Abstract
EmrD is the only structurally characterized drug/H+ antiporter of the major facilitator superfamily (MFS). It has been crystallized in a doubly-occluded conformation that is considered representative of an intermediate state in the transport cycle of MFS transporters. However, unexpected features of the crystal structure and the lack of functional information available for EmrD limit the utility of the structural data. To assess whether the crystal structure represents a stable state in a native-like environment, we used electron paramagnetic resonance (EPR) spectroscopy to determine the mobility and accessibility of spin labels at 76 positions in six transmembrane (TM) helices of EmrD reconstituted in liposomes. While the EPR data were mostly consistent with the crystal structure, they also revealed significant deviations from the predicted orientation and topology of TM helices at several locations. Additionally, we were unable to reproduce EmrD-dependent multidrug resistance phenotypes in vitro and in cell-based assays of drug transport. In spite of structural and functional discrepancies, we mapped a pH-dependent conformational change in which the cytoplasmic side of the N-terminal half opened locally in response to protonation. This conformational switch is consistent with the expected pH-dependent behavior of MFS H+-coupled antiporters.
Active efflux of cytotoxic molecules across the plasma membrane contributes to the resistance of bacteria to antibiotic treatment. Efflux of a broad spectrum of drugs is accomplished by a wide range of active transporters, including the H+-coupled major facilitator superfamily (MFS).1,2 MFS transporters have 12 transmembrane (TM) helices making up two pseudo-symmetrically related halves.3 They catalyze vectorial movement of substrates, presumably via alternating accessibility of a central binding cavity.4 Biochemical studies of two drug/H+ antiporters (DHA) from the MFS family, LmrP from Lactococcus lactis and MdfA from Escherichia coli, have demonstrated the polyspecificity of these transporters for dissimilar hydrophobic cations and begun to elucidate the mechanism of H+/substrate coupling. The proposed transport mechanism relies on several conserved acidic residues, at least one of which is membrane-embedded, involved in substrate and proton binding/coupling.5-7 The Asp residue in the highly conserved MFS signature motif (GxxxD(R/K)xG) in the loop between TMs 2 and 3 has also been shown to be important for drug transport.5,8 Additionally, sequence alignments of DHA transporters, along with biochemical and computational studies of TatA and the vesicular monoamine transporter, suggested that signature glycine-rich motifs conserved in TMs 5 and 11 have a structural role.9-11 In spite of extensive biochemical characterization, no high-resolution structural data is available for any of the above transporters.
Yin et al.12 determined the first crystal structure of a DHA family member, EmrD from E. coli. The structure revealed an architecture similar to other structurally characterized MFS transporters, e.g. LacY13 and GlpT,14 and a central cavity formed by TM helices 2, 4, 8, and 10 in the interior of the protein. Surprisingly though, the cavity appeared to be occluded to both the cytoplasmic and periplasmic sides, but no substrate was resolved. This doubly-occluded state is not expected to be stable,1,4 so it remains unclear whether EmrD was captured in a H+-bound transition state stabilized by the crystal lattice or represents a departure from the classical model of antiport. Another unexpected feature of this structure is the absence of acidic residues embedded deeply within the TM region that would be required to support H+ translocation across the membrane. Nevertheless, the crystal structure of EmrD is considered representative of an intermediate in the transport cycle3 and has been used as the basis for molecular dynamics (MD) simulations of structural flexibility15 and homology modeling of other MFS transporters.10,16
Given the unexpected features of EmrD in the crystal lattice, we sought to assess whether this structure is consistent with the conformation of EmrD in a native-like environment. We used site-directed spin labeling and electron paramagnetic resonance (EPR) spectroscopy17-20 to evaluate the topology and orientation of six of the twelve TM helices in EmrD reconstituted in liposomes. The EPR data were largely consistent with the crystal structure but also indicated that regions of EmrD reconstituted in liposomes adopt a conformation at variance with the crystal structure. A previous study16 has shown that pH differences can induce structural changes in LmrP by protonation of conserved acidic residues. Therefore, we mapped structural changes in EmrD near two of these conserved acidic residues in response to proton binding. We found that low pH results in the opening of a cytoplasmic cleft in the N-terminal half, a rearrangement of peripheral helices, and local changes in the dynamics of helices surrounding the central cavity of EmrD. These results are consistent with a protonation-dependent conformational change expected in a H+-coupled antiporter.
In addition to the uncertainties in the structure of EmrD, its drug transport activity remains ill-defined. One study21 showed that EmrD became up-regulated upon disruption of the electrochemical gradient and conferred resistance to the uncouplers carbonyl cyanide 3-chlorophenylhydrazone (CCCP) and tetrachlorosalicylanilide. A later study22 indicated that EmrD expressed in a hypersensitive ΔacrAB strain of E. coli conferred resistance to SDS and benzalkonium chloride, but not CCCP. In an effort to address the dearth of biochemical information, we conducted in vitro and cell-based functional assays but were not able to reproduce significant multidrug transport activity. These findings cast doubt on the appropriateness of EmrD as a structural and functional representative of drug/H+ antiporters of the MFS family.
EXPERIMENTAL PROCUDURES
Site-directed mutagenesis
Cys Substitution mutations were generated by PCR as previously described23using pET19b encoding WT or Cys-less EmrD as a template.
Expression and purification of EmrD
BL21 (DE3) cells were freshly transformed with a pET19b vector encoding WT or mutant EmrD with an N-terminal His-10 tag. A transformant colony was used to inoculate a 20 ml LB culture and subsequently a 1 L minimal medium A culture as previously described.24 Cultures were incubated at 37 °C until reaching an OD600 of 1.2-1.4 at which time the expression of EmrD was induced by the addition of IPTG to 1 mM. The cultures were incubated an additional 4 hours at 28 °C and then harvested and stored overnight at −20 °C. Cells pellets were resuspended in 20 ml 50 mM KH2PO4/K2HPO4, 250 mM sucrose, 150 mM choline chloride, 2.5 mM MgSO4, 0.02% NaN3, pH 8.0 and lysed by sonication. Cell debris was removed by centrifugation at 9000 g for 10 min. Membranes were collected by centrifugation at 193,000 g for 1 h. Membrane pellets were resuspended in 50 mM KH2PO4/K2HPO4, 500 mM NaCl, 10 mM imidazole, 20% w/v glycerol containing 2% β-dodecylmaltoside (β-DDM) and incubated on ice with stirring for 45 min. Insoluble material was cleared by centrifugation at 193,000 g for 30 min. Cleared extract was loaded onto a Ni affinity column equilibrated in buffer N (50 mM KH2PO4/K2HPO4, 500 mM NaCl, 20% w/v glycerol 0.05% w/v β-DDM) at a flow rate of 0.7 ml/min. After washing with >6 column volumes of Buffer N containing 30 mM imidazole, EmrD was eluted with buffer N containing 250 mM imidazole. Eluate was concentrated to 1 ml with a spin concentrator (50 kDa cutoff, Millipore) and injected onto a Sepharose 200 column equilibrated with GF buffer (50 mM HEPES, 100 mM NaCl, 20% w/v glycerol, 0.02% w/v β-DDM, pH 7.2). For EmrD preparations used for fluorescence anisotropy, glycerol was reduced to 5% w/v. During elution from gel filtration, the first third of the EmrD peak was discarded to reduce contamination by an unresolved species that appeared as a shoulder on the EmrD peak. Purified EmrD was stored on ice and used within one week. Concentration of EmrD was determined by A280 (ε = 1.45 mg−1 cm−1). Cys-substituted EmrD was spin labeled before gel filtration as previously described.18
Reconstitution of EmrD
A 3:1 (w/w) mixture of E. coli polar lipid extract (Avanti) and egg phosphatidylcholine (Avanti) was dissolved in CHCl3 at 20 mg/ml and evaporated to dryness on a rotary evaporator at room temperature. Lipids were further dried by desiccation under vacuum overnight. Multilamellar liposomes were formed by hydration of the lipids in buffer R (50 mM HEPES or MES, 50 mM NaCl, 2 mM MgCl2) at pH 7 or pH 5 at 10 mg/ml. Uniform unilamellar liposomes were formed by 5 cycles of freezing in a MeOH/CO2(s) bath and thawing in water followed by 5-10 rounds extrusion through a 100 nm membrane. Unilamellar liposomes were destabilized by the addition of β-DDM to a detergent:lipid ratio of 4:1. Purified, spin-labeled EmrD was mixed with destabilized liposomes at a protein:lipid ratio of 1:1000. For reconstitutions at pH 5, the pH of the EmrD solution was lowered with the addition of 1 M MES. The reconstitution mixture was incubated at room temperature for 1 h and diluted to 12 ml with buffer R. Detergent was removed by three additions of 100 mg Bio-Beads adsorbant (BioRad) per mg β-DDM. Bio-Beads were removed by filtration and proteoliposomes were pelleted by centrifugation at 193,000 g for 25 min. The pellet was resuspended in 3 ml reconstitution buffer and proteoliposomes were pelleted by centrifugation at 480,000 g for 10 min and resuspended in a minimal volume (~50 μl) of buffer R. Proteoliposomes were stored on ice and used within one week.
EPR spectroscopy
CW-EPR spectra were collected as previously described.18 Proteoliposomes containing spin-labeled EmrD were loaded into a 50 μl capillary tube. Spectra were collected on a Bruker EMX spectrometer at 10 mW power with a modulation amplitude of 1.6 G. Spectra were normalized to the double integral for lineshape comparisons. For accessibility measurements, proteoliposomes containing spin-labeled EmrD (with or without 50 mM NiEDDA) were loaded into a gas-permeable TPX tube and equilibrated in dry air or nitrogen. Power saturation curves were collected on a Bruker Elexys E500 spectrometer and analyzed as previously described.18
Substrate binding assays
Purified EmrD was concentrated using a spin concentrator (50 kDa cutoff). GF buffer was passed through a spin concentrator to account for concentration of empty β-DDM micelles. Dilutions of EmrD in GF buffer were mixed with a constant concentration of a test substrate in a total volume of 25 ul in a 384-well black microplate and incubated at room temperature >5 min. Fluorescence anisotropy (or intensity of Hoechst 33342) was measured using a BioTek Synergy H4 plate reader. Dissociation constants were calculated from the binding isotherm using a non-linear least-squares fit as previously described.23
Preparation of inside-out vesicles
BL21 cells overexpressing WT EmrD were prepared as above except that cultures were incubated 2-4 hours at 28 °C after induction. Cell pellets were resuspended in 20 ml 0.1 M HEPES, pH 7, and lysed by four passes through an Avestin C3 homogenizer at 15-20 kpsi. The lysate was diluted and cleared by centrifugation at 19,000 g for 20 min. The supernatant was transferred to a fresh tube taking care to avoid transfer of the loose pellet and centrifuged at 205,000 g for 1 h. Pellets, composed of inside-out vesicles, were resuspended in 3 ml 50 mM HEPES, 2 mM MgSO4, 20% w/v glycerol, 1 mM DTT, pH 7, aliquoted, and stored at −80 °C. Protein concentration in the vesicle suspension was measured using a modified Lowry assay.25
Hoechst transport assay
Fluorescence was measured using a Photon Technology International T-format fluorometer using an excitation wavelength of 355 nm and a 90°-polarized emission wavelength of 457 nm. Detector voltage was adjusted to between 750 and 850 V to maintain linear detection. An aliquot of vesicles (2 mg protein) was diluted into 3 ml HMK buffer (50 mM HEPES, 2.5 mM MgCl2, 300 mM KCl, pH 7.5) in a fluorescence cuvette with a stir bar. Background fluorescence was monitored for 30 s. Hoechst was added to 433 nM and the solution was mixed by pipetting. Hoechst fluorescence was recorded for 70 s and then ATP was added to 5 mM. Quenching of Hoechst fluorescence was monitored for 600 s and then nigericin was added to 0.5 μg/m to collapse the electrochemical gradient.
Drug resistance assay
Drug sensitive E. coli BW25113 ΔacrB ΔemrE ΔmdfA cells26 were transformed with empty pKK223-3 or pKK223-3 encoding WT EmrD. A dense overnight culture of the transformant was used to inoculate 10 ml LB broth containing 0.1 mg/ml ampicillin. Aliqouts (10 μl) of cells in the exponential growth phase (A600 = 0.6) were used to inoculate wells of a microplate containing 50% LB broth, 0.1 mg/ml ampicillin, and 0-50 μg/ml of the indicated drugs. Cell density was measured by A600 on a microplate reader after 5 h growth at 37 °C with shaking at 200 rpm. Density was normalized to the density of the control wells containing no drug.
RESULTS
Evaluation of EmrD structure by nitroxide scanning
We used site-directed mutagenesis to construct a Cys-less EmrD in which seven endogenous Cys residues (164, 172, 211, 229, 280, 281, and 379) were replaced with Ala. We subsequently introduced single Cys substitutions into this Cys-less background to generate a library of 76 EmrD mutants. Cys substitution, including removal of endogenous Cys residues and addition of novel Cys residues, and spin labeling did not alter the retention time on gel filtration chromatography relative to WT EmrD, suggesting that the folded structure was not greatly altered by these mutations or by spin label incorporation.
Sequential residues were selected for nitroxide scanning with the purpose of investigating the orientation and packing of TM helices. For each spin-labeled mutant, mobility and accessibility parameters were measured after reconstitution of EmrD into unilamellar liposomes at pH 7. Mobility, which is determined from the spectral lineshape and approximated by the mobility parameter (ΔH −10), reflects the steric restriction imposed on the label by its environment.27 Accessibility (Π) is a measure of the collision frequency of the spin label with paramagnetic relaxants, either molecular oxygen (O2) or Ni-ethylenediamine-N,N’-diacetic acid (NiEDDA).28,29 Accessibility to NiEDDA, soluble in water but not in the membrane, identifies residues exposed to water or at the water/lipid interface while accessibility to O2, most soluble in the membrane, identifies lipid-facing residues. Variation in mobility and accessibility of spin labels introduced along a TM helix should follow a helical periodicity where labels at the interface between helices should be sterically restricted and relatively inaccessible to O2. The differential solubility of NiEDDA and O2 allow the definition of membrane boundaries, as O2 concentration will be highest in the middle of the membrane and NiEDDA concentration will be highest in the surrounding aqueous solvent.30 The overall pattern of these spin label parameters enables the evaluation of the structural model of EmrD and the detection of structural changes under different biochemical conditions.
As discussed below, we were not able to reproduce assays for EmrD function, be it conferral of drug resistance to E. coli cells or ΔpH-driven extrusion of substrate from membrane vesicles. Thus it was not possible to functionally evaluate the consequences of cysteine substitutions. However, two aspects of the data presented below support the structural integrity of the spin-labeled EmrD mutants. First, the retention time of purified, spin-labeled EmrD mutants on gel filtration suggested that the protein was not aggregated. Second, the accessibility and mobility parameters of a substantial fraction of the spin-labeled mutants, often in consecutive residues, were consistent with the crystal structure.
a. TMs 2-3 are consistent with a helix-loop-helix structure
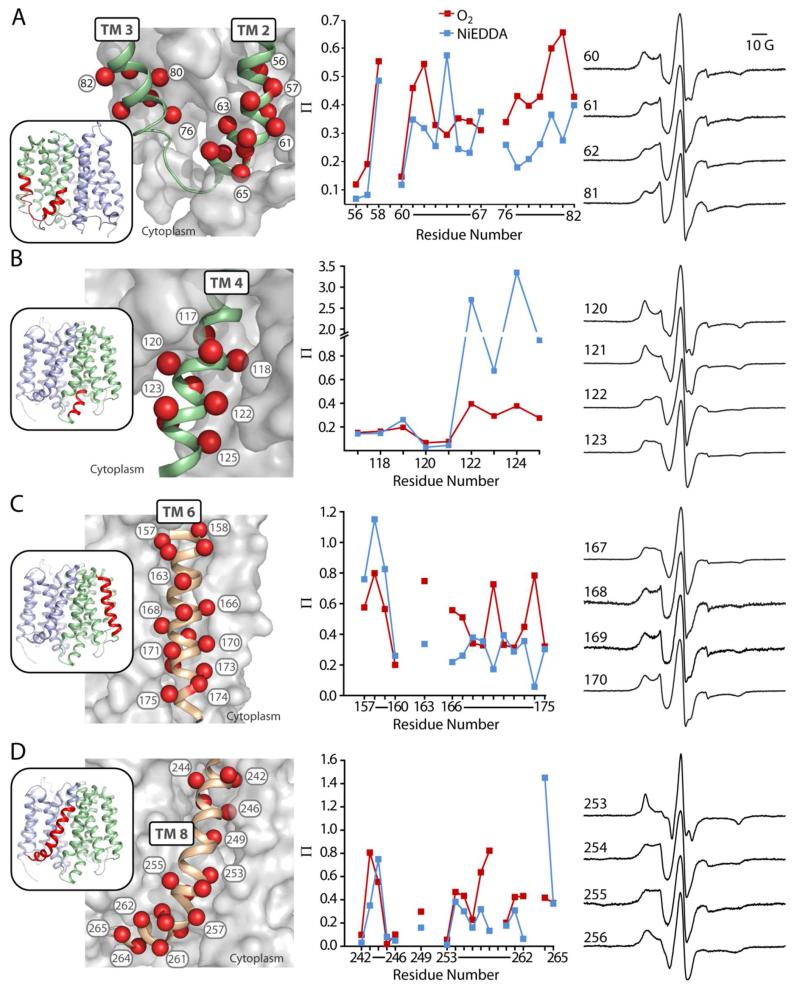
The cytoplasmic loop between TMs 2 and 3 contains the signature motif of the MFS family, including the conserved Asp68 implicated in coupling to the pH gradient.8,16,31 We examined the mobility and accessibility of spin labels at positions 56-67 (59 was excluded due to inefficient labeling) and 76-82 at the cytoplasmic ends of TMs 2 and 3 around this motif (Fig. 1, A). Positions 56, 57, and 60, which are at the interface of TM 2 and TM 4, have an immobilized component in the EPR lineshape and were inaccessible to O2 and NiEDDA (Figs. 1, A and S1). The O2 accessibility of TM 2 peaked at positions facing the lipid bilayer. NiEDDA accessibility peaked at positions 61, 64, and 67 on the face of TM 2 facing toward a cleft open to the cytoplasm. In TM 3, the trend of increasing O2 accessibility was consistent with increasing depth in the membrane. However, at variance with the crystal structure, O2 accessibility peaked at positions 80 and 81, which are predicted to be buried against TM 6. NiEDDDA accessibility in TM 3 peaked at ~0.6, which is consistent with the proximity of this region to the interface between aqueous and lipid phases.28 With the minor exception of positions 80 and 81, EPR parameters on the cytoplasmic ends of TMs 2 and 3 agree with the helix-loop-helix topology described by the crystal structure.
FIGURE 1. Accessibility of spin labels on TMs 2-3 (a), 4 (b), 6 (c), and 8 (d).
Positions on TM helices where accessibility and mobility were probed are highlighted as spheres. TM helices of interest (ribbon) are shown against a surface representation of the rest of EmrD to depict tertiary interactions. The accessibility (Π) of spin labels to O2 (red) or NiEDDA (blue) at pH 7 is plotted in sequence. CW-EPR spectra for labels in representative α-helical turns are shown to define the phase of the helices. Line shapes for all positions are found in Fig. S1.
b. TM 4 is consistent with the crystal structure
The cytoplasmic side of TM 4 extends into the aqueous phase and contains a conserved acidic residue, Asp123, as well as basic residues potentially involved in substrate binding.12 We examined positions 117-125 at the cytoplasmic end of TM 4 (Fig. 1, B). Positions 117-121 have low accessibility and immobilized spectra (Figs. 1, B and S1), consistent with their buried environment in the crystal structure, whereas positions 122-125 have high accessibility to NiEDDA and mobile spectra, which reflect their exposure to the aqueous phase near the membrane interface. The EPR parameters in this region are consistent with a buried helix emerging into the aqueous phase as described by the crystal structure.
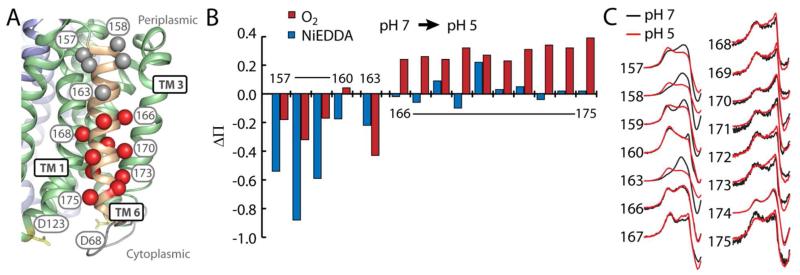
c. TM 6 is dynamic
TM 6 abuts TMs 2-4 on the periphery of the N-terminal bundle. We probed positions 157-175 along the entire length of this helix (Fig. 1, C), except several residues (161, 162, 164, and 165) in the center of the membrane that were inefficiently labeled. Throughout the length of TM6, accessibility to O2 (Fig. 1, C) showed an α-helical periodicity consistent with the orientation of this helix described by the crystal structure. NiEDDA accessibility (Fig. 1, C) was high at the periplasmic end of TM 6 and diminished in the membrane-embedded region of the helix. Interestingly, the pattern of NiEDDA accessibility on the cytoplasmic half of this helix shows a periodicity 180° out of phase with that of O2. Positions in this region where NiEDDA accessibility is higher face the cytoplasmic cleft formed by TMs 2-4. Together, data from positions in TMs 2-3 and TM 6 residues facing the interior of EmrD suggest that this cleft may be water-filled (Fig. 1, A and C). EPR lineshapes of spin labels along TM 6 showed multiple components of different mobilities (Figs. 1, C and S1) consistent with the spin labels sampling multiple environments. This pattern of multi-component spectra suggests that TM 6 is dynamic.
d. TM 8 is consistent with a kinked helix
TM 8 forms part of the interface between the two pseudo-symmetric halves of EmrD at the periphery of the cavity. Based on MD simulations of EmrD15 and EPR analysis of LmrP substrate binding,16 this helix has been proposed to open on the cytoplasmic side, acting as a gate to allow substrate entry to the cavity. We probed a turn of this helix (residues 242-246) near the periplasmic end and residues 253-265 on the cytoplasmic end of this helix (Fig. 1, D). No data was collected for 259 and 263 due to inefficient labeling and poor expression, respectively. The pattern of label mobility and accessibility (Figs. 1, D and S1) on the cytoplasmic side of TM 8 follows helical periodicity consistent with the orientation of this helix in the crystal structure. For example, the spin label at 253, buried against TM 10 in the crystal structure, is immobilized and shows little accessibility, whereas the label at 255, on the opposite face of the helix, is more mobile and accessible. The only departure from this pattern is the unexpectedly high O2 accessibility at residues 257 and 258. Given the proximity of these residues to the membrane-water interface, we would expect higher NiEDDA accessibility and lower O2 accessibility. This alteration of the expected trend likely results from a kink of TM 8 at Pro254, observed in the crystal structure of EmrD12 and other MFS transporters,3 which could shift labels at these positions deeper into the membrane. Additionally, if this end of TM 8 is dynamic, as predicted,15,16 then the high O2 accessibility could result from sampling of an environment closer to the center of the membrane bilayer.
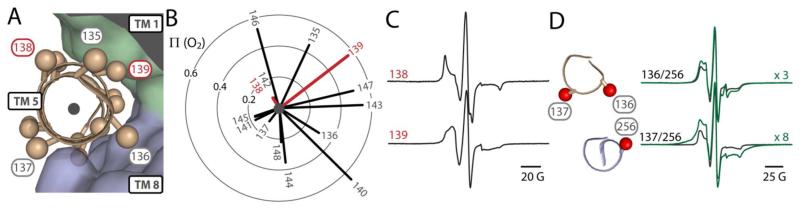
e. Orientation of TM 5 deviates from the crystal structure
TM 5, which is adjacent to TM 8, forms the other half of the interface between the two pseudo-symmetric halves of EmrD at the periphery of the cavity. The mobility and accessibility of spin labels in the membrane-embedded region (residues 135-148) of TM 5 (Fig. 2, A) indicated an orientation of this helix that conflicts with the crystal structure. Labels on a face of TM 5 predicted to be exposed to lipids on the periphery of the protein, and therefore accessible to O2, had low accessibility to O2 (Fig. 2, B). Additionally, this face of TM 5 (e.g. position 138) displayed spectral lineshapes consistent with sterically restricted, buried labels (Figs. 2, C and S1). The opposite face of TM 5, predicted to pack against TMs 1 and 8 exhibited higher O2 accessibility and mobility (Fig. 2, B and C). To confirm that the orientation of TM 5 deviates from the crystal structure, we measured dipolar coupling of spin labels at position 256 on TM 8 and at either position 136 or 137 on TM 5 (Fig. 2, D). The crystal structure predicts that labels projecting from positions 136 and 256 should be in close proximity (Cα-Cα, 10 Å; Cβ-Cβ, 7 Å), and therefore elicit a strong dipolar interaction. Experimentally, dipolar coupling, evident as broadening of the CW-EPR spectrum, was not pronounced for the 136/256 mutant (Fig. 2, D). In contrast, dipolar coupling was substantially stronger for labels at 256 and 137, which are predicted to point in opposite directions (Cα-Cα, 11 Å; Cβ-Cβ, 10 Å) and thus should be beyond the interaction range detectable by CW-EPR.32 Along with the accessibility data, this pattern of dipolar coupling supports that the helix is out of phase relative to the crystal structure.
FIGURE 2. Orientation of TM 5 conflicts with the crystal structure.
A) Positions on TM 5 where accessibility and mobility were probed are highlighted as spheres. TM 5 (ribbon) is shown against a surface representation of the rest of EmrD to depict tertiary interactions. B) Vectors indicate O2 accessibility (Π) throughout TM 5. The orientation of the plot matches the orientation of TM 5 in panel A and vectors were drawn from the center of the helix (black dot) through the β carbon of the side chain. Representative positions 138 and 139 are highlighted in red. C) CW-EPR spectra of positions 138 and 139 (red in panels A and B). D) A cross section of the TM 5-TM 8 interface indicates the predicted orientation of residues 136, 137, and 256. CW-EPR spectra of double Cys mutants (green) show strong dipolar coupling (spectral broadening) between labels at 137 and 256 relative to the sum of the spectra from single mutants (black). The y-axes of the spectra from double-mutants were scaled as indicated.
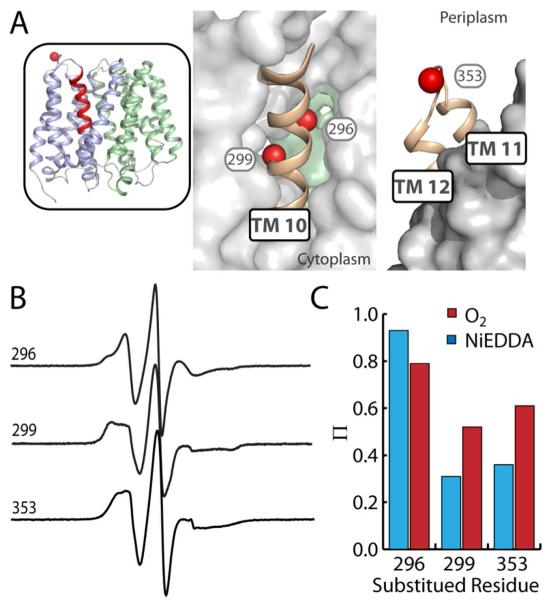
f. Topology of TMs 10 and 11 deviates from the crystal structure
We probed mobility and accessibility of spin labels at two representative positions on the periplasmic end of TM 10 (296 and 299) and one in the periplasmic loop between TMs 11 and 12 (353). In the crystal structure, positions 296 and 299 are at the edge of the interior substrate binding cavity (Fig 3, A). A label at position 299 shows an immobilized lineshape and low accessibility (Fig. 3, B and C) consistent with spin labels at cavity-facing positions in other TM helices (56, 60, 118, 249, 253, and 256; Figs. 1 and S1) and consistent with the doubly-occluded nature of the crystal structure. However, the spin label introduced at position 296 exhibits very high mobility and accessibility (Fig. 3, B and C) inconsistent with its location in the structure. The low-field resonance line indicates a degree of isotropic motion unlikely to result from a label on the face of an α-helix mostly buried in a protein environment. Furthermore, the spin label at position 353 in the 11-12 loop (Fig. 3, A), the lineshape appears more immobilized (Fig. 3, B) and substantially more accessible to O2 than NiEDDA (Fig. 3, C), parameters that are inconsistent with its location on an aqueous loop. These discrepancies suggest that the topology of the native structure deviates from the crystal structure in this region.
FIGURE 3. Topology of TMs 10-12 deviates from the crystal structure.
A) Positions on TM 10 and the periplasmic loop between TMs 11 and 12 where mobility and accessibility were probed are highlighted as spheres. Regions of interest (ribbon) are shown against a surface representation of the rest of EmrD to depict tertiary interactions and proximity of TM 10 to the central cavity. B) CW-EPR spectra showing the mobility of spin labels at the three positions. C) The accessibility (Π) of spin labels to O2 (red) or NiEDDA (blue).
pH-Sensitive Dynamics of EmrD
The inconsistencies between crystallographic data and EPR data in some regions of EmrD limited the scope of our investigation into mechanistically relevant structural dynamics. However, we focused on regions surrounding conserved acidic residues on the cytoplasmic ends of TMs 2 and 4, where EPR data agreed with the crystal structure, and explored the effects of low pH on EmrD with the purpose of identifying pH-dependent conformational changes. In some cases, differences in spin label mobility were evident in a change in the mobility parameter ΔH0−1 (Table S1). However, many of the spectra have multiple components such that ΔH0−1 does not fully reflect changes in the mobility of the spin label. Rather, these changes are only apparent in the details of the lineshape such as the intensity of the outer extrema and their separation. For this reason, we determined patterns of pH-dependent changes in structure based on the lineshapes and accessibility parameters (Π; Table S1).
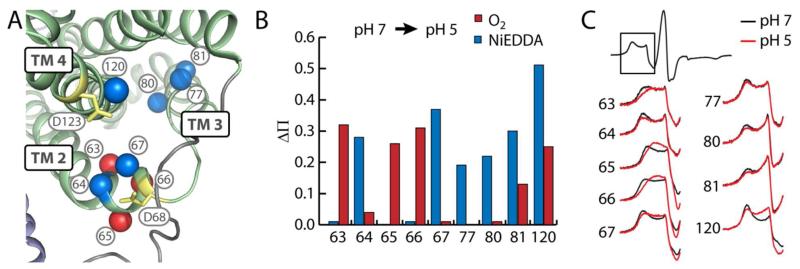
a. Opening of a cytoplasmic cleft
At pH 7, NiEDDA accessibility at positions 61, 64, and 67 in TM 2 suggested a water-filled cleft formed by TMs 2-4 (Fig. 4, A). Additionally, this region seems to have functional importance with the presence of the MFS signature motif and conserved acidic residues Asp68 and Asp123. Equivalent residues in LmrP (Asp68 and Asp128, respectively) have been shown to be essential for transport5 and lipid interactions.31 Therefore, we probed the structural response of this region to low pH by monitoring changes in the mobility and accessibility of spin labels introduced around these critical residues. At pH 5, the accessibility of spin labels at positions 64, 67, 77, 80, 81, and 120 in TMs 2-4 to water-soluble NiEDDA increased (Fig. 4, B). Additionally, O2 accessibility of spin labels at positions 65 and 66, on the opposite face from NiEDDA-accessible residues, increased at pH 5. The mobility of spin labels on the cytoplasmic end of TM 2 increases on all faces of the helix indicating a general loosening of the structure in this region (Fig. 4, C and Table S1). These changes suggest that low pH causes the 3-helix bundle to open allowing increased water penetration into the interior of the bundle and shifting residues on the exterior deeper into the membrane.
FIGURE 4. Low pH opens a cytoplasmic cleft.
A) Positions in TMs 2-4 where significant changes in Ni-EDDA (blue) or O2 (red) accessibility and/or mobility are represented as spheres on the structural model of EmrD. Conserved acidic residues are highlighted in yellow. B) Change in NiEDDA (blue) or O2 (red) accessibility from pH 7 to pH 5. C) CW-EPR spectra indicating increased mobility at pH 5 (red) versus pH 7 (black). Only the low field resonance line is shown for clarity.
b. Rearrangement of TM 6
Opening of the cytoplasmic cleft mediated by movements in TMs 2-4 would likely involve displacement of the associated peripheral helices, including TM 6. We probed the structural response of TM 6 to low pH by monitoring changes in the mobility and accessibility of spin labels introduced along the entire length of this helix (Fig 5, A). On the periplasmic end (positions 157-163), low pH caused a marked decrease in accessibility to O2 and NiEDDA (Fig. 5, B) and mobility (Fig. 5, C and Table S1). For example, ΔH0−1 of the spin label at position 158 decreases from 0.26 to 0.20 G−1. In contrast, low pH increased O2 accessibility on the cytoplasmic end (positions 166-175). This pattern suggested a tilting where the cytoplasmic side of the helix moves away from the bulk of the protein, while the periplasmic end becomes buried, likely moving toward the bulk of the protein. Outward movement of the cytoplasmic end of TM 6 is consistent with a concerted opening of the cytoplasmic side of the N-terminal bundle.
FIGURE 5. Low pH tilts TM 6.
A) Positions in TM 6 where significant increases (red) or decreases (gray) in accessibility and/or mobility are represented as spheres on the structural model of EmrD. B) Change in NiEDDA (blue) or O2 (red) accessibility. C) CW-EPR spectra (low field resonance line only) at pH 7 (black) and pH 5 (red) indicating changes in mobility.
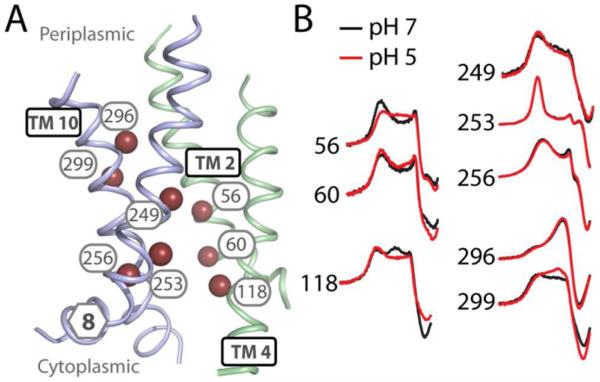
c. Changes in dynamics, but not accessibility, in the interior cavity
To probe the response of the cavity to changes in pH, we monitored spin label mobility and accessibility at several representative positions within the cavity (Fig. 6, A). Labels at positions 56 and 299 on the periplasmic side of the cavity became more mobile at low pH (Fig. 6, B and Table S1) whereas a label at 118 on the cytoplasmic side became less mobile. However, the accessibility pattern did not change in response to pH (Table S1). This pattern suggests that the lineshape changes reflect a pH-dependent alteration of the dynamics of the helices where the spin labels are incorporated but low pH did not change the exposure of the cavity.
FIGURE 6. Changes within the cavity.
A) Positions on TMs 2, 4, 8, and 10 where accessibility and/or mobility were probed are highlighted as spheres on the structural model of EmrD. B) CW-EPR spectra (low field resonance line only) showing changes in mobility at pH 5 (red) versus pH 7 (black).
d. No changes in TMs 5 and 8
TM 8 is proposed to be dynamic to allow substrate entry.15,16 Additionally, if the N- and C-terminal halves of EmrD separate upon formation of an inward-facing state, as proposed for other transporters,13,14 we would expect significant changes in mobility and accessibility of spin labels at the interface of TMs 5 and 8. However, no significant patterns of pH-dependent changes were observed for positions in these helices, although there were some local changes in accessibility (Table S1 and Fig. S1).
EmrD drug transport activity remains ill-defined
The impact and utility of the structural data on EmrD is limited by the lack of biochemical data available for this transporter. In an effort to develop the functional information available for EmrD, we have carried out the first in vitro studies of drug binding and transport by EmrD and attempted to recapitulate the previously reported EmrD-dependent drug resistance phenotype.
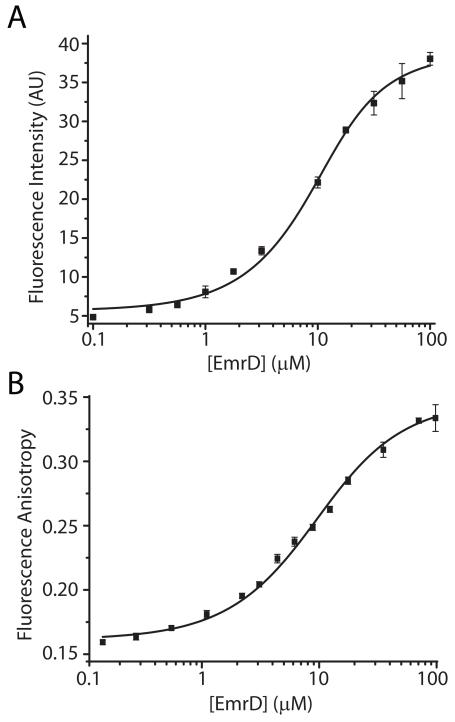
a. EmrD binds Hoechst and doxorubicin
Due to the sequence similarity and purported functional similarity of EmrD with other multidrug efflux pumps,12,33 specifically LmrP and MdfA, we tested binding of several fluorescent, hydrophobic cations that are widely used as test substrates. The binding of Hoechst33342 by EmrD was detected by monitoring fluorescence intensity and the binding of doxorubicin, ethidium, propidium, and fluorescein by monitoring fluorescence anisotropy over a range of EmrD concentrations. Fitting of the resulting curves showed that EmrD had an apparent micromolar affinity for Hoechst (Fig. 7, A; KD ≈ 3 μM) and doxorubicin (Fig. 7, B; KD ≈ 7 μM). Ethidium, propidium, and fluorescein did not bind to an appreciable extent (data not shown). The fitting of these binding curves is sub-optimal, indicating that Hoechst and doxorubicin binding may not be simple one-site binding and partitioning of these hydrophobic compounds into β-DDM micelles likely convolutes the binding analysis.23
FIGURE 7. Binding of Hoechst and Doxorubicin.
Increasing concentrations of EmrD were incubated with 5 μM substrate in GF buffer as described in Experimental Procedures. Binding was inferred from an increase in the fluorescence intensity of Hoechst (A) or the anisotropy of doxorubicin (B). Solid lines are non-linear least-squares fits with a KD values of 3 μM for Hoechst (A) and 7 μM for doxorubicin (B).
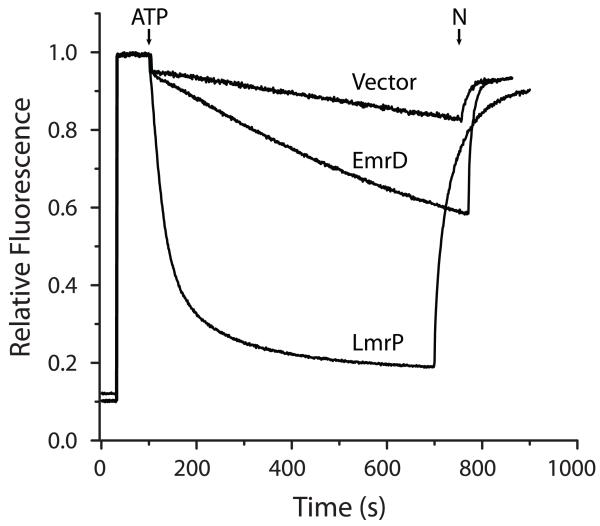
b. Minimal Hoechst transport by EmrD
The affinity of EmrD for Hoechst led us to determine whether EmrD transports Hoechst. We adapted the Hoechst transport assay previously developed34 for biochemical characterization of LmrP. We increased the sensitivity of the assay by including 300 mM KCl in the assay buffer. Chloride ions at this concentration permeate the membrane, thus removing the electrical resistance to generation of a pH gradient by F1F0 ATPase35. Inverted membrane vesicles prepared from E. coli BL21 cells expressing EmrD, LmrP, or no protein were assayed for Hoechst transport as described in Experimental Procedures. In some cases, we observed a level of transport in EmrD-containing vesicles above the level obtained with vesicles from the non-expressing strain as shown in Fig. 8. Vesicles containing no over-expressed transporter quenched 20-30% of Hoechst fluorescence. This background transport activity could be due to an endogenously expressed transporter with affinity for Hoechst or from diffusion and trapping of divalent Hoechst in the acidic lumen of the vesicles. Vesicles containing LmrP carried out robust transport as indicated by near total quenching of Hoechst fluorescence. Fluorescence quenching with vesicles containing EmrD was highly variable in the range of 20-40% of Hoechst fluorescence. Given that the fluorescence quenching observed with EmrD-containing vesicles does not differ greatly from empty vesicles, we conclude that EmrD transports Hoechst only weakly.
FIGURE 8. Minimal Hoechst transport.
A 2 mg aliquot of inside-out vesicles prepared from E. coli cells expressing EmrD or LmrP, or harboring an empty vector was diluted into HMK buffer. Vesicles were energized with ATP at the time indicated and the fluorescence of Hoechst33342 (H) was monitored for 700 s. The ΔpH was collapsed with nigericin (N) at the time indicated.
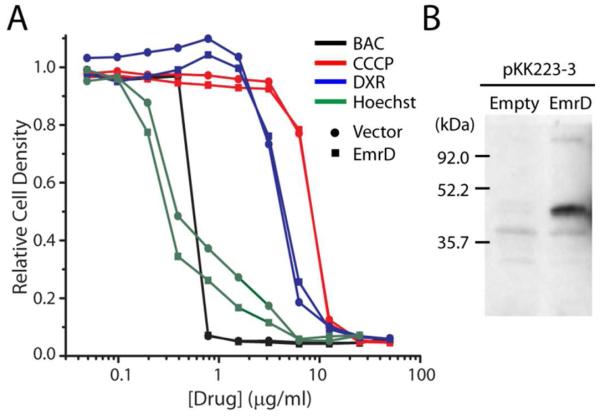
c. Lack of a drug resistance phenotype in vivo
Previous studies implicated EmrD expression in resistance of E. coli to CCCP21 and benzalkonium chloride.22 We attempted to recapitulate this resistance by measuring the growth of a drug sensitive strain of E. coli (BW25113 ΔacrAB ΔemrE ΔmdfA) 26 in drug-containing rich medium with or without non-induced expression of WT EmrD. Cells in logarithmic growth phase were exposed to increasing concentrations of benzalkonium chloride, CCCP, doxorubicin, and Hoechst3334 and growth was measured after 5 h incubation at 37 °C. While we observed cell death as a result of exposure to these compounds, expression of EmrD did not improve cell survival (Fig. 9, A). Expression of EmrD in these assays was confirmed by Western blotting of inside-out vesicles prepared from cells expressing a His-tagged construct of EmrD and grown under similar conditions (Fig. 9, B). Inclusion of a low concentration of IPTG to increase EmrD expression did not improve cell survival (data not shown).
FIGURE 9. EmrD expression did not confer multidrug resistance.
A) Drug sensitive E. coli expressing WT EmrD (squares) or harboring empty pKK223-3 vector (circles) were cultured in wells containing 50% LB, 0.1 mg/ml ampicillin, and 0-50 μg/ml benzalkonium chloride (BAC; black), CCCP (red), doxorubicin (DXR; blue), or Hoechst 33342 (green). Optical density is plotted relative to growth of the drug-free culture. B) Anti-His immunoblot of inside-out membrane vesicles isolated from drug sensitive E. coli expressing WT EmrD with an N-terminal His-10 tag or harboring empty pKK223-3 vector.
DISCUSSION
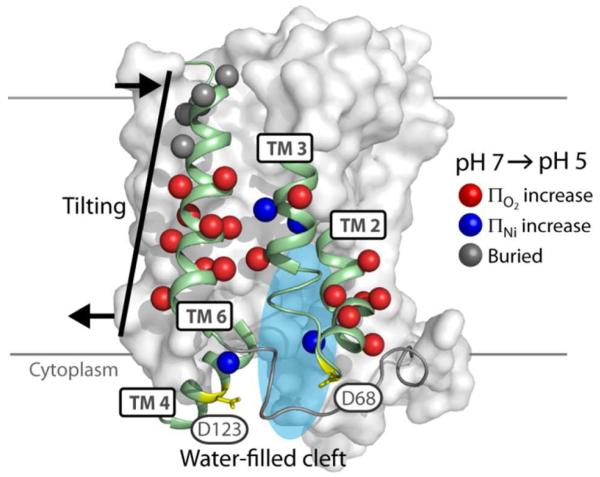
EmrD is the only structurally characterized multidrug antiporter of the MFS family. However, the moderate resolution and unexpected conformation captured in the crystal structure along with the lack of functional data for EmrD undermine its status as a model for other DHA transporters. In vitro and cell based assays reported here found minimal drug transport activity. Additionally, structural evaluation of EmrD identified significant deviations from the crystal structure in the orientation of TM 5 and the topology of the periplasmic ends of TMs 10 and 11. In spite of lingering uncertainties, EPR data confirmed the structure of several TM helices in EmrD reconstituted into liposomes and mapped a pH-dependent conformational change in EmrD involving opening of a cleft on the cytoplasmic side along with a concomitant closing of the periplasmic side (Fig. 10). This structural response to low pH is consistent with a conformational switch in the region of the conserved acidic residues and the signature MFS motif.
FIGURE 10. Summary of Structural Rearrangements.
The two regions where significant patterns of pH-dependent change were observed, TMs 2-4 and TM 6, are depicted (cartoon) against a surface representation of EmrD. For TMs 2-4, increases in accessibility to O2 (red) or NiEDDA (blue) at spin labeled positions (spheres) were interpreted as opening of a water-filled cleft at the cytoplasmic surface. For TM 6, concomitant increases in O2 accessibility (red) on the cytoplasmic end and decreases in accessibility and mobility (gray) on the periplasmic end were interpreted as tilting of this helix.
Comparison to the crystal structure
The finding that the structure of EmrD in liposomes deviates from the crystal structure obfuscates the structural context in which genetic and biochemical data can be interpreted. The pattern of mobility and O2 accessibility along the length of TM 5 and dipolar coupling between spin labels on TMs 5 and 8 indicated with a high degree of confidence that this helix is out of phase relative to the crystal structure by about 180°. This discrepancy affects which residues are exposed to the central cavity and therefore may be involved in drug recognition or proton coupling. Additionally, it affects the structural interpretation of the signature DHA1 motif36 on the periplasmic end of TM 5 (GPxxGG, APxxGG in EmrD). Mutations in this motif disrupt antiport activity in TetA, and the Pro in position 2 (Pro149 in EmrD) has been proposed to kink this helix,9 a feature not observed in the crystal structure. Two other isolated spin label positions were anomalous enough to warrant further attention. Spin labels at positions 296, predicted to lie on TM 10 facing the interior cavity, and 353, predicted to lie in the solvent-exposed loop between TMs 11 and 12, showed lineshapes indicating a mobility inconsistent with those locations. Interestingly, two recent sequence alignments of several MFS transporters10,37 shift the TM helix assignments for TM helices 9-12 of EmrD by 7-15 residues relative to the crystal structure, thus placing 296 in the loop between TMs 9 and 10 and 353 in the middle of TM 11. These alignment-based positions are more consistent with the experimental spin label mobility and accessibility at each position. Correct placement of residues in TM 11 is required to place the glycine-rich DHA2 motif (GxxxGxxxGG),36 which surrounds Leu353, in the proper structural context.
The discrepancies between the EPR data and the crystal structure could be due to errors in tracing the moderate resolution structure or to differences in the structure of EmrD in a neutral liposome environment versus an acidic crystalline environment. Evidence suggests that the discrepancies are not due simply to pH. Regions where EPR data is not consistent with the crystal structure (TMs 5, 10, and 11) showed little pH-dependent change in mobility or accessibility. Also, while the absolute values of mobility and accessibility differed between pH 7 and pH 5, the α-helical period was retained at most positions. It is important to note that the EPR dataset reported here is not exhaustive. Much of the structure of EmrD remains unevaluated, and the structure may differ from the crystal structure in other regions. Furthermore, additional dipolar coupling experiments to measure the distances between TM helices would be required to establish whether the doubly occluded state captured in the crystal structure is representative of a stable state in a liposome environment. The interface between TMs 5 and 8 is predicted to open upon formation of an open-inward state akin to that observed in LacY13,38 and GlpT.14 Dipolar coupling between spin labels on these helices indicated close proximity at pH 7 and spin labels facing the interior cavity were not particularly accessible to water-soluble Ni-EDDA at neutral or acidic pH. Both of these results are consistent with a doubly-occluded state but are not unequivocal.
Significance of pH-dependent conformational changes
Extensive characterization of MFS transporters indicate that substrates are translocated by an alternating access mechanism.4 Recently, Masureel et al.16 proposed a mechanism of H+-coupled drug antiport based on the structural response of LmrP to changes in pH and the presence of Hoechst, a hydrophobic substrate. Based on this model, we expected that low pH would induce an open-inward state in EmrD. The EPR data show that low pH causes opening, or at least loosening, of the cytoplasmic side, based on the increased mobility and aqueous accessibility in the cleft formed by TMs 2-4. This cleft contains conserved residues, including Asp68 in the MFS signature motif and Asp123 on TM 4, both of which have been implicated in a protonation-sensitive conformational switch.8,16,31,37 In LmrP, mutations in these residues abolished gradient dependent changes in the accessibility of the cavity.8 The EPR data also shows a concomitant closing of the periplasmic side, based on restriction of spin label mobility on the periplasmic side of TM 6. Indeed, the direction of tilting of TM 6 is consistent with the “swapped repeat” model of the opening of LacY to the cytoplasm.39
However, other indications are not consistent with the separation of the two halves of EmrD as expected for formation of a fully open state observed in other MFS transporters. We did not observe increases in spin label mobility or accessibility at the interface between TMs 5 and 8, which would be expected if these helices were separated to form a cytoplasmic opening. The hydrophobic nature of the substrates of DHA transporters does not necessitate a large aqueous opening to the cytoplasm since these substrates likely enter the central cavity directly from the inner leaflet of the membrane bilayer.40 The EPR study of LmrP found that the presence of substrate induced changes in the conformation of TM 8, and an MD simulation of EmrD15 predicted that TM 8 could open to allow horizontal entrance of the substrate. The EPR data at the cytoplasmic end of TM 8 does not indicate any changes in accessibility; however, the anomalous O2 accessibility and multi-component lineshapes of spin labels in this region (positions 261-265) may indicate an ensemble of states at equilibrium, even at neutral pH.
Further structural changes are likely to occur, especially in helices surrounding the cytoplasmic cleft region (e.g. TM 1). We predict, based on the pseudo-symmetry of EmrD and the placement of acidic residues, that structural rearrangements may occur on the periplasmic side of the C-terminal half. We did not address pH-induced dynamics in this region on account of the anomalous environment around positions 296 and 353.
Functional studies
Given the classification of EmrD as a DHA family transporter by the presence of conserved motifs and its similarity to other, well-characterized multidrug antiporters,12 we expected to detect in vitro drug transport activity and a drug resistance phenotype. However, we found that EmrD is not as robust a multidrug transporter as other representative members of the DHA family, namely LmrP41 and MdfA.42 While binding experiments suggest that EmrD binds Hoechst33342 and doxorubicin, minimal transport of Hoechst was detected, and expression of EmrD did not confer resistance to Hoechst or doxorubicin. We were also unable to reproduce previously reported results indicating that EmrD confers resistance against CCCP21 and benzalkonium chloride.22
Every MFS transporter for which biochemical or structural data is available contains a deeply membrane-embedded acidic residue that is thought to facilitate substrate binding and/or proton coupling. EmrD has no such residue, according to the crystal structure. The notable absence of this residue highlights the difficulty of using uncertain structural data to make functional conclusions. Does EmrD operate by a mechanism distinct from other MFS transporters, or is the missing residue simply misplaced in the structure? The alternative topology of the C-terminal half of EmrD determined by sequence alignment10 moves Glu227 from the C-terminal end of TM 7 to a membrane-embedded site on TM 7 adjacent to the central cavity. Uncertainty of the topology and orientation of TM helices in EmrD hinders the interpretation of critical residues and motifs and undermines the utility of the structure as a representative of a state in the MFS transport cycle.
Supplementary Material
Acknowledgements
We thank Dr. Richard Stein for assistance in analyzing the substrate binding and EPR data and Edward Monson for technical assistance. Prof. Shimon Schuldiner of The Hebrew University of Jerusalem generously provided the drug-sensitive strain of E. coli. We appreciate the critical reading of the manuscript by Drs. Hanane Koteiche and Brandy Verhalen.
FUNDING STATEMENT
This work was supported by National Institutes of Health grants F32 GM100569 to P.R.S. and R01 GM077659 to H.S.M.
ABBREVIATIONS
- CCCP
carbonyl cyanide 3-chlorophenylhydrazone
- CW
continuous wave
- β-DDM
β-dodecylmaltoside
- DHA
drug/H+ antiporter
- DTT
dithiothreitol
- EDDA
ethylenediamine-N, N’-diacetic acid
- EPR
electron paramagnetic resonance
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IPTG
isopropyl-β-D-1-thiogalactopyranoside
- MD
molecular dynamics
- MES
2-(N-morpholino)ethanesulfonic acid
- MFS
major facilitator superfamily
- MTSSL
1-oxyl-2,2,5,5-tetramethylpyrroline methanethiosulfonate
- NTA
nitrilotriacetic acid
- TM
transmembrane
Footnotes
Supporting Information—CW-EPR spectra and accessibility values for all spin-labeled positions at pH 5 and pH 7. Available free of charge via the internet at http://pubs.acs.org/.
REFERENCES
- 1.Law CJ, Maloney PC, Wang D-N. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fluman N, Bibi E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim. Biophys. Acta. 2009;1794:738–747. doi: 10.1016/j.bbapap.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Yan N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 2013;38:151–159. doi: 10.1016/j.tibs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Forrest LR, Krämer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta. 2011;1807:167–188. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Mazurkiewicz P, Konings WN, Poelarends GJ. Acidic residues in the lactococcal multidrug efflux pump LmrP play critical roles in transport of lipophilic cationic compounds. J. Biol. Chem. 2002;277:26081–26088. doi: 10.1074/jbc.M203141200. [DOI] [PubMed] [Google Scholar]
- 6.Adler J, Lewinson O, Bibi E. Role of a conserved membrane-embedded acidic residue in the multidrug transporter MdfA. Biochemistry. 2004;43:518–525. doi: 10.1021/bi035485t. [DOI] [PubMed] [Google Scholar]
- 7.Fluman N, Ryan CM, Whitelegge JP, Bibi E. Dissection of mechanistic principles of a secondary multidrug efflux protein. Mol. Cell. 2012;47:777–787. doi: 10.1016/j.molcel.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gbaguidi B, Mazurkiewicz P, Konings WN, Driessen a J. M., Ruysschaert JM, Vigano C. Proton motive force mediates a reorientation of the cytosolic domains of the multidrug transporter LmrP. Cell. Mol. Life Sci. 2004;61:2646–2657. doi: 10.1007/s00018-004-4298-2. [DOI] [PubMed] [Google Scholar]
- 9.Varela MF, Sansom CE, Griffith JK. Mutational analysis and molecular modelling of an amino acid sequence motif conserved in antiporters but not symporters in a transporter superfamily. Mol. Membr. Biol. 1995;12:313–319. doi: 10.3109/09687689509072433. [DOI] [PubMed] [Google Scholar]
- 10.Yaffe D, Radestock S, Shuster Y, Forrest LR, Schuldiner S. Identification of molecular hinge points mediating alternating access in the vesicular monoamine transporter VMAT2. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1332–1341. doi: 10.1073/pnas.1220497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwaki S, Tamura N, Kimura-Someya T, Nada S, Yamaguchi A. Cysteine-scanning mutagenesis of transmembrane segments 4 and 5 of the Tn10-encoded metal-tetracycline/H+ antiporter reveals a permeability barrier in the middle of a transmembrane water-filled channel. J. Biol. Chem. 2000;275:22704–22712. doi: 10.1074/jbc.m910354199. [DOI] [PubMed] [Google Scholar]
- 12.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Lemieux MJ, Song J, Auer M, Wang D-N. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 15.Baker J, Wright SH, Tama F. Simulations of substrate transport in the multidrug transporter EmrD. Proteins. 2012:1620–1632. doi: 10.1002/prot.24056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masureel M, Martens C, Stein RA, Mishra S, Ruysschaert J-M, Mchaourab HS, Govaerts C. Protonation of acidic residues triggers the conformational switch in the secondary multidrug transporter LmrP. Nat. Chem. Biol. 2013 doi: 10.1038/nchembio.1408. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mchaourab HS, Steed PR, Kazmier K. Toward the Fourth Dimension of Membrane Protein Structure: Insight into Dynamics from Spin-Labeling EPR Spectroscopy. Structure. 2011;19:1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou P, Mchaourab HS. Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J. Mol. Biol. 2009;393:574–585. doi: 10.1016/j.jmb.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amadi ST, Koteiche HA, Mishra S, Mchaourab HS. Structure, Dynamics, and Substrate-induced Conformational Changes of the Multidrug Transporter EmrE in Liposomes. J. Biol. Chem. 2010;285:26710–26718. doi: 10.1074/jbc.M110.132621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbell WL, Mchaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 21.Naroditskaya V, Schlosser MJ, Fang NY, Lewis K. An E coli gene emrD is involved in adaptation to low energy shock. Biochem. Biophys. Res. Commun. 1993;196:803–809. doi: 10.1006/bbrc.1993.2320. [DOI] [PubMed] [Google Scholar]
- 22.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steed PR, Stein RA, Mishra S, Goodman MC, Mchaourab HS. Na(+)- Substrate Coupling in the Multidrug Antiporter NorM Probed with a Spin-Labeled Substrate. Biochemistry. 2013;52:5790–5799. doi: 10.1021/bi4008935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koteiche HA, Reeves MD, Mchaourab HS. Structure of the substrate binding pocket of the multidrug transporter EmrE: site-directed spin labeling of transmembrane segment 1. Biochemistry. 2003;42:6099–6105. doi: 10.1021/bi0342867. [DOI] [PubMed] [Google Scholar]
- 25.Fillingame RH. Purification of the carbodiimide-reactive protein component of the ATP energy-transducing system of Escherichia coli. J. Biol. Chem. 1976;251:6630–6637. [PubMed] [Google Scholar]
- 26.Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 28.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. U. S. A. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altenbach C, Froncisz W, Hemker R, Mchaourab HS, Hubbell WL. Accessibility of nitroxide side chains: absolute Heisenberg exchange rates from power saturation EPR. Biophys. J. 2005;89:2103–2112. doi: 10.1529/biophysj.105.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong J, Yang G, Mchaourab HS. Structural basis of energy transduction in the transport cycle of MsbA. Science. 2005;308:1023–1028. doi: 10.1126/science.1106592. [DOI] [PubMed] [Google Scholar]
- 31.Hakizimana P, Masureel M, Gbaguidi B, Ruysschaert J-M, Govaerts C. Interactions between phosphatidylethanolamine headgroup and LmrP, a multidrug transporter: a conserved mechanism for proton gradient sensing? J. Biol. Chem. 2008;283:9369–9376. doi: 10.1074/jbc.M708427200. [DOI] [PubMed] [Google Scholar]
- 32.Rabenstein MD, Shin YK. Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulsen IT, Brown MH, Skurray R. a. Proton-dependent multidrug efflux systems. Microbiol. Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putman M, van Veen HW, Poolman B, Konings WN. Restrictive use of detergents in the functional reconstitution of the secondary multidrug transporter LmrP. Biochemistry. 1999;38:1002–1008. doi: 10.1021/bi981863w. [DOI] [PubMed] [Google Scholar]
- 35.Angevine CM, Fillingame RH. Aqueous access channels in subunit a of rotary ATP synthase. J. Biol. Chem. 2003;278:6066–6074. doi: 10.1074/jbc.M210199200. [DOI] [PubMed] [Google Scholar]
- 36.Pao SS, Paulsen IT, Saier MH. Major Facilitator Superfamily. Microbiol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang D, Zhao Y, Wang X, Fan J, Heng J, Liu X, Feng W, Kang X, Huang B, Liu J, Zhang XC. Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14664–14669. doi: 10.1073/pnas.1308127110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smirnova I, Kasho V, Choe J-Y, Altenbach C, Hubbell WL, Kaback HR. Sugar binding induces an outward facing conformation of LacY. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radestock S, Forrest LR. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J. Mol. Biol. 2011;407:698–715. doi: 10.1016/j.jmb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Bolhuis H, van Veen HW, Brands JR, Putman M, Poolman B, Driessen AJ, Konings WN. Energetics and mechanism of drug transport mediated by the lactococcal multidrug transporter LmrP. J. Biol. Chem. 1996;271:24123–24128. doi: 10.1074/jbc.271.39.24123. [DOI] [PubMed] [Google Scholar]
- 41.Putman M, van Veen HW, Degener JE, Konings WN. The lactococcal secondary multidrug transporter LmrP confers resistance to lincosamides, macrolides, streptogramins and tetracyclines. Microbiology (Reading, U. K.) 2001;147:2873–2880. doi: 10.1099/00221287-147-10-2873. [DOI] [PubMed] [Google Scholar]
- 42.Lewinson O, Adler J, Sigal N, Bibi E. Promiscuity in multidrug recognition and transport: the bacterial MFS Mdr transporters. Mol. Microbiol. 2006;61:277–284. doi: 10.1111/j.1365-2958.2006.05254.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.