Chem. 1980;255:820–823. [PubMed] [Google Scholar]
- 31.Vogel WK, Peterson GL, Broderick DJ, Mosser VA, Schimerlik MI. Double mutant cycle analysis of aspartate 69, 97, and 103 to asparagine mutants in the m2 muscarinic acetylcholine receptor. Arch Biochem Biophys. 1999;361:283–294. doi: 10.1006/abbi.1998.0985. [DOI] [PubMed] [Google Scholar]
- 32.May LT, Avlani VA, Langmead CJ, Herdon HJ, Wood MD, Sexton PM, Christopoulos A. Structure-function studies of allosteric agonism at M2 muscarinic acetylcholine receptors. Mol Pharmacol. 2007;72:463–476. doi: 10.1124/mol.107.037630. [DOI] [PubMed] [Google Scholar]
- 33.Redka DS, Pisterzi LF, Wells JW. Binding of orthosteric ligands to the allosteric site of the M(2) muscarinic cholinergic receptor. Mol Pharmacol. 2008;74:834–843. doi: 10.1124/mol.108.048074. [DOI] [PubMed] [Google Scholar]
- 34.Hulme EC, Lu ZL, Saldanha JW, Bee MS. Structure and activation of muscarinic acetylcholine receptors. Biochem Soc Trans. 2003;31:29–34. doi: 10.1042/bst0310029. [DOI] [PubMed] [Google Scholar]
- 35.Jakubik J, El-Fakahany EE, Tucek S. Evidence for a tandem two-site model of ligand binding to muscarinic acetylcholine receptors. J Biol Chem. 2000;275:18836–18844. doi: 10.1074/jbc.M000112200. [DOI] [PubMed] [Google Scholar]
- 36.Ehlert FJ, Griffin MT. Two-state Models and the Analysis of the Allosteric Effect of Gallamine at the M2 Muscarinic Receptor. J Pharmacol Exp Ther. 2008;325:1039–1060. doi: 10.1124/jpet.108.136960. [DOI] [PubMed] [Google Scholar]
- 37.Valant C, Gregory KJ, Hall NE, Scammells PJ, Lew MJ, Sexton PM, Christopoulos A. A novel mechanism of G protein-coupled receptor functional selectivity. Muscarinic partial agonist McN-A-343 as a bitopic orthosteric/allosteric ligand. J Biol Chem. 2008;283:29312–29321. doi: 10.1074/jbc.M803801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.