Structural insights into the type-VIII protein secretion system are described.
Keywords: type-VIII secretion system, curli, CsgG
Abstract
Gram-negative bacteria have eight known protein secretion systems. The type-VIII secretion system, also known as the curli biosynthesis system, is responsible for the formation of aggregative fibres known in Escherichia coli as curli. Curli are extracellular proteinaceous fibres primarily involved in bacterial biofilm formation and attachment to nonbiotic surfaces. The secretion of curli subunits depends on a dedicated lipoprotein, CsgG, which is found to form an oligomeric secretion channel in the outer membrane. A nonlipidated mutant of CsgG was expressed and crystallized in a soluble form. The crystals diffracted to 3.15 Å resolution and belong to space group P1 with a unit cell containing a predicted 16 molecules per asymmetric unit.
1. Introduction
Protein secretion plays a major role in bacterial physiology, with secreted and surface-bound proteins mediating interaction and communication with the surrounding environment, serving as catabolic machinery to liberate nutrients or as virulence factors in the case of pathogenic strains (Lee & Schneewind, 2001 ▶). In monoderm bacteria (Gram-positives), protein secretion entails translocation over the cytoplasmic membrane, which generally occurs through the SEC translocon or twin arginine transport pathway (Chatzi et al., 2013 ▶; Palmer & Berks, 2012 ▶). In diderm bacteria (Gram-negatives), however, a second bilayer, the outer membrane, needs to be overcome. This occurs through one of eight presently known protein secretion pathways which differ in the nature of the secretion cargo, the architecture of the secretion machineries and the mechanism and energy source of the secretion process (Kostakioti et al., 2005 ▶). Of these, the curli secretion pathway, or type-VIII secretion system, is dedicated to the secretion of pro-amyloid protein subunits that assemble into linear extracellular fibres implicated in biofilm formation (Chapman et al., 2002 ▶).
Curli are extracellular proteinaceous fibres of 4–7 nm in diameter and are involved in surface and cell-to-cell contacts in Gram-negative bacteria (Olsén et al., 1989 ▶; Collinson et al., 1991 ▶). Apart from their physiological importance to bacteria, curli fibres share biochemical and structural characteristics with amyloid fibres (Chapman et al., 2002 ▶). Amyloids are formed of fibrous protein aggregates with a cross-β peptide spine (Sunde et al., 1997 ▶), best known from human wasting diseases such as Alzheimer’s and Parkinson’s diseases (Dobson, 1999 ▶; Moreno-Gonzalez & Soto, 2011 ▶). In contrast to the misfolded byproducts of their pathogenic counterparts (Chiti & Dobson, 2006 ▶), curli are the result of a dedicated biosynthetic pathway and are therefore considered as ‘functional’ amyloids (Chapman et al., 2002 ▶; Desvaux et al., 2009 ▶). Curli biosynthesis is directed by two operons, which in Escherichia coli are called csgBAC and csgDEFG (curli-specific genes; Hammar et al., 1995 ▶). CsgA forms the major curli subunit, which like pathogenic amyloids shows nucleation-dependent oligomerization kinetics (Cherny et al., 2005 ▶; Wang et al., 2007 ▶). In curli, this templating role is served by the minor subunit CsgB, which acts as a nucleator for the aggregation of secreted CsgA curli subunits into curli fibres (Hammer et al., 2007 ▶). CsgA is transported across the outer membrane by a specialized outer membrane lipoprotein, CsgG, which works in coordination with the periplasmic and extracellular accessory proteins CsgE and CsgF (Robinson et al., 2006 ▶; Nenninger et al., 2009 ▶, 2011 ▶). CsgE forms a specificity factor for CsgG-mediated transport, whilst CsgF appears to couple CsgA secretion to the CsgB-templated aggregation into extracellular fibres (Robinson et al., 2006 ▶; Nenninger et al., 2009 ▶, 2011 ▶). CsgD forms a transcriptional regulator of the operon csgBAC (Hammar et al., 1995 ▶) and CsgC is a putative oxidoreductase of unknown target, as apart from CsgG the other components of curli biosynthesis lack cysteine (Taylor et al., 2011 ▶).
The bacterial curli (type-VIII) secretion system represents a unique transmembrane transport entity both from a structural and mechanistic perspective. To expand our knowledge on curli transport across the outer membrane, we initiated the structure elucidation of the E. coli curli transporter CsgG. CsgG is a 30 kDa lipoprotein targeted to the outer membrane, where it is thought to form a 2 nm translocation channel by oligomerization (Robinson et al., 2006 ▶; Taylor et al., 2011 ▶). We report the crystallization of outer-membrane-extracted CsgG as well as the isolation and crystallization of a soluble pre-channel form. Bacterial lipoproteins become triacylated at the inner membrane by the subsequent modification of the N-terminal cysteine of the mature protein with a diacylglycerol thioether and aminoacylation of the N-terminus (Narita & Tokuda, 2010 ▶; Okuda & Tokuda, 2011 ▶). Outer-membrane-destined lipoproteins are then shuttled to the outer membrane by the LOL pathway, a conserved multi-protein complex. During this transfer, they traverse the aqueous periplasm as soluble intermediates bound to the LolA shuttle protein via their lipid anchor (Narita & Tokuda, 2010 ▶; Okuda & Tokuda, 2011 ▶). Therefore, we reasoned that in the absence of the lipid anchor CsgG may be present as a soluble form analogous to the pre-channel forms seen in pore-forming proteins (PFPs), where soluble monomeric forms of the PFPs convert into the oligomeric integral membrane-bound forms (Iacovache et al., 2010 ▶).
2. Experimental procedure
2.1. Protein expression and purification
To create CsgG with a C-terminal Strep-tag II under the control of the anhydrotetracyclin-inducible tet promoter, a DNA fragment encoding the csgG gene (accession No. P0AEA2) was amplified by PCR with primers 5′-ATG GTA GGT CTC TAA TGC AGC GCT TAT TTC TTT TGG-3′ and 5′-GTC GTA GGT CTC AGC GCT GGA TTC CGG TGG AAC C-3′ from plasmid pMC2 (Robinson et al., 2006 ▶; kindly provided by Professor Scott Hultgren, Washington University in St Louis, USA). After restriction with BsaI, this fragment was ligated into the pASK-IBA3plus (IBA Life Sciences) vector by standard techniques to create pPG1. As such, pPG1 encodes the native 277-residue CsgG pre-pro-lipoprotein, additionally tagged at its C-terminal with an eight-residue Strep-tag II and a Ser-Ala linker. During maturation of the pre-pro-lipoprotein, the 15-residue leader sequence is removed and the N-terminal Cys is modified by N-palmitoylation and S-diacylglycerol addition, resulting in a 272-residue lipoprotein of approximate molecular mass 30.9 kDa (depending on the nature of the diacylglycerol) and a calculated isoelectric point of 6.3. In order to create pPG2, a DNA fragment encompassing the coding region for the mature CsgG protein in which the N-terminal cysteine was substituted by serine was PCR-amplified using the primers 5′-ATG GTA GGT CTC Agg ccT CTT TAA CCG CCC CGC CTA AAG-3′ and 5′-GTC GTA GGT CTC Agc gct GGA TTC CGG TGG AAC CGA CAT ATG-3′ and cloned into pASK-IBA2 (IBA Life Sciences) by BsaI restriction cloning for in-frame insertion behind the OmpA signal sequence. Expression of pPG2 results in a soluble, mature gene product of 272 residues (including the Strep-tag II and Ser-Ala linker) of molecular mass 30.066 kDa.
Full-length CsgG with a Strep-tag II was overexpressed in E. coli BL21 (DE3) cells transformed with plasmid pPG1. The cells were grown at 310 K to an OD600 nm of 0.6 in Terrific Broth medium. Recombinant protein production was induced with 0.0002% anhydrotetracyclin (Sigma) and the cells were grown at 298 K for a further 16 h before being harvested by centrifugation at 5500 rev min−1 for 15 min. The cell mass was resuspended in 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT and complete Protease Inhibitor Cocktail (Roche). The cells were disrupted at 193 MPa using a TS series cell disruptor (Constant Systems Ltd) and the lysed cell suspension was centrifuged at 10 000g for 15 min at 277 K to remove intact cells and debris. Total cell membranes were prepared by ultacentrifugation of the supernatant at 100 000g for 60 min. Total membrane pellets were resuspended in 25 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT (buffer A) with 1% N-lauroylsarcosine to selectively solubilize the inner membrane (Filip et al., 1973 ▶). Outer membranes were subsequently isolated by ultracentrifugation of the cell membrane suspension (100 000g, 60 min). Outer membranes were washed by resuspension and subsequent ultracentrifugation (100 000g, 60 min) in buffer A containing 2 mM lauryldimethylamine-N-oxide (LDAO; Affymetrix) and finally incubated overnight at 277 K with buffer A containing 1% n-dodecyl-β-d-maltopyranoside (DDM; Affymetrix) for outer membrane protein extraction. The supernatant was collected after a final centrifugation step at 100 000g to remove nonsolubilized membranes and was loaded onto a 5 ml streptavidin column equilibrated in buffer A and 0.01% DDM. The protein was eluted by the addition of 2.5 mM desthiobiotin. The eluted protein was concentrated using spin concentrators (100 kDa molecular-weight cutoff, Merck Millipore) to 5 mg ml−1 (determined based on calculated absorbance at 280 nm) and used for crystallization experiments after buffer exchange to 25 mM Tris–HCl pH 8.0, 500 mM NaCl, 0.01% DDM.
CsgGC1S with a Strep-tag II was produced in the periplasm of E. coli BL21 (DE3) cells. For this, bacterial cells transformed with pPG2 were grown at 310 K to an OD600 nm of 0.6 in LB medium. After lowering the temperature to 288 K, induction was performed overnight by the addition of 0.0002% anhydrotetracyclin (Sigma). Bacterial pellets were resuspended in 25 mM Tris–HCl pH 8, 200 mM NaCl, 20% sucrose, 1 mM EDTA , 0.1 mg ml−1 lysozyme and left shaking in the cold room for 30 min. The cell suspension was centrifuged at 20 000 rev min−1 at 277 K for 45 min to separate spheroplasts and the periplasmic extract. The supernatant was loaded onto a 5 ml streptavidin column pre-equilibrated with 25 mM Tris–HCl, 200 mM NaCl and the immobilized protein was eluted by the addition of 2.5 mM desthiobiotin. Fractions containing CsgGC1S were pooled and dialysed overnight against 25 mM Tris–HCl pH 8.0, 10 mM NaCl and loaded onto a 5 ml Q Sepharose anion-exchange column. Bound proteins were eluted using a linear NaCl gradient and the fractions were analysed on SDS–PAGE (Laemmli, 1970 ▶). Pure CsgGC1S fractions were pooled and dialysed against 25 mM Tris–HCl, 200 mM NaCl. Dialysed protein was concentrated using spin concentrators (10 kDa molecular-weight cutoff, Merck Millipore) to 5.3 mg ml−1 and used for crystallization trials.
2.2. Crystallization
Screening for crystallization conditions for CsgG and CsgGC1S was performed using a Phoenix crystallization robot (Art Robbins Instruments) at 293 K. MemSys, MemStart and MemGold crystallization kits purchased from Molecular Dimensions were used for the initial screening of crystallization condition for CsgG. CsgG protein solution at 5 mg ml−1 and reservoir solution were mixed in a 1:1 ratio (150 nl each) in sub-wells of Intelli-Plates (Hampton Research). Each well of the plates contained 70 µl reservoir solution and was incubated at 293 K. For CsgGC1S, crystallization kits from Jena Biosciences (JBScreen Basic HTS, JBScreen Classic HTS II and JBScreen Wizard 1 and 2) were used with a protein solution at 5.3 mg ml−1 in 25 mM Tris–HCl pH 8.0, 200 mM NaCl. 150 nl of the protein solution was mixed with 150 nl precipitant solution in the 96-well sitting-drop plates and equilibrated against 70 µl reservoir solution at 293 K. An initial hit for CsgGC1S appeared in the JBScreen Basic screen after 1 week with a precipitant solution consisting of 8% PEG 4000, 100 mM sodium acetate pH 4.2. For further optimizations, drops consisting of 0.5 µl protein solution and 0.5 µl precipitant solution were used at 293 K in 96-well sitting-drop plates.
2.3. X-ray diffraction and data processing
The crystals of CsgGC1S were cryoprotected using reservoir solution supplemented with 15% glycerol before flash-cooling in liquid nitrogen. X-ray data were collected at 100 K using an ADSC 315 detector on beamline I04 of the Diamond Light Source (Didcot, England). Data were collected over a total oscillation range of 220° using oscillation steps of 1° per image. The raw data were processed and scaled using XDS (Kabsch, 2010 ▶) and the CCP4 package (Winn et al., 2011 ▶).
3. Results
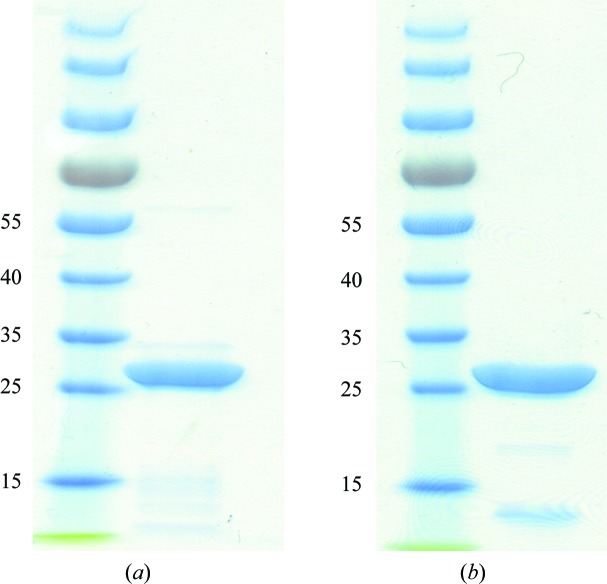
CsgG with a Strep-tag II was expressed in the bacterial outer membrane of E. coli strain BL21 (DE3). Detergent-extracted CsgG was purified using streptavidin affinity chromatography and was concentrated to 5 mg ml−1 for crystallization (Fig. 1 ▶ a). Preliminary microcrystals were obtained in a buffer condition consisting of 100 mM Tris–HCl pH 6.5, 100 mM CaCl2, 13% PEG 2000 MME (Fig. 2 ▶ a). Current efforts are focused on optimizing purification conditions and the detergents used for solubilization and crystallization in order to obtain crystals of sufficient size and diffraction quality to determine the X-ray structure of the full-length CsgG protein.
Figure 1.
SDS–PAGE of purified CsgG (a) and CsgGC1S (b) used for the crystallization trials. Molecular-weight markers are labelled in kDa.
Figure 2.
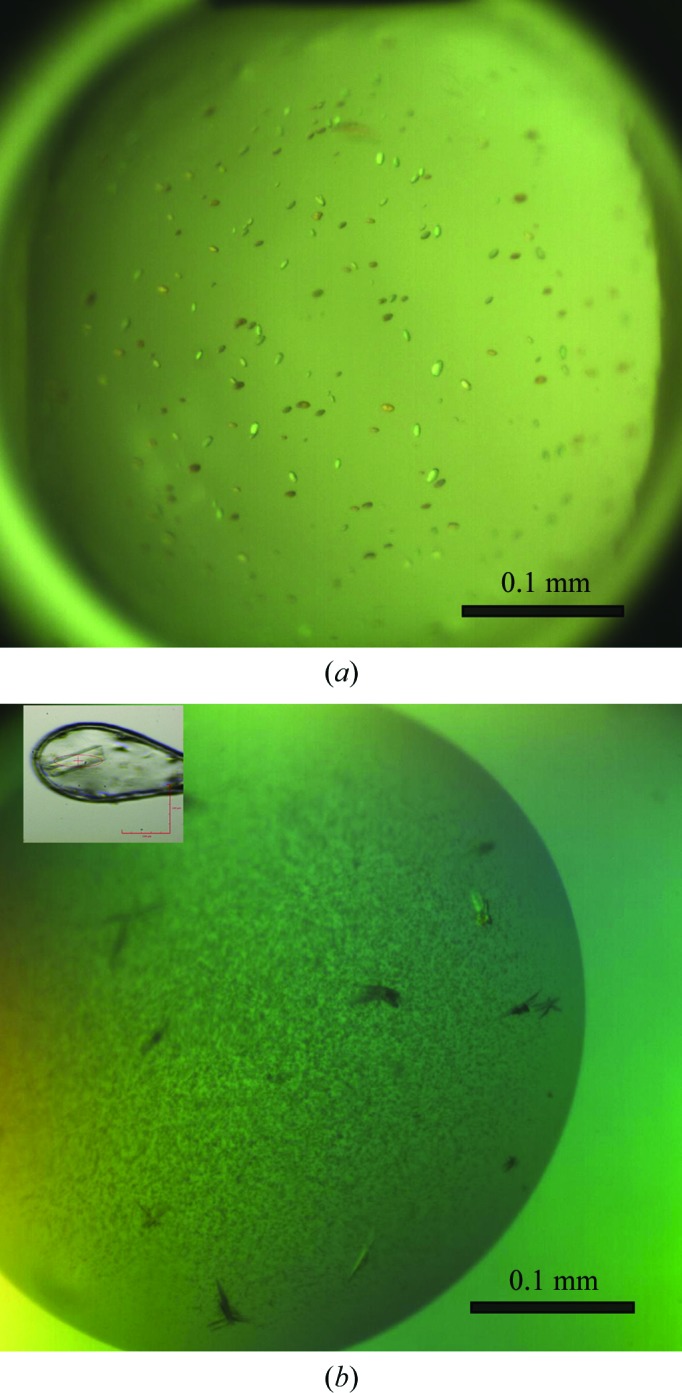
Crystals of CsgG (a) and CsgGC1S (b).
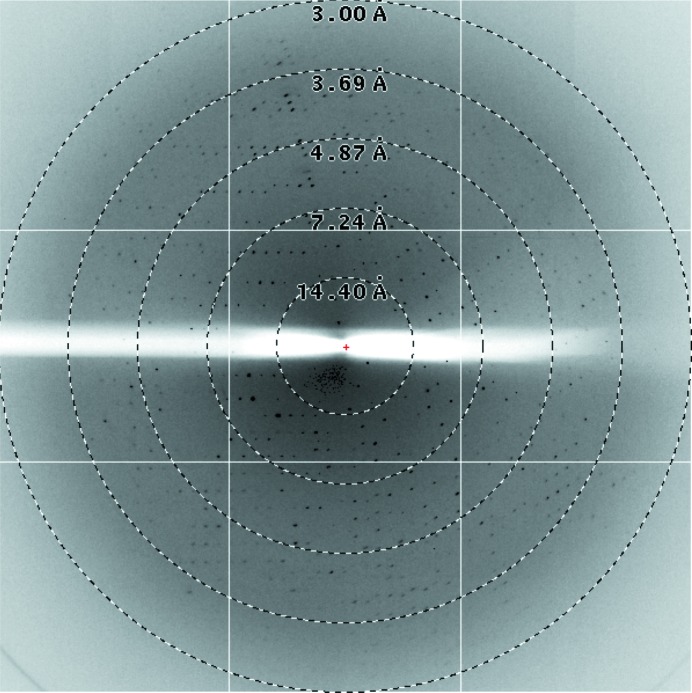
The N-terminal Cys mutant CsgGC1S from E. coli was overexpressed in soluble form in the periplasm of the expression strain. Purification of the CsgGC1S was performed using streptavidin affinity chromatography followed by ion-exchange chromatography (Fig. 1 ▶ b). The average yield of CsgGC1S was 0.2 mg per litre of E. coli culture. CsgGC1S at 5.3 mg ml−1 concentration was used for crystallization. The best diffracting crystals of CsgGC1S appeared in precipitant solution consisting of 100 mM sodium malonate pH 7.2, 8% PEG 4000, 100 mM sodium acetate pH 4 (Fig. 2 ▶ b). The crystals took 2 weeks to grow to dimensions of about 0.07 × 0.03 × 0.005 mm. A data set was collected from the best crystal on beamline I04 at the Diamond Light Source to a resolution of 3.15 Å (Fig. 3 ▶). A resolution cutoff of 3.35 Å was used during the data processing. The spots were integrated and scaled using XDS and the CCP4 suite. A total of 156 243 reflections were recorded, of which 65 368 were unique. The overall R merge is 14.6% (52.0% in the outermost resolution shell) and R p.i.m. (Weiss & Hilgenfeld, 1997 ▶) is 0.117 (0.421 in the outermost resolution shell), with a completeness of 98.6% (98.5% in the outermost resolution shell). The data-collection statistics are summarized in Table 1 ▶.
Figure 3.
X-ray diffraction pattern image of a CsgGC1S crystal.
Table 1. Data-collection statistics for CsgGC1S .
Values in parentheses are for the outermost resolution shell.
| X-ray source | I04, DLS |
| X-ray wavelength (Å) | 0.9795 |
| Temperature (K) | 100 |
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 101.37, b = 103.32, c = 141.62, α = 111.12, β = 90.70, γ = 118.13 |
| Crystal mosaicity (°) | 0.30 |
| Resolution range (Å) | 46.9–3.3 (3.53–3.35) |
| Total/unique reflections | 156243/65368 (23038/9579) |
| R merge † (%) | 14.6 (52.0) |
| R r.i.m ‡ (%) | 18.9 (67.3) |
| R p.i.m § (%) | 11.7 (42.1) |
| Data completeness (%) | 98.6 (98.5) |
| Average I/σ(I) | 6.2 (2.0) |
| Multiplicity | 2.4 (2.4) |
R
merge = 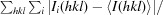
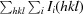 , where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry- related reflections.
, where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry- related reflections.
R
r.i.m = 
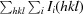 .
.
R
p.i.m = 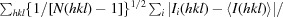
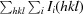 .
.
The CsgGC1S crystals belonged to space group P1, with unit-cell parameters a = 101.37, b = 103.32, c = 141.62 Å, α = 111.12, β = 90.70, γ = 118.13°. The calculated unit-cell volume is compatible with 12–20 molecules per asymmetric unit, giving a solvent content of 65–40%. Based on the average density for protein crystals, the most likely number of molecules per asymmetric unit is 16, corresponding to 50% solvent content and a Matthews coefficient of 2.48 Å3 Da−1 (Matthews, 1968 ▶). To gain further insight in the noncrystallographic symmetry in the P1 crystals, the self-rotation function was analysed using the MOLREP program (Vagin & Teplyakov, 2010 ▶). Self-rotation function peaks at κ = 180° are shown in Fig. 4 ▶. There are eight distinct peaks on the calculated self-rotation function of CsgGC1S on a twofold symmetry axis. Together with the estimated 16 molecules of CsgGC1S per asymmetric unit, the self-rotation function indicates the presence of a hexadecameric structure with D8 symmetry (dimer of octamers) in CsgGC1S crystals. Full-length CsgG has been shown to form oligomeric pores in the outer membrane; hence the octamer of soluble form CsgGC1S could represent an oligomeric pre-channel form of the transporter.
Figure 4.
κ = 180° section of the self-rotation function calculated for the CsgGC1S crystal.
No homologous structures of CsgG are presently available. Attempts to solve the phase problem are therefore focusing on the production of selenomethionine-labelled protein (the native sequence contains six methionines) for experimental phasing by single or multiple anomalous dispersion (SAD or MAD).
Acknowledgments
We are grateful to the beamline staff of I04 at the Diamond Light Source, England for support with the data collection and processing. This work was supported by VIB through grant PRJ9 and equipment grant UABR/09/005 from the Hercules Foundation.
References
- Chapman, M. R., Robinson, L. S., Pinkner, J. S., Roth, R., Heuser, J., Hammar, M., Normark, S. & Hultgren, S. J. (2002). Science, 295, 851–855. [DOI] [PMC free article] [PubMed]
- Chatzi, K. E., Sardis, M. F., Karamanou, S. & Economou, A. (2013). Biochem. J. 449, 25–37. [DOI] [PubMed]
- Cherny, I., Rockah, L., Levy-Nissenbaum, O., Gophna, U., Ron, E. Z. & Gazit, E. (2005). J. Mol. Biol. 352, 245–252. [DOI] [PubMed]
- Chiti, F. & Dobson, C. M. (2006). Annu. Rev. Biochem. 75, 333–366. [DOI] [PubMed]
- Collinson, S. K., Emödy, L., Müller, K. H., Trust, T. J. & Kay, W. W. (1991). J. Bacteriol. 173, 4773–4781. [DOI] [PMC free article] [PubMed]
- Desvaux, M., Hébraud, M., Talon, R. & Henderson, I. R. (2009). Trends Microbiol. 17, 139–145. [DOI] [PubMed]
- Dobson, C. M. (1999). Trends Biochem. Sci. 24, 329–332. [DOI] [PubMed]
- Filip, C., Fletcher, G., Wulff, J. L. & Earhart, C. F. (1973). J. Bacteriol. 115, 717–722. [DOI] [PMC free article] [PubMed]
- Hammar, M., Arnqvist, A., Bian, Z., Olsén, A. & Normark, S. (1995). Mol. Microbiol. 18, 661–670. [DOI] [PubMed]
- Hammer, N. D., Schmidt, J. C. & Chapman, M. R. (2007). Proc. Natl Acad. Sci. USA, 104, 12494–12499. [DOI] [PMC free article] [PubMed]
- Iacovache, I., Bischofberger, M. & van der Goot, F. G. (2010). Curr. Opin. Struct. Biol. 20, 241–246. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kostakioti, M., Newman, C. L., Thanassi, D. G. & Stathopoulos, C. (2005). J. Bacteriol. 187, 4306–4314. [DOI] [PMC free article] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Lee, V. T. & Schneewind, O. (2001). Genes Dev. 15, 1725–1752. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Moreno-Gonzalez, I. & Soto, C. (2011). Semin. Cell Dev. Biol. 22, 482–487. [DOI] [PMC free article] [PubMed]
- Narita, S. & Tokuda, H. (2010). Methods Mol. Biol. 619, 117–129. [DOI] [PubMed]
- Nenninger, A. A., Robinson, L. S., Hammer, N. D., Epstein, E. A., Badtke, M. P., Hultgren, S. J. & Chapman, M. R. (2011). Mol. Microbiol. 81, 486–499. [DOI] [PMC free article] [PubMed]
- Nenninger, A. A., Robinson, L. S. & Hultgren, S. J. (2009). Proc. Natl Acad. Sci. USA, 106, 900–905. [DOI] [PMC free article] [PubMed]
- Okuda, S. & Tokuda, H. (2011). Annu. Rev. Microbiol. 65, 239–259. [DOI] [PubMed]
- Olsén, A., Jonsson, A. & Normark, S. (1989). Nature (London), 338, 652–655. [DOI] [PubMed]
- Palmer, T. & Berks, B. C. (2012). Nature Rev. Microbiol. 10, 483–496. [DOI] [PubMed]
- Robinson, L. S., Ashman, E. M., Hultgren, S. J. & Chapman, M. R. (2006). Mol. Microbiol. 59, 870–881. [DOI] [PMC free article] [PubMed]
- Sunde, M., Serpell, L. C., Bartlam, M., Fraser, P. E., Pepys, M. B. & Blake, C. C. (1997). J. Mol. Biol. 273, 729–739. [DOI] [PubMed]
- Taylor, J. D., Zhou, Y., Salgado, P. S., Patwardhan, A., McGuffie, M., Pape, T., Grabe, G., Ashman, E., Constable, S. C., Simpson, P. J., Lee, W. C., Cota, E., Chapman, M. R. & Matthews, S. J. (2011). Structure, 19, 1307–1316. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wang, X., Smith, D. R., Jones, J. W. & Chapman, M. R. (2007). J. Biol. Chem. 282, 3713–3719. [DOI] [PMC free article] [PubMed]
- Weiss, M. S. & Hilgenfeld, R. (1997). J. Appl. Cryst. 30, 203–205.
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.