Abstract
The radical S-adenosylmethionine (SAM) enzyme MqnC catalyzes conversion of dehypoxanthine futalosine (DHFL) to the unique spiro-compound cyclic DHFL in the futalosine pathway for menaquinone biosynthesis. This study describes the in vitro reconstitution of [4Fe-4S]-cluster-dependent MqnC activity and identifies the site of hydrogen atom abstraction from DHFL by the adenosyl radical.
Keywords: Menaquinone, futalosine, radical SAM enzyme
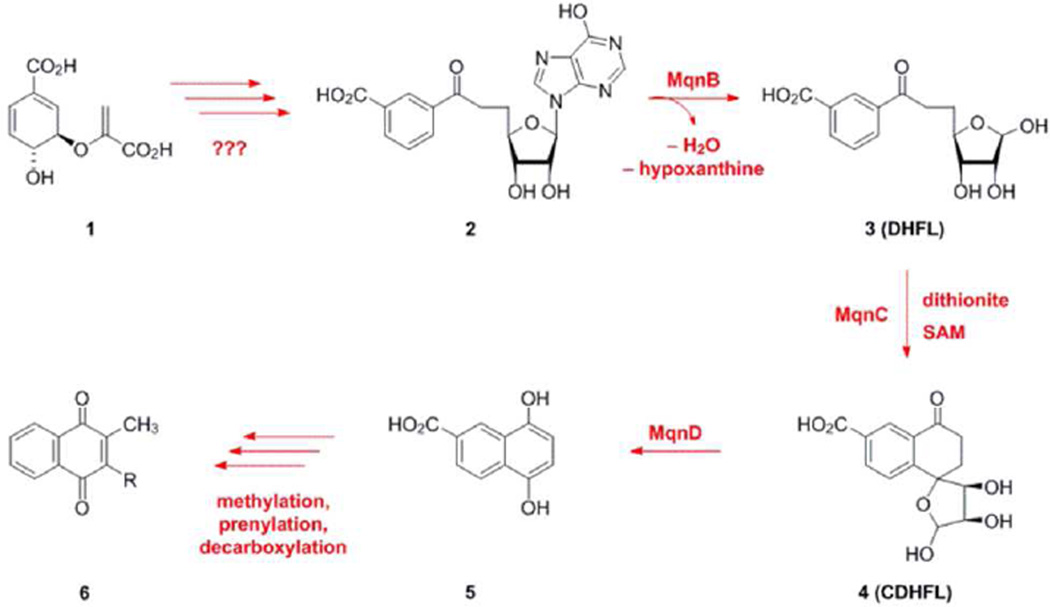
Menaquinone (MK, vitamin K2) is a lipid-soluble molecule that shuttles electrons between membrane-bound protein complexes in the respiratory chain. In the classic Escherichia coli pathway, it is biosynthesized from chorismate by eight enzymes (MenA to MenH).1–3 Recently, an alternative route for menaquinone production was discovered, termed the futalosine pathway (Scheme 1).4 Chorismate (1)-derived futalosine (2) is converted to dehypoxanthine futalosine (DHFL, 3) by futalosine hydrolase MqnB, and 3 is transformed into cyclic dehypoxanthine futalosine (CDHFL, 4) by the radical SAM enzyme MqnC. MqnD then converts 4 to 1,4-dihydroxy-6-naphthoic acid (5). In the later steps of the pathway, based on the annotation and clustering of the open reading frames in Streptomyces coelicolor A3(2), it is possible that SCO4491 (prenylation) and SCO4556 (methylation) could be involved in completing the biosynthetic pathway for production of menaquinone (6). The early and late steps in the pathway are currently unknown and under investigation. Humans and commensal intestinal bacteria lack this pathway therefore it represents an attractive target for the development of chemotherapeutic compounds.
Scheme 1.
Menaquinone biosynthesis via the futalosine pathway.
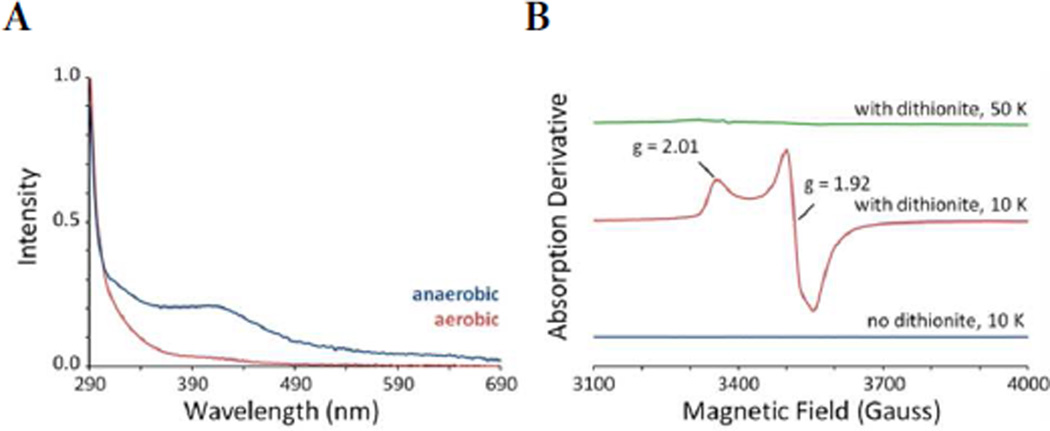
Enzymes belonging to the radical SAM superfamily are typically characterized by the presence of a cysteine-rich CX3CX2C motif that ligates a [4Fe-4S] cluster.5,6 The iron-sulfur cluster is responsible for the reductive cleavage of SAM, to generate a 5′-deoxyadenosyl (5′-dA) radical and methionine. Amino acid sequence analysis revealed that MqnC contains this characteristic CX3CX2C iron-sulfur cluster motif. Therefore, a recombinant plasmid encoding mqnC from Bacillus halodurans C-125 was introduced into E. coli BL21(DE3)-T1R containing the E. coli suf operon for co-expression of His6-MqnC with iron-sulfur cluster biogenesis proteins.7 Anaerobic purification by nickel affinity chromatography yielded pure brown protein with 3 irons and 2 sulfides per monomer, suggesting incomplete reconstitution of the cluster. The UV-Vis spectrum of MqnC had an absorbance maximum at approximately 410 nm that disappeared upon exposure of the protein to oxygen (Figure 1A).8 Electron paramagnetic resonance (EPR) spectra and g-values for MqnC treated with excess sodium dithionite were consistent with a protein-bound [4Fe-4S]1+ cluster (Figure 1B).
Figure 1.
Spectroscopic properties of MqnC. (A) UV-Vis absorbance of purified Fe/S-reconstituted MqnC (160 µM) in 100 mM phosphate buffer, pH 7.5 before (blue) and after (red) exposure to oxygen. (B) EPR spectra of purified Fe/S-reconstituted MqnC (280 µM) treated with (red and green) or without (blue) dithionite.
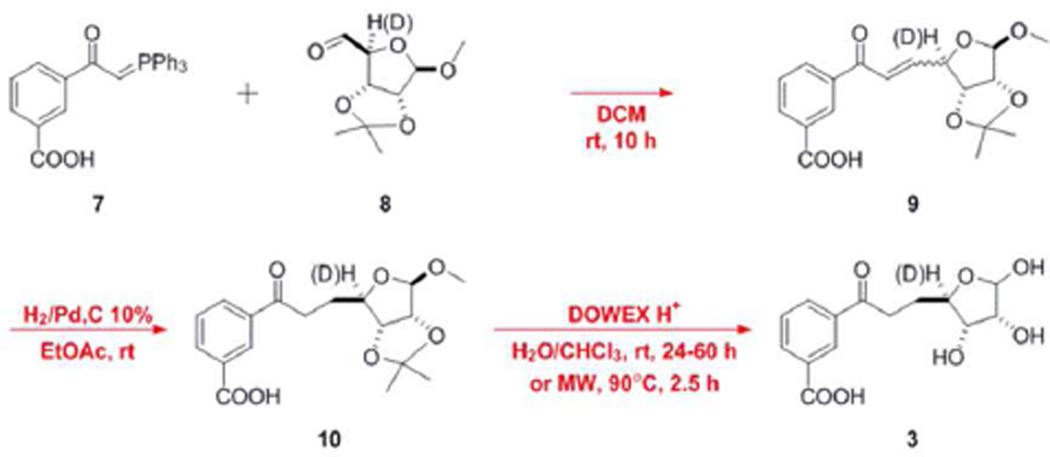
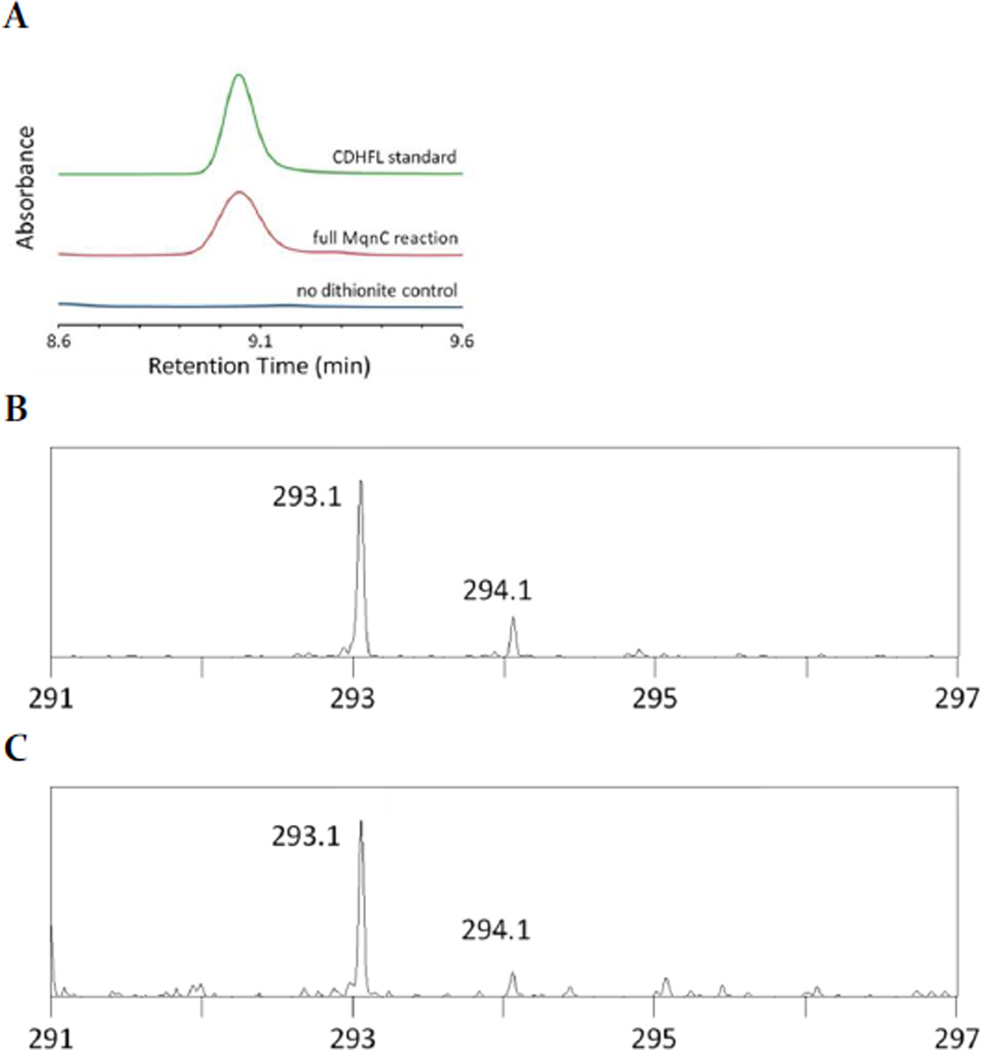
Based on gene disruption studies in S. coelicolor, MqnC is predicted to catalyze the conversion of 3 to 4.4 To test the activity of MqnC, DHFL was synthesized as illustrated in Scheme 2 (experimental methods are described in detail in the supporting information). When MqnC was incubated under anaerobic conditions with sodium dithionite, DHFL, and SAM (11) in phosphate buffer, pH 7.5, production of CDHFL and 5′-deoxyadenosine (5′-dAd) was observed by HPLC as confirmed by co-elution with authentic standards (Figure 2A). CDHFL was not observed in control reactions in which sodium dithionite, DHFL, SAM, or MqnC were omitted. Assay and control samples were also analyzed by LC-MS using an Agilent 1200 LC (ChemStation) with a Bruker Daltonics micrOTOF-Q II ESI-Qq-TOF mass spectrometer (HyStar). The observed m/z for cyclic DHFL in negative ion mode was 293.1 as compared to the theoretical exact mass of 293.1 m/z ([M-H]−, Figure 2C). The authentic standard for CDHFL was purified from the previously described S. coelicolor SCO4326-disruptant strain (observed 293.1 m/z).4 These results confirm the predicted role of MqnC as the DHFL cyclase.
Scheme 2.
Synthesis of DHFL and [4-2H]-DHFL.
Figure 2.
MqnC activity in vitro. (A) HPLC chromatograms monitoring absorbance at 250 nm. (B) Mass spectrum data for CDHLF purified from S. coelicolor mqnD-disruptant strain. (C) Mass spectrum for CDHFL from full MqnC reaction. Calculated exact mass of CDHFL: 293.1 m/z ([M-H]−).
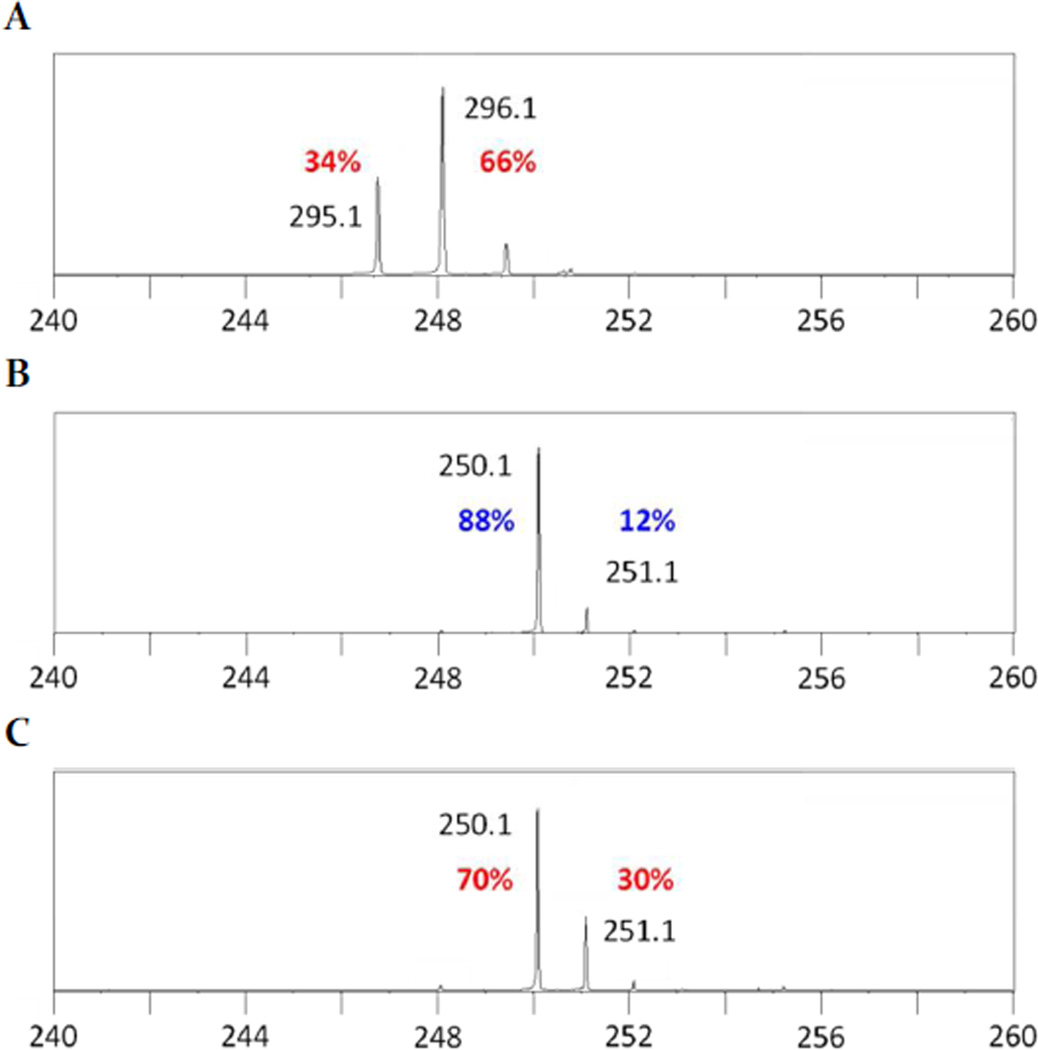
Deuterium labeling of DHFL was used to elucidate the site of hydrogen atom abstraction by the 5′-dA (13) radical during MqnC catalysis. DHFL that was site-specifically deuterated at the C4′ position was synthesized and used as a substrate for MqnC. Deuterium transfer from [4-2H]-DHFL to the 5′-dA radical (13) was monitored by LC-MS (Figure 3). The data revealed an increase of a single mass unit for 5′-dAd, thus demonstrating that the hydrogen from the C4′ position of DHFL is abstracted by the 5′-dA radical. The low efficiency of label transfer is most likely due to the substantial levels of uncoupled 5′-dAd formation (10:1, Figure S5) as well as a primary isotope effect on the initial hydrogen atom abstraction, which would favor consumption of [4-1H]-DHFL over [4-2H]-DHFL.
Figure 3.
MqnC-mediated hydrogen atom abstraction by the 5′-dA radical occurs at the C4′-position of DHFL. (A) Mass spectrum demonstrating 66% deuterated [4-2H]-DHFL. (B) 5′-dAd derived from MqnC reaction using DHFL as the substrate. (C) 5′-dAd derived from MqnC reaction using [4-2H]-DHFL as the substrate. Calculated exact mass of DHFL: 295.1; [4-2H]-DHFL: 296.1; and 5′-dAd: 250.1 m/z ([M-H]−).
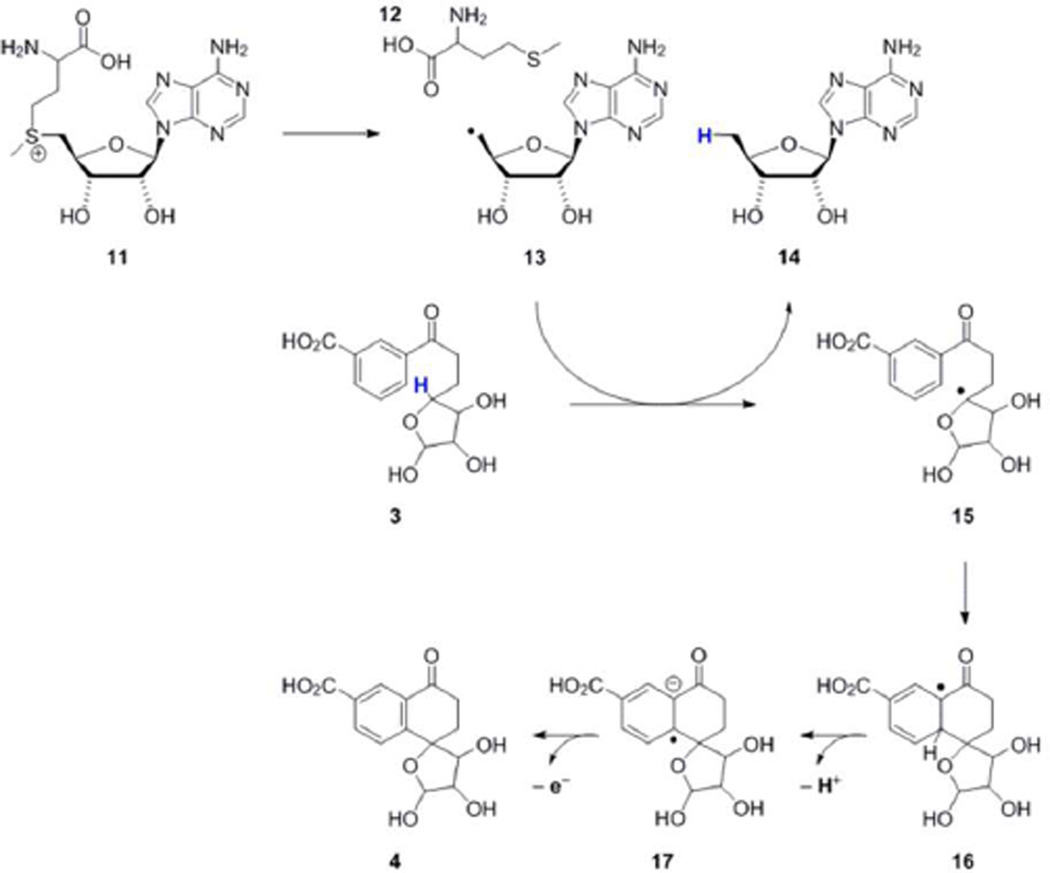
A mechanistic proposal for the MqnC catalyzed reaction is outlined in Scheme 3. Reductive cleavage of SAM generates the adenosyl radical 13. This then abstracts a hydrogen atom from DHFL 3 to give radical 15. Cyclization gives 16, which after deprotonation transfers an electron back to the [4Fe-4S]2+ cluster to give CDHFL 4.
Scheme 3.
Proposed MqnC reaction mechanism.
In summary, the activity of the radical SAM enzyme MqnC was successfully reconstituted. EPR studies confirmed that MqnC contains a [4Fe-4S] cluster and isotope labeling studies demonstrated that the C4′ hydrogen atom of DHFL 3 is abstracted by the 5′-deoxyadenosyl radical. A mechanistic proposal for the MqnC-catalyzed reaction is described. Experiments are in progress to test this proposal.
Supplementary Material
ACKNOWLEDGMENT
We thank Cynthia L. Kinsland from Cornell University for construction of pTHT-mqnC, Professor Tohru Dairi from Hokkaido University in Hokkaido, Japan for providing the mqnD-disruptant strain, and Professor Paul A. Lindahl from Texas A&M University for use of the EPR spectrometer.
Funding Sources
This research was supported by a grant from the National Institutes of Health (DK44083 to TPB) and by the Robert A. Welch Foundation (A-0034).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supporting Information Available: Detailed procedures for cloning the mqnC expression construct, MqnC expression and purification, DHFL synthesis, CDHFL isolation, and HPLC and LC-MS methods are available in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
Lisa E. Cooper was responsible for the biochemical studies. Dmytro Fedoseyenko, Sameh Abdelwahed, and Soong-Hyun Kim synthesized DHFL. Lisa E. Cooper and Tadhg P. Begley designed the experiments and prepared the manuscript.
REFERENCES
- 1.Meganathan R. Menaquinone/Ubiquinone Biosynthesis and Enzymology. In: Mander L, Liu H-W, editors. Comprehensive Natural Products II: Chemistry and Biology. Elsevier Ltd; 2010. pp. 411–444. [Google Scholar]
- 2.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 3.Dairi T, Kuzuyama T, Nishiyama M, Fujii I. Convergent strategies in biosynthesis. Nat. Prod. Rep. 2011;28:1054–1086. doi: 10.1039/c0np00047g. [DOI] [PubMed] [Google Scholar]
- 4.Hiratsuka T, Furihata K, Ishikawa J, Yamashita H, Itoh N, Seto H, Dairi T. An alternative menaquinone biosynthetic pathway operating in microorganisms. Science. 2008;321:1670–1673. doi: 10.1126/science.1160446. [DOI] [PubMed] [Google Scholar]
- 5.Duin EC, Lafferty ME, Crouse BR, Allen RM, Sanyal I, Flint DH, Johnson MK. [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry. 1997;36:11811–11820. doi: 10.1021/bi9706430. [DOI] [PubMed] [Google Scholar]
- 6.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hänzelmann P, Hernández HL, Menzel C, García-Serres R, Huynh BH, Johnson MK, Mendel RR, Schindelin H. Characterization of MOCS1A, an oxygen-sensitive iron-sulfur protein involved in human molybdenum cofactor biosynthesis. J. Biol. Chem. 2004;279:34721–34732. doi: 10.1074/jbc.M313398200. [DOI] [PubMed] [Google Scholar]
- 8.Lee KH, Saleh L, Anton BP, Madinger CL, Benner JS, Iwig DF, Roberts RJ, Krebs C, Booker SJ. Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily. Biochemistry. 2009;48:10162–10174. doi: 10.1021/bi900939w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.