Abstract
Brucellosis is a worldwide distributed zoonosis that causes important economic losses to animal production. In Brazil, information on the distribution of biovars and genotypes of Brucella spp. is scarce or unavailable. This study aimed (i) to biotype and genotype 137 Brazilian cattle isolates (from 1977 to 2008) of B. abortus and (ii) to analyze their distribution. B. abortus biovars 1, 2 and 3 (subgroup 3b) were confirmed and biovars 4 and 6 were first described in Brazil. Genotyping by the panel 1 revealed two groups, one clustering around genotype 40 and another around genotype 28. Panels 2A and 2B disclosed a high diversity among Brazilian B. abortus strains. Eighty-nine genotypes were found by MLVA16. MLVA16 panel 1 and 2 showed geographic clustering of some genotypes. Biotyping and MLVA16 genotyping of Brazilian B. abortus isolates were useful to better understand the epidemiology of bovine brucellosis in the region.
Introduction
Brucellosis is a worldwide distributed zoonosis, responsible for considerable economic losses due to abortion and culling of infected animals [1]. The genus Brucella is currently composed of ten species preferentially associated with different host: B. abortus (cattle), B. canis (canids), B. ceti (cetaceans), B. melitensis (sheep and goats), B. microti (Microtus arvalis), B. neotomae (Neotoma lepida), B. ovis (sheep), B. pinipedialis (pinnipeds), B. suis (pigs), and B. inopinata (man?) [2].
Bacteriological culture is considered the gold standard for brucellosis diagnosis. Due to its high specificity and ability to identify different species and biovars (bv), bacteriological culture is very important for epidemiological studies, allowing the determination of the way the disease spreads in a region and the possible sources of infection [3], [4].
Brucella abortus biovars have distinct geographic distributions [5] Brucella abortus bv1 and B. abortus bv2 are worldwide distributed, while B. abortus bv3 is found predominantly in Italy, India, Egypt, and Africa. B. abortus bv5 is most commonly found in Germany and United Kingdom [5], but has also been observed in France [6]. B. abortus biovars 4 and 6 have been reported on Mexico and France, though less frequently than bv1, 2, and 3 [6], [7]. B. abortus bv4 was also reported in Canada [8], Chile, Ecuador, and Cuba [9]. In India, B. abortus bv1 is the most frequent, followed by B. abortus bv3, although bv4, 6, and 9 were also found [10].
There have been just few studies concerning the identification of the biovars of Brucella spp. in Brazil describing B. abortus bv1, 2 and 3 and B. suis bv1 [9], [11], [12]. Also, B. canis and B. ovis were found infecting domestic animals [3]. Brucella melitensis was not reported in Brazil [3], [4].
Although the great homogeneity of the genus Brucella [13], conserved loci containing repeated sequences were found. Analysis of the number of repeats of these loci (Variable Number of Tandem Repeats – VNTR) is used as tool in the study of brucellosis [14]–[17]. Also, VNTR analysis of various loci (Multiple Locus VNTR Analysis – MLVA) is very important for investigations of infection by Brucella spp. and was applied to epidemiological control and surveillance of brucellosis [14], [17]–[22]. Moreover, this assay proved to be a valuable tool to determine relationships among Brucella isolates from different animal species and from humans, as well as for epidemiological trace-back studies [23], [24].
Thus, the aims of the present study were (i) to biotype and genotype Brazilian cattle B. abortus strains and (ii) to analyze their distribution to support the Programa Nacional de Controle e Erradicação de Brucelose e Tuberculose (PNCEBT) [25].
Materials and Methods
Bacterial Strains
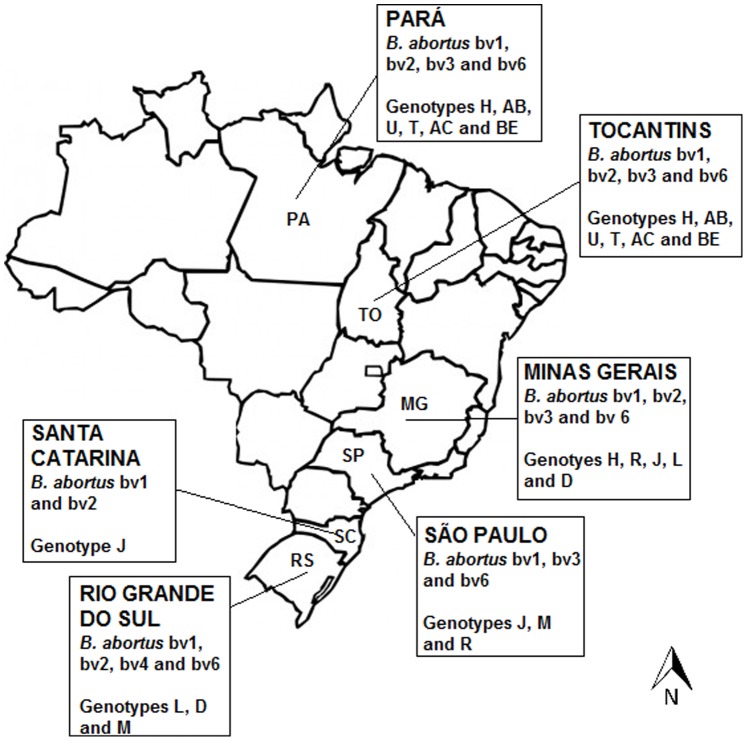
One hundred and thirty-seven Brazilian cattle B. abortus isolates, obtained from 1977 to 2008, were used. They were from the collections of Laboratório de Bacteriologia Aplicada, Escola de Veterinária, UFMG; Laboratório Nacional Agropecuário, Instituto de Pesquisas Veterinárias Desidério Finamor, and Instituto Biológico de São Paulo. These isolates were from Minas Gerais (38), Rio Grande do Sul (31), Pará (31), São Paulo (15), Tocantins (14) and Santa Catarina (8) (Figure 1). These strains represent the great majority of B. abortus isolates currently available in Brazil.
Figure 1. Sampling and distribution of biovars and MLVA16 Panel 2 genotypes of B. abortus isolates from Brazil, 1977 to 2008.
Reference strains, that were used as control in different procedures, were: B. abortus biovar (bv) 1 544 = ATCC 23448T, B. abortus bv2 ATCC 23449, B. abortus bv3 Tulya = ATCC 23450, B. abortus bv4 292 = ATCC 23451, B. abortus bv5 3196 = ATCC 23452, B. abortus bv6 870 = ATCC 23453 and B. abortus bv9 C68 = ATCC 23455, B. abortus bv1 B19 USDA, B. abortus bv1 RB51, B. melitensis bv1 16M = ATCC 23456T, B. ovis Reo 198, B. suis bv1 1330 = ATCC 23444, Escherichia coli ATCC 25922, E. coli B41, Listeria monocytogenes ATCC 19115, Pseudomonas aeroginosa ATCC 27853, Salmonella enterica biovar Typhimurium ATCC 14028 and Staphylococcus aureus ATCC 29213.
Identification and Biotyping of B. abortus
All strains were identified by routine test [26], [27]. Isolates were also tested by bcsp31-PCR [28], AMOS-PCR [28], AMOS-ERY-PCR [30] for classification of subgroups 3a and 3b, and omp2b-RFLP-PCR with restriction by TaqI to confirm the identity of B. abortus bv4 strains [31].
MLVA16 Genotyping
DNA from each strain was submitted to MLVA typing, using a subset of 16 tandem repeat loci (MLVA16) [15], [17].
Genotype Analysis
Band size estimates were converted into number of repeat units for each locus [15], with the software BioNumerics 6.1 (Applied Maths, Sint-Martens-Latem, Belgium). Clustering analysis also was performed with the software BioNumerics 6.1 based on UPGMA [15]. The diversity index HGDI was calculated [32]. Genotypes obtained were compared to those deposited in the MLVAbank for Bacterial Genotyping (http://mlva.u-psud.fr/mlvav4/genotyping/index.php). The minimum spanning tree presented is the one with the highest overall reliability score and was calculated using UPGMA associated with the priority rule and the bootstrap resampling (BioNumerics 6.1).
Results
Biotyping and Identification of B. abortus Isolates
All the 137 isolates were confirmed to be Brucella spp. by bcsp31-PCR and to be B. abortus by biochemical tests, AMOS and AMOS-ERY-PCRs. Ninety-two (67.1%) of the 137 isolates were identified as B. abortus bv1, B. abortus bv2 or B. abortus bv4 by AMOS-PCR. These were found in all studied States, being bv1 most frequent in Rio Grande do Sul (26/72), followed by Minas Gerais (23/72), São Paulo (12/72), Santa Catarina (6/72), Pará (3/72) and Tocantins (2/72). B. abortus bv2 was most frequent in Minas Gerais (10/16), followed by Santa Catarina (2/16), Rio Grande do Sul (2/16), Pará (1/16) and Tocantins (1/16), while strains of bv3 were mainly observed in Pará (16/28), followed by Tocantins (9/28), Minas Gerais (2/28) and São Paulo (1/28). Similarly, bv6 strains were also more common in Pará (11/20), followed by Minas Gerais (3/20), Rio Grande do Sul (2/20), São Paulo (2/20) and Tocantins (2/20). Interestingly, only one strain of B. abortus bv4 was observed in Santa Catarina state. AMOS-PCR-negative strains were found in all States except Santa Catarina and most frequently in Pará and Tocantins.
B. abortus biovars found by biochemical testing were 1, 2, 3, 4, and 6. The most frequent was B. abortus bv1, followed by bv3, bv6, and bv2; the least frequent was bv4. The HGDI calculated for B. abortus biovar identification was 0.65. All B. abortus bv3 strains were classified as subgroup 3b by AMOS-ERY-PCR.
The unique B. abortus bv4 isolate, strain 16/02, isolated in Rio Grande Sul, was confirmed by omp2b-RFLP-PCR [31].
MLVA16 Typing and Clustering of B. abortus Isolates
MLVA16 panel 1 identified nine genotypes (Table 1), four (28, 32, 33 and 40) previously described [15] and five new ones (I to V). These genotypes had one or more loci of panel 1 minisatellites with a number of repeat units different from reported ones [15]. Among genotypes observed for panel 1 genotyping the most frequent was genotype 28 found in all studied states, being more common in Minas Gerais (33), followed by Rio Grande do Sul (27/82), São Paulo (9/82), Santa Catarina (8/82), Tocantins (3/82) and Pará (2/82). The genotype 40 was more observed in Pará (26/45), followed by Tocantins (9/45), Minas Gerais (5/45), São Paulo (3/45) and Rio Grande do Sul (2/45). Whereas, the three strains of genotype 33 were observed in Tocantins (2/3) and São Paulo (1/3) and genotype 32 was found only in Rio Grande do Sul (1/1). Regarding new genotypes of panel 1, the profile I and II were observed only in Tocantins state, while patterns III and IV were found respectively in Pará and Rio Grande do Sul, all with one representative each. With two representative, genotype V was observed only in São Paulo state. The HGDI for panel 1 genotyping was 0.54.
Table 1. Biotyping, AMOS-PCR and genotyping based on panel 1 of MLVA16 of the strains of B. abortus isolated from cattle in Brazil, 1977 to 2008.
| Genotypea | |||||||||||
| Biovar | AMOS-PCR | 28 (%) | 32 (%) | 33 (%) | 40 (%) | I (%) | II (%) | III (%) | IV (%) | V (%) | Total (%) |
| 1 | Pos | 67 (48.9) | – | 2 (1.5) | – | – | – | – | 1 (0.7) | 2 (1.5) | 72 (52.6) |
| Neg | –b | – | – | – | – | – | – | – | – | 0 (0.0) | |
| 2 | Pos | 15 (10.9) | – | 1 (0.7) | – | – | – | – | – | – | 16 (11.7) |
| Neg | – | – | – | – | – | – | – | – | – | 0 (0.0) | |
| 3 | Pos | – | – | – | 3 (2.2) c | – | – | – | – | – | 3 (2.2) |
| Neg | – | – | – | 23 (16.8) | – | 1 (0.7) | 1 (0.7) | – | – | 25 (18.6) | |
| 4 | Pos | – | 1 (0.7) | – | – | – | – | – | – | – | 1 (0.7) |
| Neg | – | – | – | – | – | – | – | – | – | 0 (0.0) | |
| 6 | Pos | – | – | – | – | – | – | – | – | – | 0 (0.0) |
| Neg | – | – | – | 19 (13.9) | 1 (0.7) | – | – | – | – | 20 (14.6) | |
| Total | – | 82 (59.9) | 1 (0.7) | 3 (2.2) | 45 (32.8) | 1 (0.7) | 1 (0.7) | 1 (0.7) | 1 (0.7) | 2 (1.5) | 137 (100.0) |
Panel 1 of MLVA16 is based on loci Bruce06, Bruce08, Bruce11, Bruce12, Bruce42, Bruce43 and Bruce55 [15].
n = 0.
result not expected.
Panel 2A identified 13 genotypes, five (8, 30, 32, 33 and 34) previously described [15] and eight new ones (2A1 to 2A8). Panel 2A HGDI was 0.74. Genotype 33 was the most frequent (62/137), followed by 32 (28/137), 34 (13/137), 8 (10/137) and 30 (8/137). Among newly described genotypes, 2A3 was the most frequent (4/137), followed by 2A2 and 2A8 (3/137).
In panel 2B, 63 different genotypes were found, 13 of which had been previously described (18, 29, 30, 45, 52, 53, 58, 64, 66, 87, 88, 90 and 91) and 50 new genotypes, identified as 2B1 to 2B50. The HGDI for these loci was 0.97. Among these genotypes, the most frequent was genotype 91 (11/137), followed by 19 (9/137), 4 (8/137), 5 (6/137), 18 and 8 (5/137). The genotypes 30, 53, 17, 22, and 48 were found each in four of the 137 strains and genotypes 52, 58, 90, 16, 30, 44 and 46 in three of them.
Table 2 shows HGDI for the 16 loci based on biovars of studied B. abortus strains. Panel 1 had HGDIs below 0.16. Panel 2A loci with higher diversity were Bruce19 and Bruce21. HGDIs for B. abortus bv2 (0.74) and bv6 (0.73) were the highest in panel 2A. Panel 2B had the highest HGDI among all biovars (0.93 to 0.96); with B. abortus bv1 presenting the highest one. HGDI for MLVA16 (panels 1, 2A and 2B) was 0.98, differentiating 89 genotypes among the 137 studied strains (A to CY).
Table 2. Diversity index (HGDI) calculated for each locus and for panels 1, 2A, 2B and MLVA16 according to the biovars of B. abortus isolated from cattle, Brazil, 1977 to 2008.
| Group/Locus | Biovar 1 | Biovar 2 | Biovar 3 | Biovar 6 | ||||
| Typesa | HGDIb | Types | HGDI | Types | HGDI | Types | HGDI | |
| Panel 1 | 5 | 0.16 | 2 | 0.07 | 3 | 0.14 | 2 | 0.1 |
| Bruce06 | 2 | 0.05 | 2 | 0.12 | 2 | 0.07 | 2 | 0.09 |
| Bruce08 | 2 | 0.03 | 1 | 0.00 | 2 | 0.07 | 1 | 0.00 |
| Bruce11 | 2 | 0.03 | 1 | 0.00 | 1 | 0.00 | 2 | 0.09 |
| Bruce12 | 3 | 0.08 | 1 | 0.00 | 1 | 0.00 | 2 | 0.09 |
| Bruce42 | 1 | 0.00 | 1 | 0.00 | 1 | 0.00 | 1 | 0.00 |
| Bruce43 | 1 | 0.00 | 1 | 0.00 | 1 | 0.00 | 1 | 0.00 |
| Bruce45 | 2 | 0.03 | 1 | 0.00 | 1 | 0.00 | 2 | 0.09 |
| Bruce55 | 1 | 0.00 | 1 | 0.00 | 2 | 0.07 | 1 | 0.00 |
| Panel 2A | 7 | 0.42 | 4 | 0.74 | 4 | 0.62 | 7 | 0.73 |
| Bruce18 | 3 | 0.18 | 1 | 0.00 | 1 | 0.00 | 4 | 0.27 |
| Bruce19 | 2 | 0.13 | 2 | 0.50 | 3 | 0.37 | 3 | 0.40 |
| Bruce21 | 2 | 0.26 | 3 | 0.62 | 2 | 0.35 | 5 | 0.54 |
| Panel 2B | 38 | 0.96 | 10 | 0.93 | 18 | 0.95 | 12 | 0.95 |
| Bruce04 | 4 | 0.49 | 2 | 0.50 | 9 | 0.88 | 8 | 0.87 |
| Bruce07 | 3 | 0.46 | 2 | 0.12 | 2 | 0.07 | 3 | 0.33 |
| Bruce09 | 4 | 0.08 | 1 | 0.00 | 2 | 0.07 | 2 | 0.09 |
| Bruce16 | 6 | 0.70 | 3 | 0.59 | 2 | 0.35 | 2 | 0.18 |
| Bruce30 | 6 | 0.62 | 4 | 0.67 | 3 | 0.20 | 2 | 0.18 |
| MLVA16 | 48 | 0.93 | 14 | 0.98 | 21 | 0.97 | 16 | 0.98 |
Number of genotypes;
Diversity index calculated according to Hunter and Gaston (1988) [32].
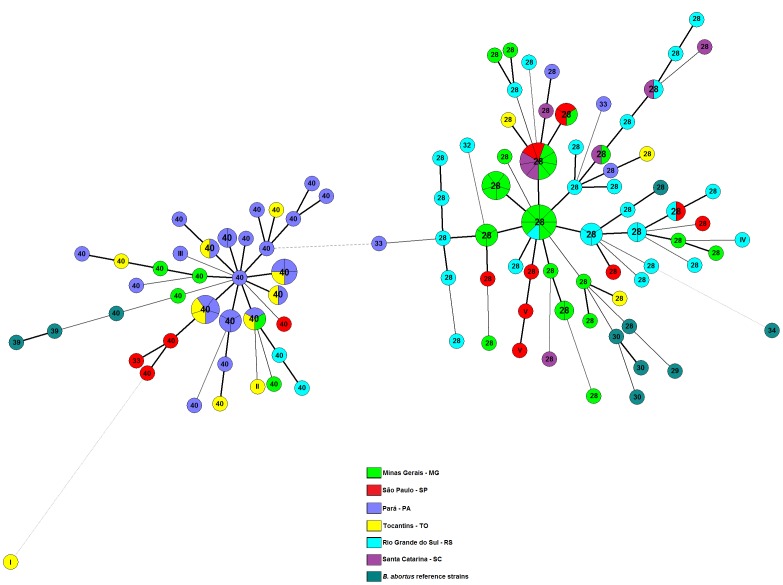
The generated Minimum Spanning Tree (MST), using categorical and parsimonious coefficients, clearly separated the two groups (Figure 2).
Figure 2. Minimum Spanning Tree (MST) analysis of B. abortus isolates using the MLVA16 data.
The minimum spanning tree presented is the one with the highest overall reliability score and was calculated using UPGMA associated with the priority rule and the bootstrap resampling for strains of Brucella abortus isolated from cattle in Brazil, 1977 to 2008, the reference strains for each biovar of B. abortus and the vaccine strains S19 and RB51. Numbers in each clonal complex represent the genotype on Panel 1. Information on the origin of the isolates was color labeled.
Among MLVA16 genotypes found, the most frequent was genotype J (9/137; 6.6%), found in Minas Gerais, São Paulo, and Santa Catarina. Other frequent genotypes were D (5.8%; 8/137), found in Minas Gerais and Rio Grande do Sul; AB (3.6%; 5/137) and U (2.9%; 4/137), both found in Pará and Tocantins; H and R (2.2%; 3/137) from Minas Gerais, Pará and Tocantins, and Minas Gerais and São Paulo, respectively. Genotypes T, AC and BE (1.5%; 2/137) were found in Pará and Tocantins, and genotypes L and M (2.2%) in Minas Gerais and Santa Catarina, and in Rio Grande do Sul and São Paulo, respectively. Table S1 summarize the genotypic profile in MLVA16, genotypes in panel 1, alleles in each MLVA16 loci, biovar and field information of 137 cattle B. abortus isolates.
All new genotypes found in this study were included in MLVAbank (http://mlva.u-psud.fr/mlvav4/genotyping/index.php) in a cooperative database called Brucella_Brazil.
Comparison among Typing Methods
Comparison of biochemical test results with panel 1 genotyping and AMOS-PCR showed compatible results by the three techniques in 68.6% (94/137) of the cases (Table 1). Twenty-three strains (16.8%) biochemically classified as B. abortus bv3 were from genotype 40 and negative by AMOS-PCR. Three strains biochemically classified as B. abortus bv3 were positive by AMOS-PCR and also from genotype 40. One strain phenotypically identified as B. abortus bv4 was AMOS-PCR positive and from genotype 32. Five strains (10.9%) that were AMOS-PCR positive B. abortus bv2 were typed as genotype 28. Another AMOS-PCR positive B. abortus bv2 was from genotype 33.
Discussion
Brucella abortus biovars found in this study were 1, 2, 3, 4, and 6 (Table 1), the latter two being reported for the first time in Brazil. B. abortus bv1 was the most frequent biovar isolated, as also described by previous studies in Brazil [3], [12], [33] and most regions of the world [4], [18]. Interestingly, our B. abortus bv2 isolates did not need addition of serum to culture medium for their isolation, as it is usually described for strains of this biovar [27]. This phenomenon was also observed for other B. abortus bv2 strains from Brazil [12]. Of the two biovars that were described for the first time in Brazil (bv4 and bv6), only one strain of B. abortus bv4 was isolated from Rio Grande do Sul. Conversely, B. abortus bv6 was observed in all the States sampled except Santa Catarina.
In our study, the high isolation frequency of B. abortus bv 1 and bv2 could be explained by the continuous importation of cattle, since the beginning of colonization [34], from regions where these two biovars are or were endemic [1], [4], [10]. As all isolates of B. abortus bv3 were from sub-group 3b, they were more closely related to B. abortus bv3 of European than of African origin [30], [35], which is in accordance to the origin of most cattle imported into Brazil, that came from Europe and India [10], [34].The unique B. abortus bv4 isolate were from Rio Grande do Sul, at the border with Argentina, where this biovar has already been described [4].
B. abortus bv6 was not previously reported in South America. The highest frequency of this biovar was found in Pará, where 55% of the isolates were of B. abortus bv6. Although the way this biovar was introduced into Brazil is not clear, B. abortus bv6 is one of the most frequent biovars of B. abortus in cattle in India, which is origin of the Zebu breeds that predominate in Pará and most of the country [10], [34].
Although MLVA16 could correctly group the strains by species of Brucella, it was not able to identify Brucella spp. biovars, probably due to the high rates of mutation at some loci. Therefore, this technique is not an adequate substitute for classic biovar identification [15], which is supported by this study, as three genotypes (28, 33 and 40) were found in B. abortus from more than one biovar (Table 1).
The most frequent genotype among the studied Brazilian B. abortus isolates was genotype 28, found in all six States. In Santa Catarina, it was the only genotype found among the eight isolates of B. abortus. Genotype 28, generally reported to be associated with strains of B. abortus bv1 and 4 from USA, Costa Rica, Switzerland, France, Italy, and Germany, was observed in strains of B. abortus bv1 and in almost all (15/16) of B. abortus bv2 strains examined.
Genotype 40, first reported from strains of B. abortus bv6 from Africa, was also very frequently found in Brazil and is the most common among strains of B. abortus bv6 (95.0%) and B. abortus bv3 (92.9%). This explains the geographic distribution of this genotype, as these two biovars were also the most common in Pará and Tocantins.
The unique B. abortus bv4 strain, isolated from Rio Grande do Sul, was from genotype 32, previously reported among German B. abortus bv1 strains.Genotype 33 strains, described in B. abortus bv1 strains from France, were observed in this study in B. abortus bv3 strains from Pará (2) and São Paulo (1). The lack of sound epidemiological data on those isolates precludes any deeper inference on the way this genotype circulates in Brazil.
Among the newly described panel 1 genotypes, the major differences of genotype I are the presence of 11 repeat units at locus Bruce12 and six repeat units at locus Bruce45, whereas genotype II had only one repeat unit at locus Bruce55. Genotype III had as leading feature the presence of just four repeat units at locus Bruce08 and genotype IV, observed in a B. abortus bv1 isolated from Rio Grande do Sul, was mainly characterized by four repeat units at locus Bruce45. Genotype V strains are characterized by the presence of 13 repeat units at locus Bruce12. All of these new genotypes were still represented by just one strain each, exception for genotype V, which was observed in two strains from São Paulo. Additional systematic studies in the regions where those genotypes were observed could help to understand their epidemiological importance.
The HGDI calculated for MLVA16 panel 1 (0.54) was lower than that calculated for biotyping (0.65), indicating a lower discriminatory efficacy of MLVA16 panel 1, due to the presence of more than one biovar in the same genotype by this panel (Table 1). The HGDI calculated for MLVA16 panel 2A (0.74) is similar to that found by Al Dahouk et al. (2007) [17], who analyzed 128 human isolates of B. melitensis and found that the mean diversity index in panel 2A was less than 0.75. This panel has mainly helped in the differentiation of strains of bv2 and 6 (Table 2). The high HGDI calculated for MLVA16 panel 2B, which has the most variable loci, with diversity indexes generally higher than 0.80, demonstrates that it can be used, together with MLVA16 panel 2A, for identifying strains associated with disease outbreaks [14], [15], [17]. Comparing HGDI for MLVA16 panels 2A and 2B calculated in the present study and those described in the literature [15], [17], [19]–[21] leads to the conclusion that these loci were effective for differentiating the Brazilian B. abortus isolates, especially when they are compared to biotyping.
Genotyping based on MLVA16 identified 89 previously undescribed genotypes (http://mlva.u-psud.fr/mlvav4/genotyping/index.php) among Brazilian B. abortus strains. The difference between the number of genotypes identified by MLVA16 panel 1 and by MLVA16 panel 2 is a consequence of the hypervariability of the loci in this second panel [15], [17].
The results of genotyping by MLVA16 show that the same genotypes tend to circulate in neighboring States, such as genotypes AB, U, T, AC, and BE in Pará and Tocantins in North Region, and genotypes J, D, L, and M in Minas Gerais, São Paulo, Santa Catarina, and Rio Grande do Sul in Southeast and South regions. However, some genotypes, as genotype H, found in North (Pará and Tocantins) and Southeast (Minas Gerais) regions, were present in distant and unrelated regions, thus requiring newer studies to clarify their epidemiology.
The MST (Figure 2) showed that the B. abortus isolates from six Brazilian States were displayed into two well-defined clusters, with 51.7% similarity. Cluster I included B. abortus strains of genotypes 40, 33 (2/3), II and III, distributed in five of the six States, except Santa Catarina. Cluster I also included reference strains of B. abortus bv5 3196, B. abortus bv6 870 and B. abortus bv9 C68. Cluster II included B. abortus strains from genotypes 28, IV, 33 (1/3) and V, as well as reference strains of B. abortus bv1 544, B. abortus bv2 86/8/59 and B. abortus bv4 292 and vaccine strains B. abortus bv1 S19 and B. abortus bv1 RB51. Clustering was performed basically from locus Bruce04, which was the most variable, followed by locus Bruce19 and then by locus Bruce30, first grouping strains with higher number of repeat units and then those that had fewer repeat units. Similarly, MLVA analysis of the Spanish human B. melitensis strains showed a high diversity of genotypes with locus Bruce04 featuring the highest HGDI [20].
Two strains appeared apart from the main clusters: B. abortus bv3 reference strain Tulya, isolated from Africa, which is MLVA16 genotype 207, and B. abortus bv6 strain 95, isolated from Tocantins, which is MLVA16 genotype AR. This shows that these strains are more distantly related to the other Brazilian B. abortus isolates (Figure S1). Likewise, this can be observed in MST (Figure 2), which also shows that strain Tulya is distantly linked to cluster II (gathered around genotype 28) and that strain 95 is distantly linked to cluster I (gathered around genotype 40).
Typing the strains by biotyping, genotyping by MLVA16 panel 1 and AMOS-PCR gave compatible result in the majority of the cases. Most of the genotypes that were observed in the present study and were not previously described in a particular biovar (Table 1) are probably due to small numbers of isolates from these biovars that were already analyzed by MLVA16, being the present ones new descriptions of these genotypes in different biovars.
B. abortus bv3 is a heterogeneous biovar, divided into subgroups 3a and 3b [15], [30] and the MST (Figure 2) depicts these differences. All Brazilian B. abortus bv3 strains were closer to B. abortus bv6 reference strain 870 than to the B. abortus bv3 reference strain Tulya, which did not cluster with other strains. These results agreed to published reports, which showed that strains of B. abortus bv3 subgroup 3b present a pattern similar to that of strains of B. abortus bv5, B. abortus bv6 and B. abortus bv9 [30].
Interestingly, three strains of B. abortus bv3, genotype 40, isolated from Pará, showed positive results in AMOS-PCR, which was described as able of detecting only biovars 1, 2, and 4 of B. abortus [29]. However, occasionally, atypical isolates of Brucella spp., that do not match identification tables and keys [36], has been described. Banai et al. (1990) [37] found B. mellitensis bv1 strains that grew with acid fuchsin and thionine in two regions of Israel, suggesting evolution of a new variant. Corbel (1991) [38] also found isolates of B. mellintensis in Italy, Kuwait, Saudi Arabia, Zimbabwe, India, and Germany that were sensitive to thionine (20 µg/mL). Garcia et al. (1988) [39] isolated atypical strains of B. abortus from seven infected cattle herds in Ontario (Canada) that were sensitive to thionine and fuchsin, similar to B. abortus bv2, but they agglutinated in anti-M serum, which is characteristic of B. abortus bv4.
When these atypical isolates appear, it is necessary to determine if their appearance is a reproducible phenomenon, indicating a local taxonomic pattern, and not a laboratory error. If reproducibility is proven, this can be used as an epidemiological marker [27]. Bricker (2002) [36] reported that due to the small number of differences among species and biovars of Brucella spp., the smallest mutation could result in conflicting data, making interpretation of the characterization of the isolates difficult. The three strains of B. abortus bv3, genotype 40, that were reactive in AMOS-PCR were isolated from the municipality of Conceição do Araguaia, Pará, within an interval of four years between the isolation of the first strain and the other two; which were isolated within an interval of five months. This could be an indication that these markers are characteristic of atypical strains from this region.
Biotyping and MLVA16 genotyping of Brazilian isolates of B. abortus will allow the construction of a database of genotypes of Brucella spp. from Brazil, and, in the future, for Latin America, which certainly will contribute to a better understanding of the epidemiology and control of bovine brucellosis in the region.
Supporting Information
(TIF)
(DOCX)
Funding Statement
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq, Fundação de Amparo à Pesquisa do Estado de Minas Gerais – Fapemig and Fundação de Estudo e Pesquisa em Medicina Veterinária e Zootecnia – FEP-MVZ. Silvia Minharro, Juliana P. S. Mol, Elaine M. S. Dorneles, Rebeca B. Pauletti, Marcos B. Heinemann, Renato L. Santos and Andrey P. Lage had fellowships from CNPq. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Corbel MJ, Elberg SS, Cosivi O (2006) Brucellosis in humans and animals. Geneva: WHO Press. 89p.
- 2. Whatmore AM (2009) Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect Genet Evol 9: 1168–1184. [DOI] [PubMed] [Google Scholar]
- 3. Poester FP, Gonçalves VSP, Lage AP (2002) Brucellosis in Brazil. Vet Microbiol 90: 55–62. [DOI] [PubMed] [Google Scholar]
- 4. Lucero NE, Ayala SM, Escobar GI, Jacob NR (2008) Brucella isolated in humans and animals in Latin America from 1968 to 2006. Epidemiol Infect 136: 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsh DC, Zee YC (2003) Microbiologia Veterinária. Rio de Janeiro: Guanabara Koogan. p. 191–195.
- 6. Garin-Bastuji B (1993) Brucelloses bovine, ovine et caprine: Contrôle et prevention. Le point Veterinaire 25: 107–114. [Google Scholar]
- 7. Luna-Martínez JE, Mejía-Terán C (2002) Brucellosis in Mexico: current status and trends. Vet Microbiol 90: 19–30. [DOI] [PubMed] [Google Scholar]
- 8. Ragan V (2002) The Animal and Plant Health Inspection Service (APHIS) Brucellosis eradication program in the Unites States. Vet Microbiol 90: 11–18. [DOI] [PubMed] [Google Scholar]
- 9. Garcia-Carillo D, Szyfres B, Gonzales-Tome J (1972) Tipificação de brucelas isoladas del hombre y los animales en América Latina. Rev Lat-amer Microbiol 14: 117–125. [PubMed] [Google Scholar]
- 10. Renukaradhya GJ, Isloor S, Rajasekhar M (2002) Epidemiology, zoonotic aspects, vaccination and control/eradication of brucellosis in India. Vet Microbiol 90: 183–195. Giorgi W, Pestana De Castro AF, Portugal MASC (1972) Tipificação de amostras de Brucella isoladas no Estado de São Paulo, Brasil. Rev Microbiol 3: 39–44. [DOI] [PubMed] [Google Scholar]
- 11. Giorgi W, Pestana De Castro AF, Portugal MASC (1972) Tipificação de amostras de Brucella isoladas no Estado de São Paulo, Brasil. Rev Microbiol 3: 39–44. [Google Scholar]
- 12. Megid J, Albert D, Fagliari AC, Paes AC, Listonis FP, et al. (2005) Isolation of Brucella abortus from cattle and water buffalo in Brazil. Vet Rec 156: 147–148. [DOI] [PubMed] [Google Scholar]
- 13. Verger JMF, Grimont PAD, Grimont Grayon M (1985) Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int J Syst Bacteriol 35: 292–295. [Google Scholar]
- 14. Whatmore AM, Shankster SJ, Perrett LL, Murphy TJ, Brew SD, et al. (2006) Identification and characterization of variable-number of tandem-repeat markers for typing of Brucella spp. J Clin Microbiol 44: 1982–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Flèche P, Jacques I, Grayon M, Al Dahouk S, Bouchon P, et al. (2006) Evaluation and selection of tandem repeat Loci for a Brucella MLVA typing assay. BMC Microbiol 6. [DOI] [PMC free article] [PubMed]
- 16. García-Yoldi D, Leflèche P, Marín CM, De Miguel MJ, Munoz PM, et al. (2007) Assessment of genetic stability of Brucella melitensis Rev 1 vaccine strain by multiple-locus variable-number tandem repeat analysis. Vaccine 25: 2858–2862. [DOI] [PubMed] [Google Scholar]
- 17. Al Dahouk S, Le Flèche P, Nöckler K, Jacques I, Grayon M, et al. (2007) Evaluation of Brucella MLVA typing for human brucellosis. J Microbiol Methods 69: 137–145. [DOI] [PubMed] [Google Scholar]
- 18.Bricker BJ, Ewalt DR (2005) Evaluations of the HOOF-print assay for typing Brucella abortus strains isolated from cattle in the United States: results with four performance criteria. BMC Microbiol 5. [DOI] [PMC free article] [PubMed]
- 19.Her M, Kang SI, Cho DH, Cho YS, Hwang IY, et al. (2009) Application and evaluation of the MLVA typing assay for the Brucella abortus strains isolated in Korea. BMC Microbiol 9. [DOI] [PMC free article] [PubMed]
- 20. Valdezate S, Navarro A, Villalón P, Carrasco G, Saéz-Nieto JA (2010) Epidemiological and phylogenetic analysis of Spanish human Brucella melitensis strains by multiple-locus variable-number tandem-repeat typing, hypervariable octameric oligonucleotide fingerprinting, and rpoB typing. J Clin Microbiol 48: 2734–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferreira AC, Chambel L, Tenreiro T, Cardoso R, Flor L, et al. (2012) MLVA16 Typing of Portuguese Human and Animal Brucella melitensis and Brucella abortus Isolates. PloS One 7: e42514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Céspedes S, Salgado P, Valenzuela P, Vidal R, Oñate AA (2011) Characterization of Genomic Island 3 and Genetic Variability of Chilean Field Strains of Brucella abortus . J Clin Microbiol 49: 2461–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marianelli C, Petrucca A, Pasquali P, Ciuchinia F, Papadopouloub S, et al. (2008) Use of MLVA-16 typing to trace the source of a laboratory-acquired Brucella infection. J Hosp Infect 68: 274–276. [DOI] [PubMed] [Google Scholar]
- 24. Alvarez J, Sáez JL, García N, Serrat C, Pérez-Sancho M, et al. (2011) Management of an outbreak of brucellosis due to B. melitensis in dairy cattle in Spain. Res Vet Sci 90: 208–211. [DOI] [PubMed] [Google Scholar]
- 25. Brasil Secretaria de Defesa Agropecuária, Ministério da Agricultura, Pecuária e Abastecimento. Instrução Normativa No 6 de 8 de janeiro de 2004. Aprova o Regulamento Técnico do Programa Nacional de Controle e Erradicação da Brucelose e Tuberculose Animal. Diário Oficial da União, Brasília, 12 jan. 2004, Seção 1, p. 6–10: 2001. [Google Scholar]
- 26.Mac Faddin JF(1980) Pruebas bioquimicas para la identificacion de bacterias de importancia clinica. Buenos Aires: Panamericana. 301 p. [Google Scholar]
- 27.Alton GG, Jones LM, Angus RD, Verger JM (1988) Techniques for the brucellosis laboratory. Paris: INRA. 189 p. [Google Scholar]
- 28. Baily GG, Krahn JB, Drasar BS, Stocker NG (1992) Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg 95: 271–275. [PubMed] [Google Scholar]
- 29. Bricker BJ, Halling SM (1994) Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv 1 by PCR. J Clin Microbiol 32: 2660–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ocampo-Sosa AA, Balbín JA, Garcia-Lobo JM (2005) Development of a new PCR assay to identify Brucella abortus biovars 5, 6 and 9 the new subgroup 3b of biovar 3. Vet Microbiol 110: 41–51. [DOI] [PubMed] [Google Scholar]
- 31. Cloeckaert A, Verger JM, Grayon M, Grepinet O (1995) Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella . Microbiol 141: 2111–2121. [DOI] [PubMed] [Google Scholar]
- 32. Hunter PR, Gaston MA (1988) Numeric Index of the discriminatory ability of typing systems: in application of Simpson?s Index of Diversity. J Clin Microbiol 26: 2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langoni H, Ichihara SM, Silva AV, Pardo RB, Tonin FB, et al. (2000) Isolation of Brucella spp. from Milk of Brucellosis positive cows in São Paulo and Minas Gerais States. Braz J Vet Res Anim Sci 6: 444–448. Vercesi Filho AE, Carvalho Faria FG, Madalena FE (2002) Estrutura populacional do rebanho indubrasil registrado no Brasil. Arch Latinoam Prod Anim 10: 86–92. [Google Scholar]
- 34. Vercesi Filho AE, Carvalho Faria FG, Madalena FE (2002) Estrutura populacional do rebanho indubrasil registrado no Brasil. Arch Latinoam Prod Anim 10: 86–92. [Google Scholar]
- 35. Cloeckaert A, Grayon M, Grepinet O, Boumedine KS (2003) Classification of Brucella strains isolated from marine mammals by infrequent restriction site-PCR and development of specific PCR identification tests. Microbes Infect 5: 593–602. [DOI] [PubMed] [Google Scholar]
- 36. Bricker B (2002) Diagnostic strategies used for the identification of Brucella . Vet Microbiol 90: 433–434. [DOI] [PubMed] [Google Scholar]
- 37. Banai M, Mayer I, Cohen A (1990) Isolation, identification and characterization in Israel of Brucella melitensis biovar 1 atypical strains susceptible to dyes and penicillin, indicating the evolution of a new variant. J Clin Microbiol 28: 1057–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Corbel MJ (1991) Identification of dyes-sensitive strains of Brucella mellitensis . J Clin Microbiol 29: 1066–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia MM, Brian B, Gerda M, Ruckerbauer GM (1988) Characterization of an atypical biotype of Brucella abortus . Can J Vet Res 52: 338–342. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOCX)