Abstract
Brown adipose tissue (BAT) dissipates chemical energy in the form of heat as a defense against hypothermia and obesity. Current evidence indicates that brown adipocytes arise from Myf5+ dermotomal precursors through the action of PRDM16 (PR domain containing protein16) transcriptional complex 1,2. However, the enzymatic component of the molecular switch that determines lineage specification of brown adipocytes remains unknown. Here we show that EHMT1 (euchromatic histone-lysine N-methyltransferase 1) is an essential BAT-enriched lysine methyltransferase in the PRDM16 transcriptional complex and controls brown adipose cell fate. Loss of EHMT1 in brown adipocytes causes a severe loss of brown fat characteristics and induces muscle differentiation in vivo through demethylation of histone 3 Lys 9 (H3K9me2 and 3) of the muscle-selective gene promoters. Conversely, EHMT1 expression positively regulates the BAT-selective thermogenic program by stabilizing the PRDM16 protein. Notably, adipose-specific deletion of EHMT1 leads to a marked reduction of BAT-mediated adaptive thermogenesis, obesity, and systemic insulin resistance. These data indicate that EHMT1 is an essential enzymatic switch that controls brown adipose cell fate and energy homeostasis.
Obesity develops when energy intake chronically exceeds total energy expenditure. All anti-obesity medications currently approved by the FDA act to repress energy intake, either by suppressing appetite or by inhibiting intestinal fat absorption. However, due to their side effects including depression, oily bowel movements, and steatorrhea, there is an urgent need for alternative approaches. BAT is specialized to dissipate energy via uncoupling protein 1 (UCP1). Recent studies with 18Fluoro-labeled 2-deoxy-glucose positron emission tomography (18FDG-PET) scanning demonstrated that adult humans have active BAT deposits3–6 and that its amount inversely correlate with adiposity and body mass index (BMI)4,5, indicating that BAT plays an important role in energy homeostasis in adult humans. Hence, a better understanding of the molecular control of BAT development may lead to an alternative approach to alter energy balance by increasing energy expenditure.
It has been reported that brown adipocytes in the interscapular and perirenal BAT arise from Engrailed-1+ and Myf5+ dermotomal precursors1,7,8. The PRDM16-C/EBP-β complex in the myogenic precursors activates the brown adipogenic gene program through inducing PPARγ expression1,2,9; however, the mechanism by which the PRDM16-C/EBP-β complex functions as a fate switch to control brown adipocyte versus myocyte lineage remains unexplored.
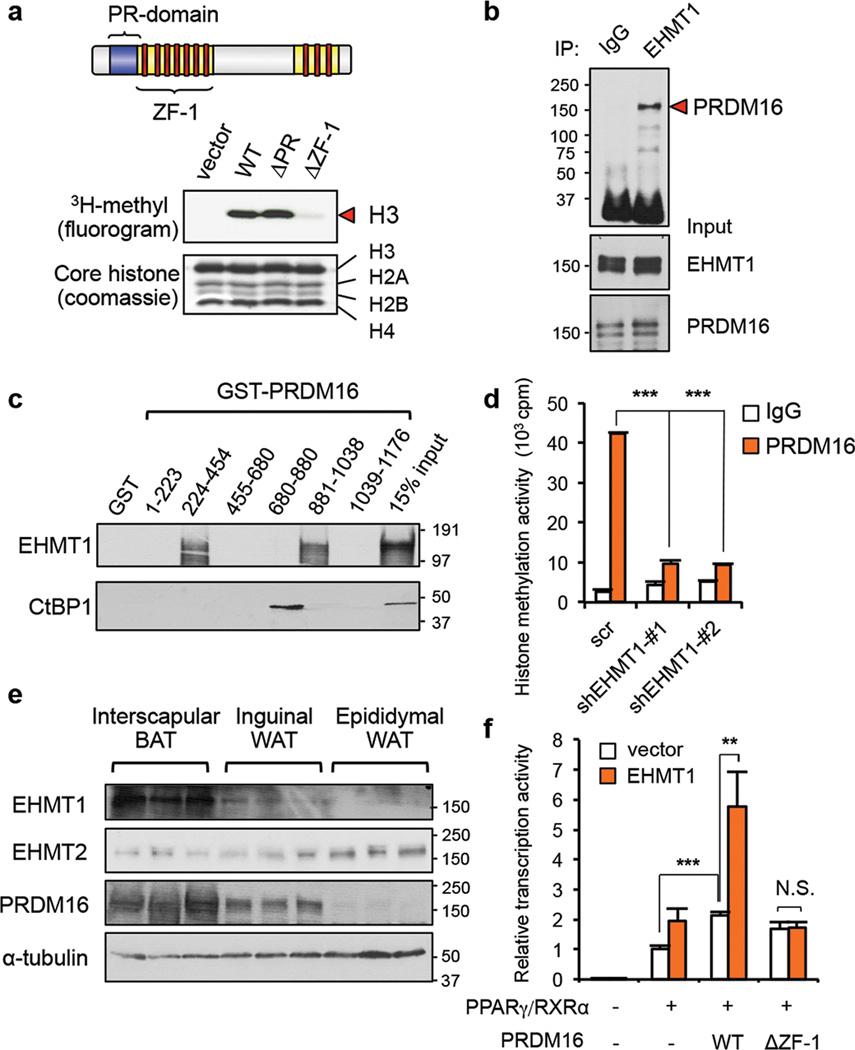
Previously we determined the essential domains of PRDM16 for converting myoblasts into brown adipocytes by generating two deletion mutants of PRDM16: a mutant lacking the PR-domain (∆PR), a domain which shares high homology with methyltransferase SET domains10,11, and a mutant lacking the zinc finger domain-1 (∆ZF-1) (Fig. 1a, upper panel). Wild-type (WT) and the ∆PR mutant, but not the ∆ZF-1 mutant, were able to convert myoblasts into brown adipocytes, suggesting that the ZF-1 domain is required2. Consistent with the results, the PRDM16 complex purified from brown adipocytes expressing wild-type and ∆PR, but not ∆ZF-1, had significant methyltransferase activities on H3 (Fig.1a, bottom panel). Since this effect was independent of its SET domain, we searched for methyltransferases that were associated with differentiation-competent PRDM16 proteins (i.e., WT and ∆PR), but not with differentiation-incompetent PRDM16 (∆ZF-1). By employing high-resolution liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS), we found EHMT1 as the only methyltransferase that was co-purified preferentially with the differentiation-competent PRDM16 complexes2. EHMT1 has enzymatic activity on H3K9 mono or di-Me12. Notably, haploinsufficiency of the EHMT1 gene, due to 9q34.3 microdeletions or point mutations in humans13 is associated with clinical phenotypes including mental retardation. Importantly, 40–50% of the patients with EHMT1 mutations develop obesity14,15; however, the underlying mechanism remains completely unknown. Given the essential role of the PRDM16 complex for BAT development, we hypothesized that EHMT1 is a key enzymatic component that controls the lineage specification and thermogenic function of BAT.
Figure 1. Identification of EHMT1 in the PRDM16 transcriptional complex.
a, Up: schematic illustration of PRDM16. Bottom: PRDM16 complex purified from brown adipocytes were subjected to in vitro histone methylation assay. b, Immunoprecipitation of EHMT1 protein followed by Western blotting to detect PRDM16. Input was shown in lower panels. c, In vitro binding assay of 35S-labeled EHMT1 or CtBP1 and purified PRDM16 fragments. d, Histone methylation assay of PRDM16 complex from brown adipocytes expressing indicated constructs. n=3–4. e, Western blotting for indicated proteins in adipose tissues. f, Transcriptional activities of PRDM16 using a PPAR-γ responsive luciferase reporter. n=3. Error bars are s.e.m.; ** P <0.01, *** P <0.001.
To test this hypothesis, we first confirmed the PRDM16-EHMT1 interaction by immunoprecipitation followed by Western blotting in brown adipocytes (Fig.1b and Supplementary Fig.1). The purified ZF-1 (224–454) and ZF-2 (881–1038) domains of GST-PRDM16 protein bound to the in vitro -translated EHMT1 protein, while the 680–1038 region of PRDM16 bound to CtBP1 as previously reported16 (Fig.1c and Supplementary Fig.2). These results indicate that EHMT1 directly interacts with PRDM16. EHMT1 is the major methyltransferase of the PRDM16 complex in brown adipocytes, because the histone methyltransferase activity of the PRDM16 complex was largely lost when EHMT1 was depleted using two short hairpin RNAs targeted to EHMT1 (Fig.1d and Supplementary Fig.3). Furthermore, expression of EHMT1 protein was highly enriched in BAT and in cultured brown adipocytes, correlating well with PRDM16 (Fig.1e and Supplementary Fig.4). In contrast, EHMT2 protein levels were higher in WAT than in BAT. To test if EHMT1 modulates the PRDM16 transcriptional activity, we performed luciferase assays using a luciferase reporter gene containing PPAR-γ binding sites1. As shown in Fig.1f, co-expression of EHMT1 and PRDM16 synergistically increased the reporter gene activity, whereas this induction was completely lost when the ∆ZF-1 mutant was expressed. These data indicate that EHMT1 forms a transcriptional complex with PRDM16 and regulates its activity through direct interaction.
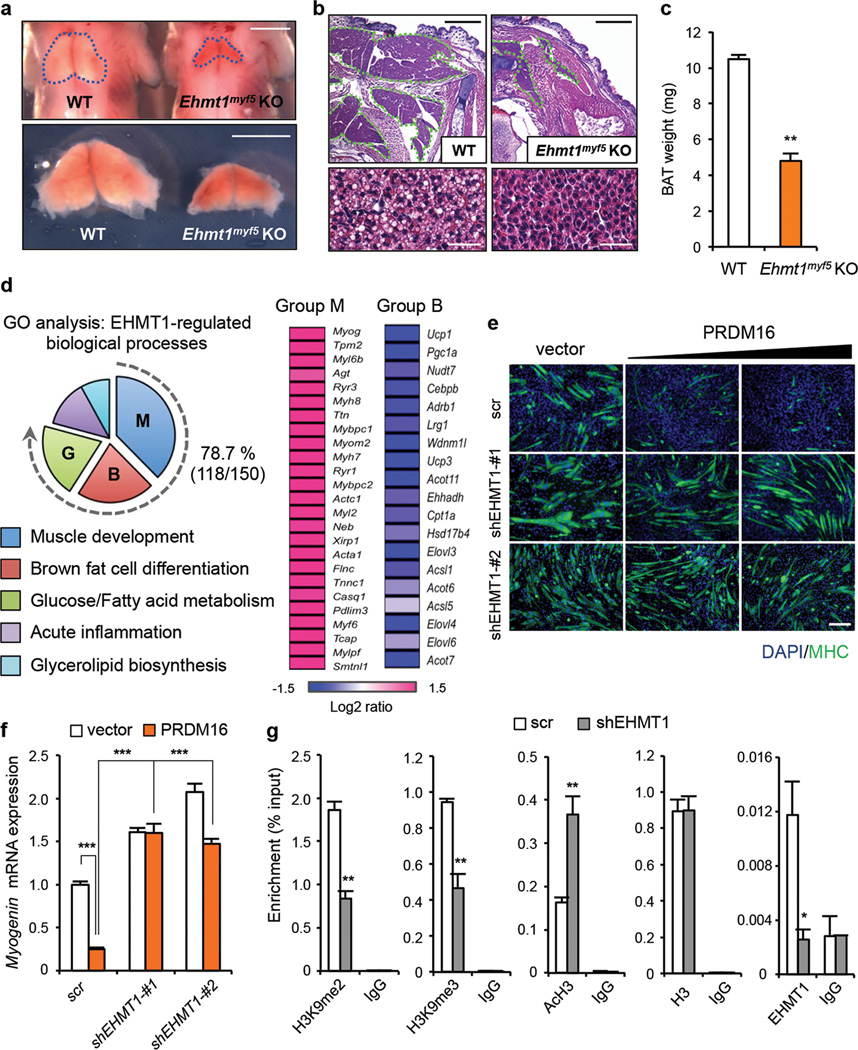
Next, we investigated the genetic requirement for EHMT1 in BAT development in vivo. Since a whole-body knock out of the Ehmt1 gene causes embryonic lethality before the emergence of brown adipocytes12, the Ehmt1 gene was deleted in brown adipocyte precursors by crossing Ehmt1flox/flox mice17 to Myf5-Cre mice1. As shown in Fig. 2a–c, the interscapular BAT of Ehmt1myf5 KO mice was substantially smaller than in wild-type mice at postnatal stage (P)1. Haematoxylin and eosin (H&E) staining showed that brown adipocytes in Ehmt1myf5 KO mice were significantly smaller and contained less lipids than in wild-type mice, whereas other tissues near the BAT including skin appeared normal (Fig. 2b and Supplementary Fig.5). Similar results were observed in embryos at E18.5 (Supplementary Fig.6). Subsequently, we analyzed the global gene expression of BAT from the wild-type and Ehmt1myf5 KO embryos by RNA-sequencing. The following gene ontology (GO) analysis found that the gene expression pattern in the Ehmt1myf5 KO BAT exhibited a skeletal-muscle phenotype; i.e., a broad activation of the skeletal muscle-selective genes, and a broad reduction of the BAT-selective genes. Strikingly, 78.7 % of the differentially expressed genes (118/150 genes) between wild-type and KO mice were stratified into categories of skeletal muscle development, BAT development, and BAT function (glucose/fatty acid metabolism). Specifically, 77.5% of the ectopically activated genes in the KO BAT were related to skeletal muscle development, including Myogenin and Myosin heavy chains. On the other hand, 80.0 % of the reduced genes in the KO BAT were involved in BAT development and fatty acid/glucose metabolism, including Ucp1, Pgc1a, Cebpb, Cpt1a, and Elovl3 (Fig. 2d and Supplementary Fig.7). These results indicate that EHMT1 is absolutely required for the cell fate specification between BAT-versus -muscle.
Figure 2. EHMT1 is required for BAT-versus-muscle lineage specification.
a, Morphology of BAT from wild-type and Ehmt1 myf5 KO embryos at P1. Scale bar, 2.5 mm. b, H&E staining of wild-type and Ehmt1 myf5 KO BAT. Scale bar, 600 µm. Bottom: high magnification images. Scale bar, 30 µm. c, BAT weight from wild-type (n=14) and KO embryos (n=8). d, GO analyses of RNA-sequencing data. The Log2-fold changes in the expression of skeletal muscle (group M) and BAT (group B) genes are shown. e, Immunocytochemistry for MHC in C2C12 cells expressing indicated constructs under pro-myogenic culture conditions. Scale bar, 200 µm. f, Myogenin mRNA expression in e. n=3. g, ChIP assays using indicated antibodies. n=3. Error bars are s.e.m.; * P < 0.05, ** P <0.01, *** P <0.001.
To investigate the mechanisms by which EHMT1 determines BAT lineage, retroviruses expressing a scrambled control RNA (scr) or shRNAs targeting EHMT1 (shEHMT1-#1 and -#2) were transduced into C2C12 myoblasts together with PRDM16 (Supplementary Fig.8a). As shown in Fig.2e (upper panels), PRDM16 expression powerfully blocked myogenic differentiation in a dose-dependent fashion, as shown by immunohistochemistry using a pan-skeletal myosin heavy chain (MHC) antibody. In contrast, EHMT1 depletion significantly impaired the PRDM16-mediated repression on myogenesis (Fig.2e, lower panels). Gene expression analysis showed that the repression on muscle-selective genes such as Myogenin was near completely abolished when EHMT1 was depleted (Fig.2f and Supplementary Fig.8b). This repressive effects was mediated through the EHMT1’s methyltransferase activity, because ectopic expression of the EHMT1 mutant (N1198L;H1199E) that lacks methyltransferase activity18 significantly blunted the PRDM16-mediated repression on myogenesis (Supplementary Fig.9a). Additionally, two chemical inhibitors of EHMT1/2, BIX-01294 and UNC0638, blocked the repressive effects of PRDM16 (Supplementary Fig.9b–c). BIX-01294 treatment in brown adipocytes also significantly reduced the expression of BAT-selective genes (Supplementary Fig.9d). Consistent with these data, ChIP assays found that EHMT1 depletion robustly reduced levels of H3K9me2 and me3 at the proximal region of the Myogenin gene promoter on which EHMT1 was recruited (Fig.2g). On the contrary, the H3K9/14ac levels were significantly increased by EHMT1 depletion without any effect on total H3 levels. Similar changes were observed at the promoter regions of other muscle-selective genes including Acts, Ryr1 , and Mylpf1, where EHMT1 was recruited (Supplementary Fig. 10). Conversely, under pro-adipogenic culture conditions, knockdown of EHMT1 largely blocked the PRDM16-induced brown adipogenesis in C2C12 cells (Supplementary Fig.11). All together, these results indicate that EHMT1 determines BAT-versu s-muscle cell lineage via PRDM16 by controlling H3K9 methylation status of the muscle-selective gene promoters.
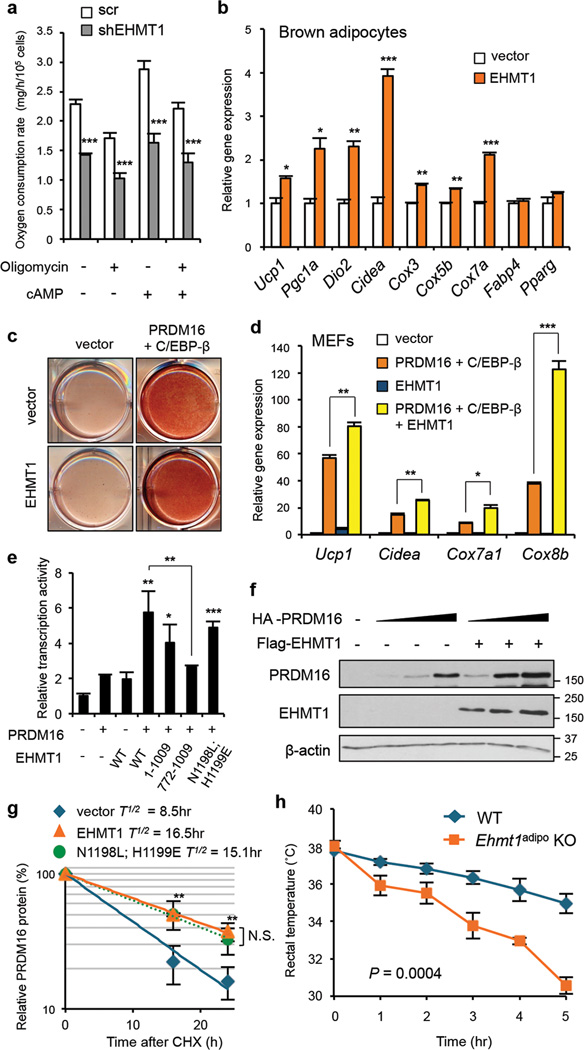
To investigate the role of EHMT1 in BAT thermogenesis, EHMT1 was depleted in immortalized brown adipocytes by retrovirus-mediated shRNA knockdown (Supplementary Fig.12a–b). Total and uncoupled (oligomycin-insensitive) O2 consumption rate in the EHMT1-depleted brown adipocytes was significantly reduced both at the basal and cAMP-stimulated states (Fig.3a). Conversely, EHMT1 overexpression significantly increased mRNA levels of BAT-selective thermogenic genes, including Ucp1, Pgc1a , and Dio2 (Fig.3b), and O2 consumption rate (Supplementary Fig.12c). To further test if this EHMT1 action requires PRDM16, EHMT1 was ectopically introduced into mouse embryonic fibroblasts (MEFs) that did not express endogenous PRDM16. As shown in Fig.3c, MEFs expressing PRDM16 and C/EBP-β uniformly differentiated into lipid-containing adipocytes as previously reported2. While EHMT1 alone did not stimulate brown adipogenesis, combination of EHMT1 with PRDM16 and C/EBP-β synergistically increased mRNA levels of the BAT-selective genes, including Ucp1, Cidea, Cox7a and Cox8b (Fig. 3d). These data indicate that EHMT1 positively regulates the BAT-selective thermogenic gene program through PRDM16.
Figure 3. EHMT1 controls BAT thermogenesis through stabilizing PRDM16 protein.
a, Cellular respiration in brown adipocytes expressing indicated constructs. n=6. b, BAT-selective gene expression in brown adipocytes expressing indicated constructs. n=3. c, Oil-Red-O staining of MEFs expressing indicated constructs under pro-adipogenic culture conditions. d, BAT-selective gene expression in c. n=3. e, Effects of EHMT1 mutants on PRDM16 transcriptional activites. n=3. f, PRDM16 protein levels in COS7 cells expressing indicated constructs. g, Regression analysis of the PRDM16 protein stability. n=3. h, Changes in rectal temperature during a cold challenge. n=4–5. Error bars are s.e.m.; * P < 0.05, ** P <0.01, *** P <0.001.
To determine which domains of EHMT1 are required for the induction of PRDM16 transcriptional activity we tested a series of EHMT1 mutants for their ability to interact and co-localise with PRDM16 (Supplementary Fig.13 and 14). A deletion mutant of EHMT1 (772–1009), which failed to interact with PRDM16 was unable to increase PRDM16 reporter gene activity (Fig.3e). EHMT1 with mutations in the SET domain (N1198L; H1199E) had no methyltransferase activity, but was still able to bind to and activate PRDM16. Even a mutant that lacked the SET-domain all together (1–1009) interacted with and activated PRDM16. Thus an interaction between EHMT1 and PRDM16 seems required to activate PRDM16 transcriptional activity. Notably, expression of EHMT1 robustly increased PRDM16 protein level, independent of its mRNA expression (Fig.3f and Supplementary Fig.15). This effect was due to changes in the rate of protein degradation, because cycloheximide chase experiments showed that expression of EHMT1 extended the half-life of PRDM16 protein from 8.5 hours to 16.5 hours. The N1198L;H1199E mutant also extended the half-life of PRDM16 protein as potently as the wild-type form (Fig.3g). PRDM16 protein accumulation was induced only by the EHMT1 mutants that bind to PRDM16 (Supplementary Fig.16). Importantly, EHMT1 regulates endogenous PRDM16 protein levels in vivo (Extended Data1). EHMT1 protein stability was not affected by PRDM16 (Supplementary Fig.17). These results collectively suggest that EHMT1 has dual functions, i.e., repressive effects on the muscle-selective gene program through its methyltransferase activity, and activation of the BAT-selective gene program through stabilization of PRDM16 protein via direct association.
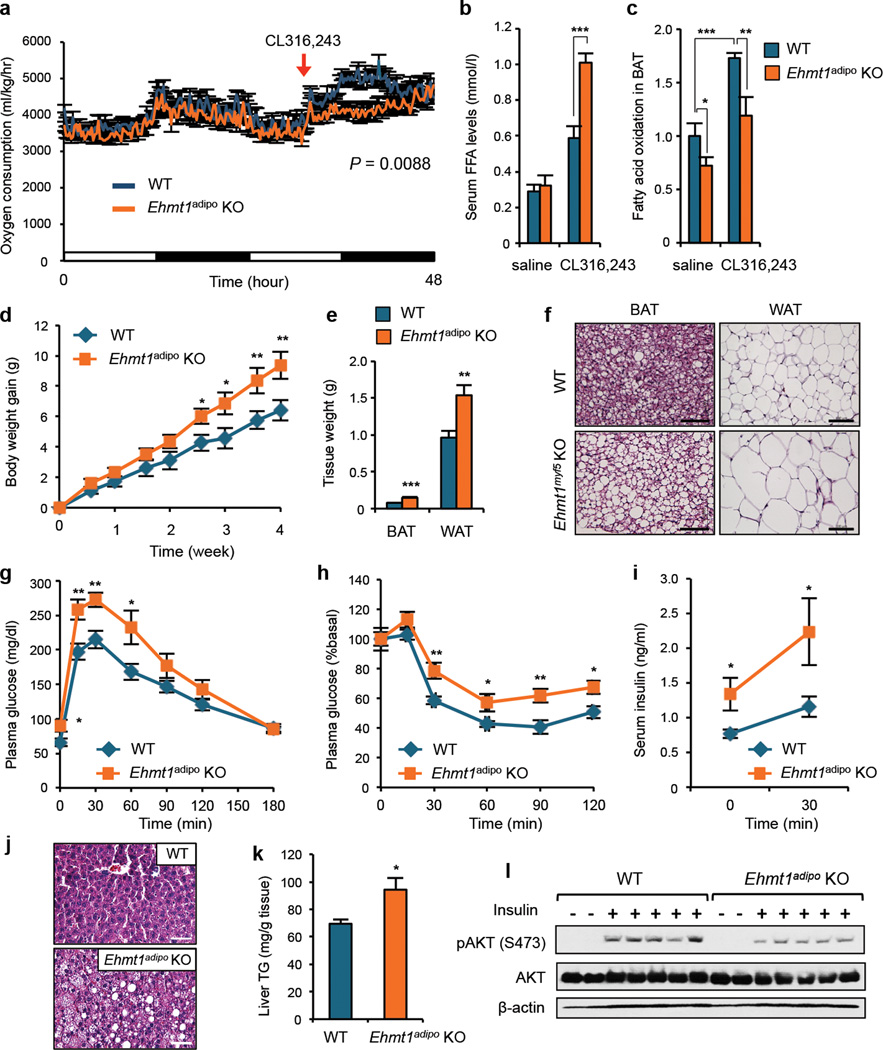
Next, we examined the requirement for EHMT1 in adaptive thermogenesis in vivo. To exclude potential defects in the skeletal muscle of Ehmt1myf5 KO mice, we generated adipose tissue-specific Ehmt1 knockout mice (Ehmt1adipo KO) using Adiponectin-Cre mice19. Of note, 62.3% of the differentially expressed muscle/BAT-selective genes in the Ehmt1myf5 mice were similarly dysregulated in the Ehmt1adipo KO mice (Extended Data2). While Adiponectin-Cre is expressed both in BAT and WAT, expression of EHMT1 is highly enriched in BAT as compared to WAT (Fig.1e). Furthermore, lipolysis capacity in the WAT of Ehmt1adipo KO mice was indistinguishable from wild-type mice (Supplementary Fig. 18). Importantly, EHMT1 is required for beige/brite cell development (Extended Data3). Hence, the Ehmt1adipo KO mice allow us to examine the role of EHMT1 in BAT/beige fat-mediated thermogenesis in vivo. As shown in Fig.3h, rectal temperature of Ehmt1adipo KO mice strikingly dropped within one hour after a cold challenge to 4°C, whereas control mice remained a constant level. Expression of BAT-selective genes in skeletal muscle20 was not altered in Ehmt1adipo KO mice (Supplementary Fig. 19). We subsequently measured oxygen consumption rate (VO2) at thermoneutrality (29–30°C)21 in response to an activation of the β3-adrenoceptor pathway. As shown in Fig.4a, VO2 of wild-type mice was significantly increased after administering CL316,243 whereas this induction was completely lost in KO mice. The impaired thermogenesis in KO mice was accompanied by higher serum levels of free fatty acids (FFAs) (Fig.4b). This is consistent with previous findings that BAT serves as a major sink of FFAs for heat generation22, and that reduced FA oxidation in BAT lead to an increase in serum FFA levels23. Indeed, FA oxidation capacity in the KO BAT was significantly lower than in wild-type mice at the basal state and after administering CL316,243 (Fig. 4c). Additionally, FA uptake in the KO BAT was reduced (Supplementary Fig. 20). These results suggest that EHMT1 is absolutely required for BAT-mediated adaptive thermogenesis and FA metabolism in vivo.
Figure 4. EHMT1 deficiency in BAT causes obesity and insulin resistance.
a, VO2 of wild-type and Ehmt1 adipo KO mice treated with CL316,243 (0.5 mg/kg) at thermoneutrality. n=6. b, Serum FFA levels in mice treated with saline or CL316,243. c, FA oxidation in BAT. n=6–10. d, Body weight change under a high-fat diet at thermoneutrality. n=16. e, Adipose tissue weight after 4-week high-fat diet. n=16. f, H&E staining of adipose tissues. Scale bar, 100 µm. g, GTT in 9-week high-fat diet-fed mice. n=9. h, ITT in 10-week high-fat diet-fed mice. n=9. i, Serum insulin levels at the fasted and glucose-stimulated states. n=9. j, H&E staining of liver in d. Scale bar, 50 µm. k, Liver triglyceride levels in j. n=9. l, Hepatic insulin signaling as assessed by phosphorylated (S473) and total Akt levels. Error bars are s.e.m.; * P < 0.05, ** P <0.01, *** P <0.001.
Lastly, we tested if EHMT1 deficiency in BAT affects the propensity for weight gain in response to an obesogenic diet at thermoneutrality (29–30°C), since an obesity phenotype in UCP1 KO mice was observed only at thermoneutrality24. As shown in Fig.4d and Supplementary Fig.21, Ehmt1adipo KO mice gained significantly more body weight than wild-type mice without any change in food intake (Supplementary Table S1). KO mice had higher amounts of epididymal WAT and interscapular BAT that contained substantially larger lipid droplets than wild-type mice (Fig.4e–f). Glucose tolerance test (GTT) found that KO mice displayed significantly higher blood glucose concentrations than wild-type mice (Fig.4g). Similarly, KO mice exhibited impaired responses to insulin during insulin tolerance test (ITT) (Fig.4h) and higher serum levels of insulin at the fasted and glucose-stimulated states (Fig.4i). KO mice showed an insulin resistance phenotype even at ambient temperature, while no statistically significant difference was observed in body weight (Supplementary Fig. 22). Notably, the liver from KO mice contained higher amounts of lipids and triglyceride (Fig.4j and k) and showed impaired insulin signaling as assessed by phosphorylation of Akt in response to insulin (Fig.4l and Supplementary Fig. 23). All together, these results indicate that EHMT1 deficiency in BAT leads to obesity, systemic insulin resistance, and hepatic steatosis under a high-fat diet.
In conclusion, we identified EHMT1 as an essential BAT-enriched methyltransferase that controls brown adipose cell fate, adaptive thermogenesis, and glucose homeostasis in vivo. While presence of BAT in adult humans is now widely appreciated, no mutation that causes defects in human BAT development and thermogenesis had been described except polymorphisms in UCP1 and β3-adrenoceptor genes25. Delineating the causal link between EHMT1 mutations and BAT thermogenesis will provide a new perspective in understanding the molecular control of energy homeostasis through the epigenetic pathways, which may lead to effective therapeutic interventions for obesity and metabolic diseases.
METHODS
Animals
All animal experiments were performed according to procedures approved by UCSF's Institutional Animal Care and Use Committee for animal care and handling. Ehmt1flox mice and AdiponectinCre/+ mice were kindly provided by Dr. Tarakhovsky at Rockefeller University and by Dr. Rosen at Harvard Medical School, respectively. Myf5Cre/+ mice were obtained from the Jackson Laboratory26. To analyze embryonic BAT development, Ehmt1flox/flox or Myf5-Cre+/−; Ehmt1flox/flox embryos at embryonic stage E18.5 or newborn mice at P1 were harvested and fixed in 4% paraformaldehyde for histological analyses. The presumptive BAT depots in the interscapular region were micro-dissected for histological and RNA expression analyses. For cold exposure experiments, male Ehmt1adipo KO mice (Adipo-Cre+/−; Ehmt1flox/flox ) and body-weight-matched control mice (Ehmt1flox/flox ) at 10 weeks of age were single-caged and exposed to 4 °C for 5 hours. Rectal temperatures were monitored every hour using a TH-5 thermometer (Physitemp).
Metabolic studies
Whole body energy expenditure of Ehmt1adipo KO mice or body-weight-matched control mice at 14 weeks of age was measured at thermoneutrality (29–30 °C) using a Comprehensive Lab Animal Monitoring System (CLAMS, Columbus Instruments). The mice were injected intraperitoneally with a β3-adrenergic receptor-specific agonist CL316,243 at a dose of 0.5 mg/kg. For diet-induced obesity studies, male mice at 6–7 weeks of age were fed a high-fat diet (D12492, Research Diet Inc.) for 4 weeks at thermoneutrality. At the end of the experiments, serum samples were collected. Serum levels of insulin (Millipore), triglyceride (Thermo), and free fatty acid (Wako) were measured using commercially available kits. For GTT experiments, male mice were fed a high-fat diet for 9 weeks. After an overnight fast, the mice were injected intraperitoneally with glucose (1 g/kg). For ITT experiments, male mice under a high-fat diet for 10 weeks were used. After an overnight fast, the mice were injected intraperitoneally with insulin (0.75 U/kg). Blood samples were collected at indicated time points and glucose levels were measured using blood glucose test strips (Abbott). To measure liver triglyceride contents, the liver tissue (25 mg) was homogenized in 1.25ml of Folch solution (chloroform / methanol, 2:1, v/v). Subsequently, equal amounts (0.4ml) of chloroform and water were added to the lysate. After centrifugation at 3,000 rpm for 3min, the chloroform phase was harvested and dried. The pellet was dissolved in isopropanol. Triglycerides levels were determined by Infinity Triglycerides kit (Thermo).
Fatty acid oxidation assay
Wild-type and Ehmt1adipo KO mice at 11-weeks old were intraperitoneally injected with saline or CL316,243. Five hours after the injection, the interscapular BAT depots were isolated. Fatty acid oxidation assay was performed according to the protocol described by Mao et al.27 Briefly, the adipose tissues were minced to small pieces and incubated with DMEM supplemented with 1 mM pyruvate, 1% FFA-free BSA, and 0.5 mM oleate. 14C-Oleic acid at 1 µCi/µl was added for 2 hours at 37 °C. After adding 70% perchloric acid into each well, CO2 was captured by Whatman paper soaked in 3 M NaOH solution for 1 hour. 14C radioactivity was measured by liquid scintillation counter and normalized to tissue weight. To assess FA uptake, BAT (approximately 100 mg) were isolated from wild-type and Ehmt1adipo KO mice and incubated in DMEM containing oleic acid (250 µM, NU-CHEK PREP, INC.) supplemented with 14C-Oleic acid (0.25 µCi/ml) and 10% FBS for 15min. 14C radioactivity in the BAT explants was measured by liquid scintillation counter and normalized to the total protein content.
In vivo insulin stimulation assay
Mice were anesthetized with Tribromoethanol (Avertin®). Insulin (5U) was injected into the inferior venae cavae. Livers were removed 2 minutes after the injection and lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% (w/v) glycerol, 100 mM NaF, 10 mM EGTA, 1 mM Na3VO4, 1% (w/v) Triton X-100, 5 µM ZnCl2, 2 mM, with protease inhibitor cocktail (cOmplete®, Roche). The lysates were isolated and separated by SDS-page. Akt (Pan) antibody (Cell Signaling) and Phospho-Akt (Ser473) Antibody (Cell Signaling) were used for Western blotting.
Fat lipolysis assay
Epididymal fat pads were harvested and digested in a digestion buffer (121mM NaCl, 4.9mM KCL, 1.2mM MgSO4, 0.33mM CaCl2, 12mM HEPES) containing collagenese D (1.5U/ml), dipase II (2.4U/ml), 3mM glucose, and fatty acid free 1% BSA (Akron Biotech). After an hour digestion at 37 °C with gentle shaking, adipocytes were filtrated through nylon mesh and centrifuged at 200 rpm for 5minutes. Floating adipocytes were harvested and incubated in DMEM containing 10% FBS in the presence or absence of isoproterenol (1 µM) for 1.5 hour at 37 °C. Glycerol release into the media was determined using a free glycerol reagent (Sigma). Glycerol levels were normalized to the total protein content of the primary adipocytes using Pierce BCA Protein Assay reagent (Thermo Scientific).
Cell Culture
Immortalized brown fat cells were isolated from the interscapular BAT of wild-type mice at P1–P3. MEFs have been described previously2. HEK293 cells and C2C12 cells were obtained from ATCC. Adipocyte differentiation in C2C12 cells was induced by treating confluent cells with DMEM containing 10% FBS, 0.5 mM isobutylmethylxanthine, 125 µM indomethacin, 2 µg/ml dexamethasone, 850 nM insulin, 1 nM T3, and 0.5 µM rosiglitazone. Two days after induction, cells were switched to the maintenance medium containing 10% FBS, 850 nM insulin, 1 nM T3, and 0.5 µM rosiglitazone. For cAMP treatment, cells were incubated with 10 µM forskolin for 4 hours. Myocyte differentiation in C2C12 myoblasts was induced by treating cells in DMEM containing 2% horse serum. For beige cell differentiation in culture, the SV fraction was isolated from Ehmt1flox/flox mice and plated in collagen coated plates (BD Biosciences). Cells were differentiated in the absence or presence of rosiglitazone at 0.5 µM according to the previous paper28.
DNA constructs and viruses production
Deletion mutants of flag-tagged PRDM16 and GST-fused PRDM16 fragments (1–223, 224–454, 455–680, 680–880, 881–1038, and 1039–1176) were described previously16. EHMT1 expression constructs were kind gifts from Dr. Y. Shinkai (Riken institute)18 and from Dr. E. Hara (Japanese Foundation for cancer research)29. EHMT1 was cloned to pMSCV-puro vector for retroviral expression. The sequences used for retroviral shRNA expression vectors targeting EHMT1 were 5’-CGC TAT GAT GAT GAT GAA TAA-3’ (shEHMT1-#1) and 5’-GAG GAT AGT AGG ACT TCT AAA-3’ (shEHMT1-#2). The corresponding double-stranded DNA sequences were ligated into pSUPER-Retro (GFP-Neo) (Oligoengine) for retroviral expression. For retrovirus production, Phoenix packaging cells were transfected at 70% confluence by calcium phosphate method with 10 µg retroviral vectors. After 48 hours, the viral supernatant was harvested and filtered. Cells were incubated overnight with the viral supernatant and supplemented with 6 µg/ml polybrene. Subsequently, puromycin (PRDM16 and EHMT1), hygromycin (C/EBP-β) or G418 (shRNAs) were used for selection.
Gene expression analysis
Total RNA was isolated from tissues using Trizol (Invitrogen) or RiboZol reagents (AMRESCO) following the manufacturer’s protocol. Quality of RNA from all the samples was checked by spectrophotometer. Reverse transcription reactions were performed using IScript cDNA synthesis kit (Bio-Rad). The sequences of primers used in this study can be found in Supplementary table 2. Quantitative reverse transcriptase PCR (qRT-PCR) was performed with SYBR green fluorescent dye using an ABI ViiA™7 PCR machine. Relative mRNA expression was determined by the ΔΔ-Ct method using TATA-binding protein (TBP) as an endogenous control to normalize samples.
RNA-sequencing and gene ontology (GO) analysis
Total RNA was isolated from the presumptive interscapular BAT depots of wild-type and Ehmt1myf5 KO mice at P1 or from the interscapular BAT depots of wild-type and Ehmt1adipo KO mice at 12-week-old. RNA-sequencing libraries were constructed from 50 ng of total RNA from the Ehmt1adipo KO and Ehmt1myf5 KO BAT using Ovation RNA-sequencing system v2 kit (NuGEN). mRNA was reverse transcribed to cDNAs using a combination of random hexameric and a poly-T chimeric primer. The cDNA libraries were subsequently amplified by single primer isothermal amplification (SPIA) method 30 using Ultralow DR library kit (NuGEN) according to manufacturer’s instruction. Qualities of the libraries were determined by Bioanalyzer (Agilent Technologies). Subsequently, high-throughput sequencing was performed using a HiSeq 2500 instrument (Illumina) at the UCSF genomics core facility. RNA-sequencing reads for each library were mapped independently using TopHat version 2.0.8 against the UCSC mouse genome build mm9 indexes, downloaded from TopHat website (http://tophat.cbcb.umd.edu/igenomes.shtml). The mapped reads were converted to FPKM (fragments per kilobase of exon per million fragments mapped) by running Cuffdiff 2 31 on the alignments from TopHat and the UCSC coding genes to estimate gene and isoform expression levels. Based on the list of genes that showed significant difference (P <0.05, the Delta method-based hypothesis test) from the RNA-sequencing data, enrichment of the Gene Ontology (GO) biological process terms (GO FAT category) was analyzed using the Gene Set Enrichment Analysis (GSEA) program, according to the method described by the previous paper 32. RNA-sequencing reads have been deposited in ArrayExpress (www.ebi.ac.uk) under accession number E-MTAB-1704.
Immunocytostaining
Differentiated C2C12 myotubes or COS7 cells expressing GFP-PRDM16 and EHMT1 constructs were fixed with 4% paraformaldehyde for 10 minutes at RT, rinsed with PBS, and then exposed to 0.2% Triton X-100 in PBS for 5 minutes. The cells were subsequently incubated with anti-MF20 mouse antibody (DSHB, 1:50) for myosin heavy chain (MHC) and with flag antibody (M2, 1:200) for EHMT1. After washing with PBS, Alexa594-labeled anti-mouse IgG (1:800) was added as a secondary antibody.
Protein interaction analyses
Immortalized brown fat cells stably expressing flag-tagged wild type, PR-domain deletion mutant, and ZF-1 deletion mutant of PRDM16 or an empty vector were grown to confluence2. Nuclear extracts were isolated from these cells and incubated with flag M2 agarose beads, washed in a binding buffer (180 mM KCl), and subsequently eluted by either 3x or 1xflag peptides (0.2 µg/ml). The eluted proteins were subjected to histone methyltransferase assay or to reverse-phase liquid chromatography with tandem mass spectrometry (LC-MS/MS) for peptide sequencing using a high-resolution hybrid mass spectrometer (LTQ-Orbitrap, Thermo Scientific) with TOP10 method. Data obtained was annotated using the IPI mouse database33. Proteins were considered significantly identified with at least two unique valid peptides, and the false discovery rate was estimated to be 0% using the target-decoy approach34. To confirm the interaction between PRDM16 and EHMT1 in brown adipocytes, the immunopurified complex was purified using anti-EHMT1 (R&D Systems) or flag antibody (M2) and subjected to 4–12% SDS-PAGE. Rabbit polyclonal PRDM16 antibody35 or EHMT1 antibody (R&D Systems) was used for Western blotting. COS7 cells expressing HA-tagged PRDM16 or deletion fragments of flag-tagged EHMT129 were harvested 48 hours after transfection. Total cell lysates were incubated overnight at 4 °C with flag M2 agarose beads, washed and eluted by boiling. The immunoprecipitants were analyzed by Western blot analysis using HA antibody (Roche). For in vitro binding assays, various fragments of the GST–fusion PRDM16 fragments were purified as previously described16. [35S]-labeled proteins (EHMT1, EHMT2, CtBP1, C/EBP-β) were prepared with a TNT reticulocyte lysate kit (Promega). Equal amounts of GST fusion proteins (2 µg) were incubated overnight at 4°C with in vitro translated proteins in a binding buffer containing 20mM HEPES pH 7.7, 300 mM KCl, 2.5 mM MgCl2, 0.05% NP40, 1 mM DTT, and 10% glycerol. The sepharose beads were then washed five times with the binding buffer. Bound proteins were separated by SDS-PAGE and analyzed by autoradiography.
Histone methylation assay
The PRDM16 transcriptional complex was immunopurified from nuclear extracts of brown adipocytes using flag M2 agarose or IgG (negative control). The immunoprecipitants were incubated with 2 µg of core histone (Millipore) with 3H-labeled S-adenosyl−methionine at 30 °C for 1 h. Subsequently, the reaction was stopped by addition of sample buffer. Core histone was resolved by 4–12% SDS-PAGE and detected by autoradiography or by scintillation counter.
ChIP assay
After cross-linking with 1% formaldehyde at RT for 10 minutes, total cell lysates from brown adipocytes were sonicated to shear the chromatin, and immunoprecipitated overnight at 4°C using antibodies for H3 di-methyl and tri-methyl K9 (Abcam), acetyl-H3K9/K14 (Millipore), pan-H3 (Cell Signaling), EHMT1 (R&D Systems), or IgG (Santa Cruz). After extensive washing, the immunoprecipitants were eluted with 2% SDS in 0.1 M NaH2CO3. Cross-linking was reversed by heating at 65°C overnight. Input DNA and immunoprecipitated DNA were purified by PCR purification kit (Qiagen) and analyzed by qRT-PCR using SYBR green fluorescent dye (Bio-Rad). Enrichment of each protein was calculated as a ratio to input DNA. Primer sequences used in the ChIP assays were provided in Supplementary table2.
Protein stability assay
COS7 cells expressing HA-tagged PRDM16 and EHMT1 or vector control were incubated with a medium containing 60 µg/ml cycloheximide for up to 24 hours. Total cell lysates were isolated and separated by SDS-page. HRP-conjugated HA antibody (Sigma) and β-actin (Sigma) were used for Western blotting. Image J software was used to quantify the intensity of signals. Half-life of the protein was estimated by regression analysis.
Reporter gene assay
A luciferase reporter gene controlled by PPAR-γ binding sites (3xDR1-Luciferase) was transiently transfected with PPAR-γ/RXR-α, PRDM16 and EHMT1 expression plasmids in COS7 cells using Lipofectamine 2000 (Invitrogen). Forty-eight hours after the transfection, cells were harvested and reporter gene assays were carried out using the Dual Luciferase Kit (Promega). Transfection efficiency was normalized by measuring expression of Renilla Luciferase.
Cellular respiration assay
Immortalized brown adipocytes were transduced with retroviral shEHMT1 (shEHMT1-#1) or scramble control and induced to differentiate. Brown adipocytes expressing EHMT1 or vector control were also differentiated under a pro-adipogenic condition. At day 6 of differentiation, oxygen consumption was measured as previously described36. Oligomycin was used to determine uncoupled respiration. In addition, antimycin A was added in the end of experiments to determine non-mitochondrial cellular respiration. For cAMP-induced respiration assays, fully differentiated brown adipocytes were incubated with 0.5 mM dibutyryl cyclic AMP for 12 hours prior to measuring oxygen consumption.
Statistical analyses
Statistical analysis was performed using JMP version 9.0 (SAS Institute). Two-way repeated-measures ANOVA (analysis of variance) was applied to determine the statistical difference in GTT, ITT, body weight gain and rectal temperatures between genotypes. Effect size and power analysis was done by pwr.t.test function of R statistics package. Other statistical comparison was assessed by an unpaired Student’s t test. P values less than 0.05 were considered significant throughout the study.
Supplementary Material
Acknowledgements
We are grateful to Drs. A. Tarakhovsky (Rockefeller University), E. D. Rosen (Harvard Medical School), Y. Shinkai (Riken institute), and E. Hara (Japanese Foundation for cancer research) for providing mice and plasmids. We would like to thank our colleagues in UCSF, including Y. Qiu, A. Chawla, C. Paillart, S. Koliwad, M. Robblee, D. Scheel, S. Ohata, L. Mera, D. Lowe, S. Sonne, S. Keylin, I. Luijten, H. Hong, and E. Tomoda for their assistance. This work was supported by grants from the NIH (DK087853 and DK97441) to S.K. We acknowledge supports from the DERC center grant (DK63720), UCSF PBBR program, the Pew Charitable Trust, and PRESTO from Japan Science and Technology Agency to S.K. H.O. is supported by Manpei Suzuki Diabetes Foundation.
Footnotes
Author Contributions. S.K. and H.O. conceived and designed the experiments. All authors performed the experiments and analyzed the data. S.K. and H.O. wrote the paper.
REFERENCE
- 1.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kajimura S, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. American journal of physiology. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 4.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. The New England journal of medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 5.Saito M, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. The New England journal of medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 7.Atit R, et al. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Developmental biology. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- 8.Timmons JA, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell metabolism. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shing DC, et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. The Journal of clinical investigation. 2007;117:3696–3707. doi: 10.1172/JCI32390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinheiro I, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. 2012;150:948–960. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 12.Tachibana M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes & development. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleefstra T, et al. Disruption of the gene Euchromatin Histone Methyl Transferase1 (Eu- HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. Journal of medical genetics. 2005;42:299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormier-Daire V, et al. Cryptic terminal deletion of chromosome 9q34: a novel cause of syndromic obesity in childhood? Journal of medical genetics. 2003;40:300–303. doi: 10.1136/jmg.40.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willemsen MH, et al. Update on Kleefstra Syndrome. Molecular syndromology. 2012;2:202–212. doi: 10.1159/000335648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kajimura S, et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes & development. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. The EMBO journal. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eguchi J, et al. Transcriptional control of adipose lipid handling by IRF4. Cell metabolism. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almind K, Manieri M, Sivitz WI, Cinti S, Kahn CR. Ectopic brown adipose tissue in muscle provides a mechanism for differences in risk of metabolic syndrome in mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2366–2371. doi: 10.1073/pnas.0610416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannon B, Nedergaard J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. The Journal of experimental biology. 2011;214:242–253. doi: 10.1242/jeb.050989. [DOI] [PubMed] [Google Scholar]
- 22.Ouellet V, et al. Outdoor temperature, age, sex, body mass index, and diabetic status determine the prevalence, mass, and glucose-uptake activity of 18F-FDG-detected BAT in humans. The Journal of clinical endocrinology and metabolism. 2012;96:192–199. doi: 10.1210/jc.2010-0989. [DOI] [PubMed] [Google Scholar]
- 23.Wu Q, et al. Fatty acid transport protein 1 is required for nonshivering thermogenesis in brown adipose tissue. Diabetes. 2006;55:3229–3237. doi: 10.2337/db06-0749. [DOI] [PubMed] [Google Scholar]
- 24.Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell metabolism. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Yoneshiro T, et al. Impact of UCP1 and beta3AR gene polymorphisms on age-related changes in brown adipose tissue and adiposity in humans. International journal of obesity (2005) 2013;37:993–998. doi: 10.1038/ijo.2012.161. [DOI] [PubMed] [Google Scholar]
- 26.Huh MS, Parker MH, Scime A, Parks R, Rudnicki MA. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. The Journal of cell biology. 2004;166:865–876. doi: 10.1083/jcb.200403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao X, et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nature cell biology. 2006;8:516–523. doi: 10.1038/ncb1404. [DOI] [PubMed] [Google Scholar]
- 28.Liisberg Aune U, Ruiz L, Kajimura S. Isolation and differentiation of stromal vascular cells to beige/brite cells. Journal of visualized experiments : JoVE. 2013 doi: 10.3791/50191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi A, et al. DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Molecular cell. 2012;45:123–131. doi: 10.1016/j.molcel.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Kurn N, et al. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clinical chemistry. 2005;51:1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 31.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Kersey PJ, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 34.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in largescale protein identifications by mass spectrometry. Nature methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 35.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists Induce a White-to-Brown Fat Conversion through Stabilization of PRDM16 Protein. Cell metabolism. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.