Abstract
CrataBL, a protein isolated from Crataeva tapia bark, which is both a serine protease inhibitor and a lectin, has been previously shown to exhibit a number of interesting biological properties, including anti-inflammatory, analgesic, anti-tumor, and insecticidal activities. Using a glycan array we have now shown that only sulfated carbohydrates are effectively bound by CrataBL. Since this protein was recently shown to delay clot formation by impairing the intrinsic pathway of coagulation cascade, we considered that its natural ligand might be heparin. Heparin is a glycosaminoglycan (GAG) that interacts with a number of proteins including thrombin and antithrombin III, which have a critical, essential pharmacological role in regulating blood coagulation. We have thus employed surface plasmon resonance to better understand the binding interaction between the heparin polysaccharide and CrataBL. Kinetic analysis shows that CrataBL displays strong heparin-binding affinity (KD= 49 nM). Competition studies using different size heparin-derived oligosaccharides showed that the binding of CrataBL to heparin is chain length dependent. Full chain heparin with 40 saccharides, or large oligosaccharides, having 16 to 18 saccharide residues, show strong binding affinity for CrataBL. Heparin-derived disaccharides through tetradecasaccharides show considerably lower binding affinity. Other highly sulfated GAGs including chondroitin sulfate E and dermatan 4,6-disulfate showed comparable CrataBL binding affinity to heparin. Less highly sulfated GAGs, heparan sulfate, chondroitin sulfate A and C and dermatan sulfate displayed modest binding affinity as did chondroitin sulfate D. Studies using chemically modified heparin show that N-sulfo and 6-O-sulfo groups on heparin are essential for CrataBL- heparin interaction.
Keywords: glycosaminoglycans, lectin, heparin, interaction, surface plasmon resonance
It is well known that glycanscan mediate a wide variety of specific physiological or pathophysiological functions by interacting with glycan-binding proteins (GBPs). These interactions are broadly classified into two major groups—lectins and glycosaminoglycan (GAG)-binding proteins.1 Interaction of lectins with glycans on different cellular surfaces can promote hemagglutination and other responses, such as antimicrobial, mitogenic, anti-tumor and insecticidal activities.2,3 Interactions between heparin, heparan sulfate (HS), and other GAGs with proteins mediate diverse biological processes, such as blood coagulation, cell growth and differentiation, host defense and viral infection, lipid transport and metabolism, cell-to-cell and cell-to-matrix signaling, inflammation, and cancer.4–8 From the molecular structural view of the interactions, most lectins have defined “carbohydrate-recognition domains” (CRDs) with specific features of amino acid sequence or three-dimensional structure.1 These CRDs bind to their carbohydrate ligands primarily through hydrogen bonding interactions. In contrast, most of protein interactions with sulfated GAGs seem to involve surface clusters of positively charged amino acids that align to bind the anionic GAG chains primarily through ion-pairing interactions.4,9,10 Thus, an understanding of glycan-protein interactions at the molecular level is of fundamental importance to biology and to development of highly specific therapeutic agents.4,10
CrataBL (see structure in Figure 1), a lectin from the bark of Crataeva tapia tree (belonging to the Capparaceae family, found in northeastern Brazil) has been purified using reversed micelles in isooctane11 and through chromatographic processes.12 Previous research demonstrated that CrataBL shows some specificity for binding glucose and galactose.12 A number of biological properties associated with CrataBL have been characterized, including anti-inflammatory, analgesic, anti-tumor,13 and insecticidal activities.12 A recent study showed that CrataBL delayed clot formation by impairing the intrinsic pathway of coagulation cascade, suggesting this protein might interact with heparin.14 Heparin and heparin-derived low molecular weight heparins (LMWHs) are the most widely used clinical anticoagulants. The heparins bind specifically and with high affinity (KD ~20 nM) to the serine protease inhibitor antithrombin III (AT), resulting in its conformational activation and leading to the inhibition of major coagulation cascade proteases, including thrombin (factor (F) IIa) and FXa. In this study we have shown, using a lectin array, that CrataBL binds effectively only sulfated oligosaccharides, thus confirming the prediction that it might be a heparin-binding protein. We have further analyzed molecular interactions of heparin and other GAGs with CrataBL in order to understand the possible mechanism of the disruption of the intrinsic pathway of the coagulation cascade by CrataBL. This was accomplished with the BIAcore system (BIAcore 3000), which utilizes the surface plasmon resonance (SPR) and allows a direct quantitative analysis of the label-free molecular interactions in real time.
Figure 1.
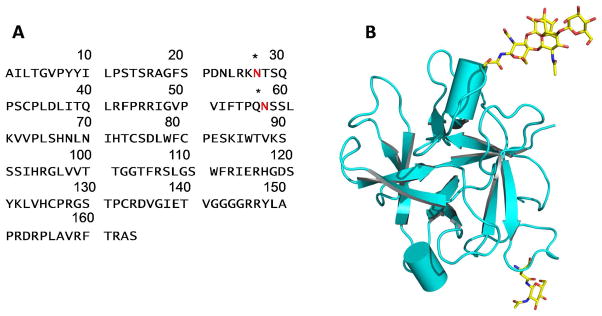
Structure of CrataBL. A. The amino acid sequence of CrataBL. Asterisks indicate glycosylation sites. B. A chain tracing (prepared with PyMol30) showing the three-dimensional structure of a CrataBL monomer. Two glycosylated residues and the attached carbohydrates are shown in sticks.
EXPERIMENTAL PROCEDURES
Isolation of CrataBL
Isolation of the protein was performed according to the procedure of Araújo et al.12 Briefly, extracts from Crataeva tapia bark 10% (w/v) were fractionated with 30–60% ammonium sulfate. The fraction was dialyzed against 10 mM sodium citrate/sodium phosphate buffer pH 5.5 and applied to a column of CM-cellulose equilibrated with the same buffer, also containing 0.5 M NaCl. Adsorbed protein was eluted with equilibration buffer containing 0.5 M NaCl. The single peak from ion exchange chromatography was submitted to size exclusion chromatography on Superdex 75 column, equilibrated in 0.15 M NaCl using an ÄKTA Purifier (GE Healthcare, Uppsala, Sweden), followed by high performance liquid chromatography (HPLC) on a C18 column, to confirm the homogeneity of the sample. Elutions were monitored at 280 nm. This glycosylated protein has a molecular mass of 20.2 kDa, including ~1170 Da N-linked carbohydrates bound to two asparagines residues (Ferreira et al., submitted).
Carbohydrate array studies
Purified CrataBL was labeled with Alexa Fluor488. The protein (781 μg/mL) was dissolved in 0.1 M NaHCO3 containing 50 mM GlcNAc, 50mM glucose, and Alexa Fluor® 488 carboxylic acid, 2,3,5,6-tetrafluorophenyl ester (Life Technologies) at a final concentration of 0.3 mM. After 1 h at room temperature, the reaction mixture was applied to a Bio-Gel P2 column (5 mL) and eluted with phosphate buffered saline (PBS) in 0.5 mL fractions to separate the labeled protein from unreacted dye and the monosaccharides, which were included to protect the glycan binding site during the labeling process. The fraction containing the highest concentration (estimated using a calculated extinction coefficient) of protein with a molar ratio of dye to protein of 0.6 was used to interrogate version 5.0 of the glycan microarray produced by the Consortium for Functional Glycomics (http://www.functionalglycomics.org) at concentrations of 200 and 20 μg/mL, according to the standard protocol.15,16 Key sulfated glycans in composition of the array were synthesized as described.17,18 After processing the microarray slide was scanned using a PerkinElmer ProScanArray for detecting AlexaFluor488, and the data were displayed as histograms of relative fluorescence units (RFU) corresponding to individual glycan structures (see supplemental data).
GAGs and heparin oligosaccharides
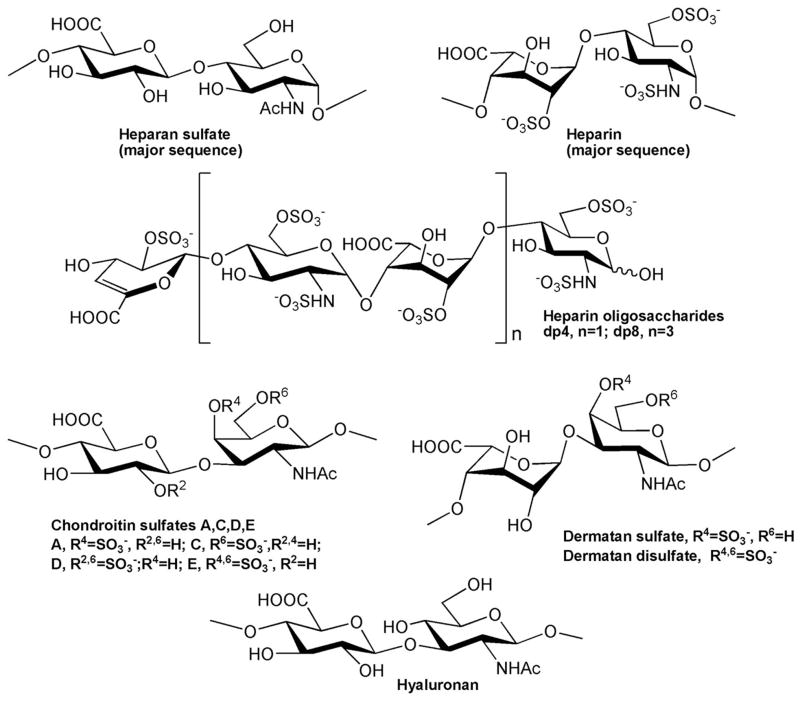
The GAGs used in this study were porcine intestinal heparin (16 kDa) (Celsus Laboratories, Cincinnati, OH), low molecular weight (LMW) heparin (4.8 kDa) (Celsus); and porcine intestinal heparan sulfate (HS) (Celsus); chondroitin sulfate A (CS-A) (20 kDa) from porcine rib cartilage (Sigma, St. Louis, MO), dermatan sulfate (DS) (also known as chondroitin sulfate B, 30 kDa, from porcine intestine, Sigma), dermatandisulfate (Dis-DS) (4,6 disulfo DS, 33 kDa, Celsus) prepared through the chemical 6-O-sulfonation of dermatan sulfate,19 chondroitin sulfate C (CS-C) (20 kDa, from shark cartilage, Sigma), chondroitin sulfate D (CS-D) (20 kDa, from whale cartilage, Seikagaku, Tokyo, Japan), chondroitin sulfate E (CS-E) (20 kDa from squid cartilage, Seikagaku), and hyaluronic acid sodium salt (100 kDa, from Streptococuszooepidemicus, Sigma). Fully desulfated heparin (14 kDa), N-desulfated heparin (14 kDa) and 2-O-desulfated heparin (13 kDa) were all prepared based on Yates et al.20 The 6-O-desulfated heparin (13 kDa) was a generous gift from Dr. Lianchun Wang of the Complex Carbohydrate Research Center, University of Georgia. Heparin oligosaccharides included disaccharide (dp2) tetrasaccharide (dp4), hexasaccharide (dp6), octasaccharide (dp8), decasaccharide(dp10), dodecasaccharide (dp12), tetradecasaccharide (dp14), hexadecasaccharide (dp16) and octadecasaccharide (dp18) and were prepared from controlled partial heparin lyase 1 treatment of bovine lung heparin (Sigma) followed by size fractionation.21 Chemical structures of these GAGs and heparin oligosaccharides are showed in Figure 2.
Figure 2.
Chemical structures of GAGs and heparin-derived oligosaccharides.
Preparation of heparin biochip
The biotinylated heparin was prepared by reaction of sulfo-N-hydroxysuccinimide long-chain biotin (Piece, Rockford, IL) with free amino groups of unsubstituted glucosamine residues in the polysaccharide chain following a published procedure.22 The biotinylated heparin was immobilized to streptavidin (SA) chip (GE Healthcare, Uppsala, Sweden) based on the manufacturer’s protocol. In brief, 20-μl solution of the heparin-biotin conjugate (0.1 mg/mL) in HBS-EP running buffer was injected over flow cell 2 (FC2) of the SA chip at a flow rate of 10μL/min. The successful immobilization of heparin was confirmed by the observation of a ~250 resonance unit (RU) increase in the sensor chip. The control flow cell (FC1) was prepared by 1 min injection with saturated biotin. SPR measurements were performed on a BIAcore 3000 (GE Healthcare, Uppsala, Sweden) operated using BIAcore 3000 control and BIA evaluation software (version 4.0.1).
Measurement of interaction between heparin and CrataBL using BIAcore
The protein samples were diluted in HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4). Different dilutions of protein samples were injected at a flow rate of 30 μL/min. At the end of the sample injection, the same buffer was flowed over the sensor surface to facilitate dissociation. After a 3 min dissociation time the sensor was regenerated by injecting with 30 μL of 2 M NaCl. The response was monitored as a function of time (sensorgram) at 25 °C.
Competition between heparin on chip surface and heparin-derived oligosaccharides
CrataBL protein (125nM) mixed with 1000 nM of heparin oligosaccharides, including disaccharide (dp2), tetrasaccharide (dp4), hexasaccharide (dp6), octasaccharide (dp8), decasaccharide(dp10), dodecasaccharide (dp12), tetradecasaccharide (dp14), hexadecasaccharide (dp16), and octadecasaccharide (dp18) in HBS-EP buffer were injected over heparin chip at a flow rate of 30 μL/min, respectively. After each run, the dissociation and the regeneration were performed as described above. For each set of competition experiments on SPR, a control experiment (only CrataBL protein without any heparin or oligosaccharides) was performed to make sure the surface was completely regenerated and that the results obtained between runs were comparable.
Competition between heparin on chip surface and GAGs
For testing the ability of other GAGs and chemically modified heparins to inhibit the CrataBL-heparin interaction, CrataBL at 125 nM was pre-mixed with 1000 nM of GAG or chemically modified heparin and injected over the heparin chip at a flow-rate of 30 μL/min. After each run, a dissociation period and regeneration protocol was performed as described above.
RESULTS AND DISCUSSION
Glycan array studies
The results of binding Alexa Fluor 488-labeled CrataBL to the glycan targets on the CFG glycan microarray indicated that this lectin preferred sulfated oligosaccharides (see supplemental data). Of the 611 glycans on this array 67 of the glycans are sulfated and are primarily di-, tri- and tetra-saccharides. The results of CrataBL binding to selected sulfated and related non-sulfated disaccharides are shown in Table 1. Among this group the strongest binding glycans were di- and tri-sulfated derivatives of Galβ1-3/4GlcNAc (glycans 1–7, 18) and LacdiNAc (glycan 2a). All binding was lost when the sulfates were not present on these structures (glycans 13 and 20) and addition or substitution of a different negatively charged group, such as sialic acid (glycans 14–17, 19) or phosphate (glycans 11, 12), did not enhance binding. With the limited number of sulfated structures on the CFG array it is difficult to identify an obvious binding motif, but the preference for binding to sulfated derivatives of lactosamine strongly suggests that CrataBL may be a GAG-binding protein as has been observed previously in many studies using the CFG glycan array.
Table 1.
Best binding glycans found using the glycan array (20 μg/ml CrataBL – see supplementary material for details)
| Glycan | Structure on Master List | Average RFU | St. Dev. | %CV | Chart Number |
|---|---|---|---|---|---|
| 1 | (6S)(3S)Galβ1–4(6S)GlcNAc | 9604 | 427 | 4 | 22 |
| 2 | (3S)Galβ1–4(6S)GlcNAc | 4868 | 1065 | 22 | 35 |
| 2a | (6S)(4S)GalNAcβ1–4(6S)GlcNAc | 4834 | 377 | 8 | 512 |
| 3 | (6S)Galβ1–4(6S)GlcNAc | 3119 | 1673 | 54 | 298 |
| 4 | (3S)Galβ1–4(6S)GlcNAc | 2917 | 178 | 6 | 34 |
| 5 | (6S)(3S)Galβ1–4GlcNAc | 2776 | 189 | 7 | 23 |
| 6 | (6S)(4S)Galβ1–4GlcNAc | 2191 | 362 | 17 | 39 |
| 7 | (4S)(3S)Galβ1–4GlcNAc | 1405 | 87 | 6 | 297 |
| 8 | (3S)Galβ1–4GlcNAc | 242 | 52 | 21 | 36 |
| 9 | (6S)Galβ1–4GlcNAc | 143 | 34 | 24 | 44 |
| 10 | (4S)Galβ1–4GlcNAc | 109 | 13 | 12 | 40 |
| 11 | Galβ1–4(6P)GlcNAc | 287 | 24 | 8 | 518 |
| 12 | (6P)Galβ1–4GlcNAc | 34 | 16 | 48 | 519 |
| 13 | Galβ1–4GlcNAc | 202 | 46 | 23 | 169 |
| 14 | Neu5Acα2–3Galβ1–4(6S)GlcNAc | 1294 | 83 | 6 | 252 |
| 15 | Neu5Acα2–6Galβ1–4(6S)GlcNAc | 466 | 36 | 8 | 268 |
| 16 | Neu5Acα2–3Galβ1–4GlcNAc | 39 | 14 | 35 | 260 |
| 17 | Neu5Acα2–6Galβ1–4GlcNAc | 54 | 14 | 27 | 269 |
| 18 | (6S)Galβ1–3(6S)GlcNAc | 7347 | 2689 | 37 | 445 |
| 19 | Neu5Acα2–3Galβ1–3(6S)GlcNAc | 454 | 38 | 8 | 239 |
| 20 | Galβ1–3GlcNAc | 17 | 9 | 54 | 150 |
The disaccharide (6S)(3S)Galβ1–4(6S)GlcNAc which exhibited the best binding to CrataBL, in the form of -OCH2CH2CH2NH2β-glycoside, was utilized in experiments aimed at creating ligand-carbohydrate complexes suitable for crystallographic investigations. Although crystals could be grown from a mixture of CrataBL and the disaccharide, analysis of the electron density has not shown any appreciable binding. This result, most likely due to the still relatively low binding constant for a short oligosaccharide, prompted us to search for other, longer sulfated carbohydrates that that might exhibit higher avidity of binding.
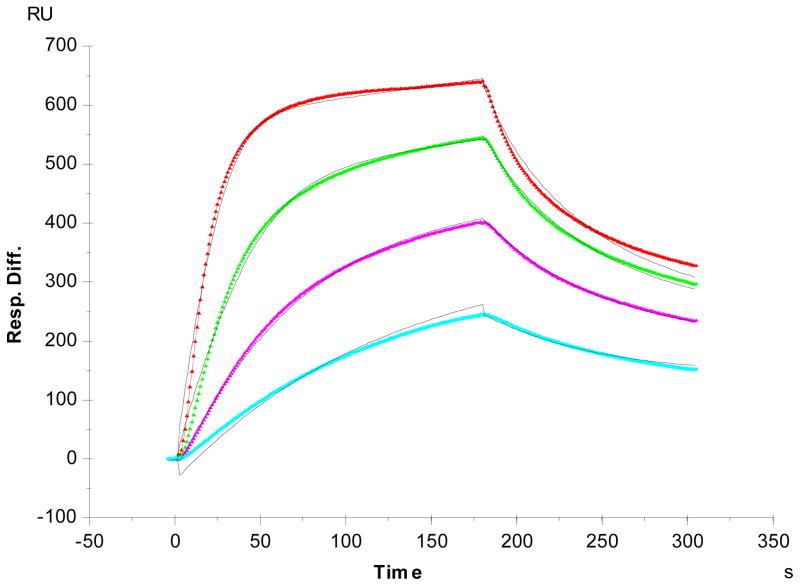
Kinetics measurement of CrataBL-heparin interactions
Previous research on CrataBL bioactivities showed that it delayed formation by impairing the intrinsic pathway of coagulation cascade indicating this lectin might be a heparin-binding protein.14 SPR was used to characterize the binding of CrataBL to heparin. Heparin was immobilized on SA SPR sensor chips and sensorgrams of CrataBL-heparin interaction were obtained (Figure 3). These sensorgrams fit well to a Langmuir 1:1 binding model, and the binding kinetics are presented in Table 2. The SPR data show that CrataBL binds to heparin with dissociation constants KD of ~49nM. Although lectins were first discovered in plants, they are now known to be present throughout the organisms i.e., plant, animal, bacteria, and virus. Most lectins have defined “carbohydrate-recognition domains” (CRDs) with specific features for binding specific sequence of glycans. In general, single-site-binding affinities in many lectins appear to be low (with KD values in the micromolar range), although some lectins recognize glycans with much higher affinity (with KD values in the nanomolar range).1 Despite the numerous lectins that exit throughout nature, only a few heparin-binding proteins, from human placenta, fish, clams and cloned thymic myoid cells have been reported in the literature.23–26 The affinity of interactions between these lectins and heparin have not been rigorously determined. The kinetics data of CrataBL-heparin interaction could be used for explaining some of the CrataBL bioactivities.
Figure 3.
SPR sensorgrams of CrataBL-heparin interaction. Concentrations of CrataBL (from top to bottom): 250, 125, 63 and 32 nM, respectively. The black curves are the fitting curves using models from BIAevaluate 4.0.1.
Table 2.
Summary of kinetic data of CrataBL- heparin interactions*
| Interaction | ka(1/MS) | kd(1/S) | KD (M) |
|---|---|---|---|
| CrataBL/Heparin | 6.3 × 105 (±3.5 × 105) | 0.03 (±0.02) | 4.9 × 10−8 (±0.8 × 10−8) |
The data with (±) in parentheses are the standard deviations (SD) from triplicate binding experiments.
Solution competition study on the interaction between heparin (on surface) with protein to heparin-derived oligosaccharides (in solution) using SPR
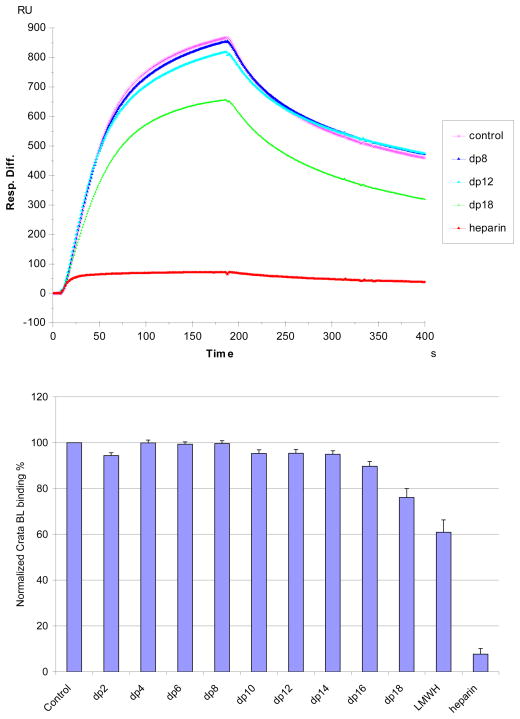
Solution/surface competition experiments were performed by SPR to examine the effect of saccharide chain size of heparin on the heparin-protein interaction. Different size heparin-derived oligosaccharides (from dp2 to dp18) were used in the competition study. The same concentration (1000 nM) of heparin-derived oligosaccharides were present in the CrataBL (125 nM) /heparin interaction solution. Only negligible competition was observed when 1000nM of heparin-derived oligosaccharides (from dp 2 to dp14) present in the CrataBL protein solution. In contrast, heparin-derived oligosaccharides dp 16 and dp18, LMWH (average dp ~20), full chain heparin (average dp ~40), showed competition with immobilized heparin that increased with increasing oligosaccharide size (Figure 4). These results suggest that the interaction between CrataBL and heparin is chain-length dependent and CrataBL requiring a relatively long heparin chain (>dp16) to interact. While chain length requirements for heparin protein interactions were reported for binding to other proteins, including: Shh-heparin, FGF-heparin and IL-7-heparin interactions,27–29 this chain size requirement is larger than those observed previously. The large minimum heparin chain length requirement for CrataBL binding appears to be critical for accommodation of the protein as shorter saccharide chains may not suffice in neutralizing the positive charged heparin binding regions of the protein.
Figure 4.
Top: Sensorgrams of solution heparin oligosaccharides/surface heparin competition. CrataBL concentration was 125 nM, and concentrations of heparin oligosaccharides in solution were 1000 nM). Bottom: Bar graphs (based on triplicate experiments with standard deviation) of normalized CrataBL binding preference to surface heparin by competing with different size of heparin oligosaccharides in solution.
SPR solution competition study of different GAGs
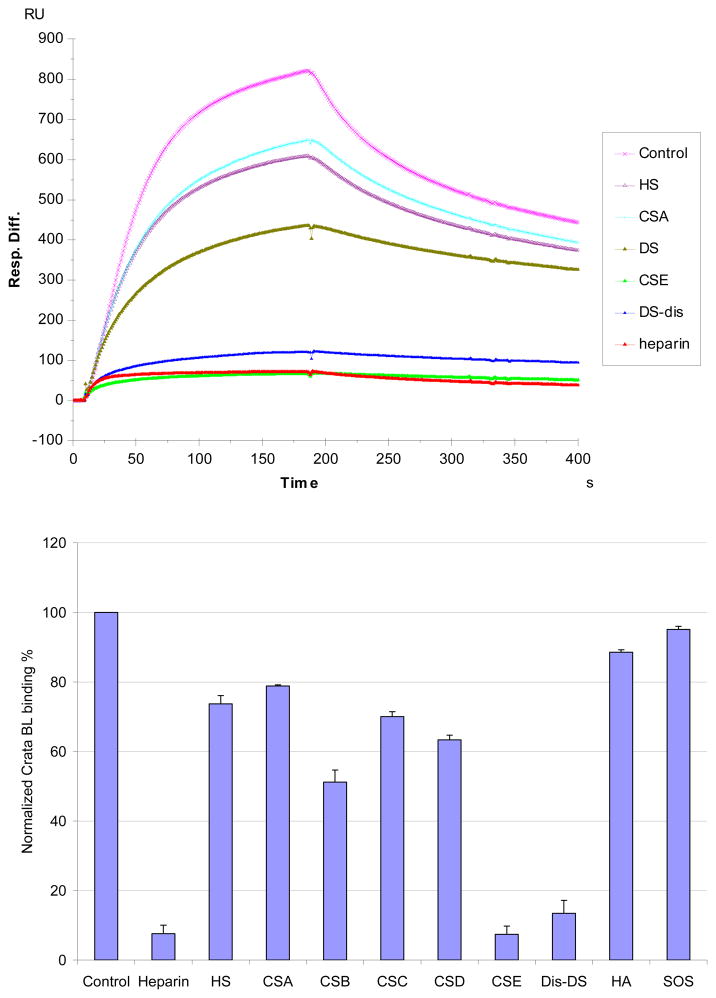
The SPR competition assay was also utilized to determine the binding preference of CrataBL to various GAGs (Figure 2). SPR competition sensorgrams and bar graphs of the GAG competition levels are displayed in Figure 5. Heparin, CSE and DiS-DS produced the strongest inhibition in CrataBL binding of immobilized heparin by competing >80% of the CrataBL binding to immobilized heparin on the chip surface. Modest inhibitory activities were observed for HS, CSA, DS, CSC and CSD. Weak inhibitory activities were observed for hyaluronan (HA), and sucrose octasulfate (SOS, a persulfated disaccharide). The data suggests that the binding interactions of CrataBL to GAGs appear structure-dependent and highly influenced by the level of GAG sulfation.
Figure 5.
Top: Sensorgrams of solution GAGs/surface heparin competition. CrataBL concentration was 125 nM, and concentrations of GAGsin solution were 1000 nM). Bottom: Bar graphs (based on triplicate experiments with standard deviation) of CrataBL binding RU to surface heparin by competing with different GAGs.
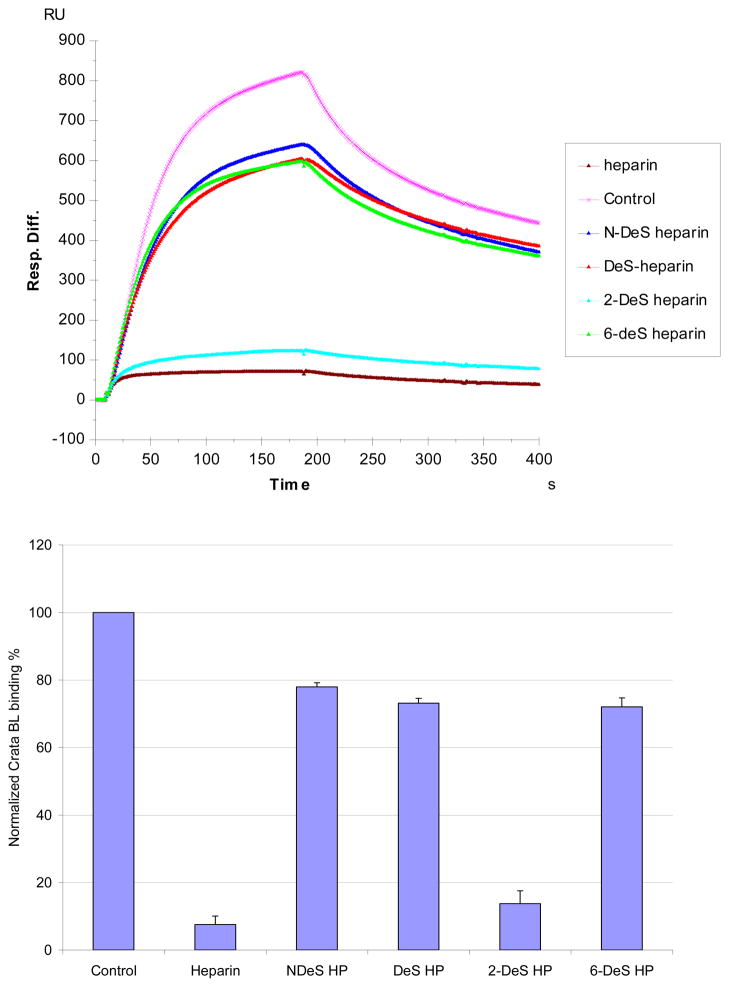
SPR solution competition study of different chemically modified heparins
SPR competition experiments using chemically modified heparins competition are displayed in Figure 6. The results show that all the four chemically modified heparins (fully desulfated heparin, N-desulfated heparin, 2-O-desulfated heparin and 6-O-desulfated heparin) showed reduced inhibitory activities. Removal of N-sulfo groups and 6-O-sulfo groups from heparin markedly reduced the ability of these chemically modified heparins to compete with surface immobilized heparin for CrataBL binding. In contrast, removal of 2-O-sulfo groups afforded a chemically modified heparin that competed well with surface immobilized heparin for CrataBL binding.
Figure 6.
Top: Sensorgrams of solution chemical modified heparin/surface heparin competition. CrataBL concentration was 125 nM, and concentrations of chemical modified heparin in solution were 1000nM). Bottom: Bar graphs (based on triplicate experiments with standard deviation) of CrataBL binding RU to surface heparin by competing with different chemical modified heparin in solution.
SPR competition experiments with different GAGs and chemically modified heparin clearly showed an inhibitory activity that was dependent on the level of GAG sulfation and fine structure. The inhibitory effects of soluble GAGs to the CrataBL/immobilized heparin interaction was greatest for heparin with 2.8 moles of sulfate per disaccharide repeating unit, followed by CS-E and Dis-DS with 1.5–2 moles of sulfate per disaccharide, then HS with 1.2 moles of sulfate per disaccharide, and finally by DS, CS-A and CS-C with < 1 mole sulfate per disaccharide. Surprisingly, CS-D with 1.5–2 mole per sulfate per disaccharide showed a comparable level of competition with GAGs having < 1 mole sulfate per disaccharide. Combining the structural requirements in the heparin and chondroitin/dermatan series of the GADs, suggests a CrataBL binding motif having eight repeating disaccharide units having two sulfo groups on a hexosamine residue linked to an uronic acid having a carboxyl group (Figure 7). These results suggest that GAG fine structure plays a prominent role in the CrataBL interaction. Two charge groups per disaccharide repeating unit (N-sulfo and6-O-sulfo groups) on heparin, or the 4-O-sulfo and 6-O-sulfo on CS-E or on Dis-DS appear to be critical for the CrataBL- GAG interaction. These results are similar to the GAG sulfation preference of other proteins, such as Shh, Ihog, FGF1, and FGF2, by interacting with more highly sulfated heparin than the less sulfated HS.27,28
Figure 7.
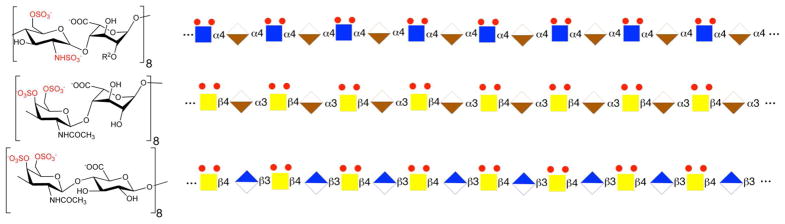
Proposed CrataBL binding GAG motif: eight repeating disaccharide units having two sulfo groups on a hexosamine residue linked to an uronic acid having a carboxyl group. Top: heparin dp 16 Middle: Dis-DS dp 16; Bottom: CS-E dp 16. Sugar symbols: Blue square, GlcNAc; Tan lower segment diamond, IdoA; Yellow square, GalNAc; Blue upper segment diamond, GlcA.
In conclusion, SPR analysis shows that CrataBL is a heparin-binding lectin with high affinity (KD~ 49 nM). The SPR solution competition study shows that CrataBL binding to heparin is chain length dependent. CrataBL prefers to bind full chain heparin or large oligosaccharides, having 16 to 18 sugars. Higher sulfation levels of GAGs enhance their binding affinities, i.e., CSE and DiS-DS showed comparable binding affinity to CrataBL as heparin, and low sulfated GAGs(HS, CSA, DS, CSC and CSD) having modest binding affinity. N-sulfo and 6-O-sulfo groups on heparin are required for the CrataBL- heparin interaction.
Supplementary Material
Acknowledgments
Funding
This work was supported in part by grants from the National Institutes of Health: GM-38060 to R.J.L, in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and in part by grant Molecular and Cell Biology (Presidium RAS) to N.V.B. We are also grateful to CAPES, CNPq and FAPESP (Process 09/53766-5) for providing financial support. The authors would like to acknowledge The Consortium for Functional Glycomics funded by the NIGMS GM62116 and GM98791 for services provided by the Glycan Array Synthesis Core (The Scripps Research Institute, LaJolla, CA) that produced the mammalian glycan microarray and the Protein-Glycan Interaction Core (Emory University School of Medicine, Atlanta, GA) that assisted with analysis of samples on the array.
ABBREVIATIONS
- SPR
surface plasmon resonance
- GAG
glycosaminoglycan
- HS
heparan sulfate
- HP
heparin
- LMWH
low molecular weight heparin
- CS-A
chondroitin sulfate A
- DS
dermatan sulfate
- Dis-DS
dermatan disulfate
- CS-C
chondroitin sulfate C
- CS-D
chondroitin sulfate D
- CS-E
chondroitin sulfate E
- HA
hyaluronic acid
- SA
streptavidin
- FC
flow cell
- RU
resonance unit
Footnotes
The authors declare no competing financial interest.
Additional experimental data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology. 2. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 2.Correia MTS, Coelho LCBB, Paiva PMG. Lectins, carbohydrate recognition molecules: are they toxic? In: Siddique YH, editor. Recent Trends in Toxicology. Vol. 37. Transworld Research Network; Kerala: 2008. pp. 47–59. [Google Scholar]
- 3.Li YR, Liu QH, Wang HX, Ng TB. A novel lectin with potent antitumor, mitogenic and HIV-1 reverse transcriptase inhibitory activities from the edible mushroom Pleurotus citrinopileatus. Biochim Biophys Acta: Gen Subjects. 2008;1780:51–57. doi: 10.1016/j.bbagen.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Capila I, Linhardt RJ. Heparin-protein interactions. AngewandteChemie-Intern Ed. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 5.Hacker U, Nybakken K, Perrimon N. Heparansulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Bio. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 6.Parish CR. The role of heparansulphate in inflammation. Nat Rev Immunol. 2006;6:633–643. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 7.Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 8.Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure-function relationships of glycosaminoglycans. Ann Rev Biomed Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 9.Hileman RE, Jennings RN, Linhardt RJ. Thermodynamic analysis of heparin interaction with a basic cyclic protein using isothermal titration calorimetry. Biochemistry. 1998;37:15231–15237. doi: 10.1021/bi980212x. [DOI] [PubMed] [Google Scholar]
- 10.Hileman RE, Fromm JR, Weiler JM, Linhardt RJ. Glycosaminoglycan-protein interaction: definition of consensus sites in glycosaminoglycan binding proteins. BioEssays. 1998;20:156–167. doi: 10.1002/(SICI)1521-1878(199802)20:2<156::AID-BIES8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Nascimento CO, Costa RMPB, Araújo RMS, Chaves MEC, Coelho LCBB, Paiva PMG, Teixeira JA, Correia MTS, Carneiro-da-Cunha MG. Optimized extraction of a lectin from Crataeva tapia bark using AOT in isooctane reversed micelles. Process Biochem. 2008;43:779–782. [Google Scholar]
- 12.Araújo RMS, Ferreira RS, Napoleão TH, Carneiro-da-Cunha MG, Coelho LCBB, Correia MTS, Oliva MLV, Paiva PMG. Crataeva tapia bark lectin is an affinity adsorbent and insecticidal agent. Plant Sci. 2012;183:20–26. doi: 10.1016/j.plantsci.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Araújo RMS, Vaz AFM, Aguiar JS, Coelho LCBB, Paiva PMG, Melo AMM, Silva TG, Correia MTS. Lectin from Crataeva tapia bark exerts antitumor, anti-inflammatory and analgesic activities. Nat Prod Bioprospect. 2011;1:97–100. [Google Scholar]
- 14.Araújo RMS, Vaz AFM, Santos ME, Zingali RB, Coelho LCBB, Paiva PMG, Oliva MLV, Ferreira RS, Correia MTS. A new exogen anticoagulant with high selectivity to intrinsic pathway of coagulation. Thrombosis Res. 2011;128:395–397. doi: 10.1016/j.thromres.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heimburg-Molinaro J, Song X, Smith DF, Cummings RD. Preparation and analysis of glycan microarrays. Curr Protoc Protein Sci. 2011;Chapter 12(Unit 12) doi: 10.1002/0471140864.ps1210s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pazynina GV, Severov VV, Maisel ML, Belyanchikov IM, Bovin NV. Synthesis of mono-, di- and tri-O-sulfated N-acetyllactosamines in a form suitable for glycochip printing. Mendeleev Commun. 2008;18:238–240. [Google Scholar]
- 18.Pazynina GV, Sablina MA, Nasonov VV, Pustovalova YE, Belyanchikov IM, Bovin NV. Synthesis of GalNAcβ1-4GlcNAcβ (LacdiNAc) O-sulfates. Mendeleev Commun. 2010;20:316–317. [Google Scholar]
- 19.Brister SJ, Buchanan MR, Griffin CC, Van Gorp CL, Linhardt RJ. Dermatandisulfate, an inhibitor of thrombin and complement activation. #5. US Patent. 1999;922:690.
- 20.Yates EA, Santini F, Guerrini M, Naggi A, Torri G, Casu B. 1H and 13C NMR spectral assignments of the major sequences of twelve systematically modified heparin derivatives. Carbohydrate Res. 1996;294:15–27. doi: 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]
- 21.Pervin A, Gallo C, Jandik KA, Han XJ, Linhardt RJ. Preparation and structural characterization of large heparin-derived oligosaccharides. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 22.Hernaiz M, Liu J, Rosenberg RD, Linhardt RJ. Enzymatic modification of heparan sulfate on a biochip promotes its interaction with antithrombin III. Biochem Biophys Res Commun. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 23.Kohnke-Godt B, Gabius HJ. Heparin-binding lectin from human placenta: further characterization of ligand binding and structural properties and its relationship to histones and heparin-binding growth factors. Biochemistry. 1991;30:55–65. doi: 10.1021/bi00215a009. [DOI] [PubMed] [Google Scholar]
- 24.Dam TK, Bandyopadhyay P, Sarkar M, Ghosal J, Bhattacharya A, Choudhury A. Purification and partial characterization of a heparin-binding lectin from the marine clam Anadaragranosa. Biochem Biophys Res Commun. 1994;203:36–45. doi: 10.1006/bbrc.1994.2145. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa H, Yamaguchi C, Sakai H, Kanemaru K, Hayashi H, Araki Y, Tomihara Y, Shinohara M, Ohura K, Kitagawa H. Biochemical and physiological properties of pedicellariallectins from the toxopneustid sea urchins. J Nat Toxins. 1999;8:297–308. [PubMed] [Google Scholar]
- 26.Kamo I, Furukawa S, Akazawa S, Fujisawa K, Tada-Kikuchi A, Nonaka I, Satoyoshi E. Mitogenic heparin-binding lectin-like protein from cloned thymicmyoid cells. Cell Immunol. 1986;103:183–190. doi: 10.1016/0008-8749(86)90079-1. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F, McLellan JS, Ayala AM, Leahy DJ, Linhardt RJ. Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry. 2007;46:3933–3941. doi: 10.1021/bi6025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang F, Zhang Z, Lin X, Beenken A, Eliseenkova AV, Mohammadi M, Linhardt RJ. Compositional analysis of heparin/heparan sulfate interacting with fibroblast growth factor. fibroblast growth factor receptor complexes. Biochemistry. 2009;48:8379–8386. doi: 10.1021/bi9006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang F, Liang X, Pu D, Walsh S, Linhardt RJ. Biophysical characterization of glycosaminoglycan-IL-7 interactions using SPR. Biochimie. 2012;94:242–249. doi: 10.1016/j.biochi.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.