A comparative study of vapor diffusion versus liquid–liquid diffusion methods used for the crystallization of EndoS is reported. X-ray diffraction data to 2.6 and 1.9 Å resolution were collected for wild-type endoglycosidase and glycosynthase constructs of EndoS, respectively.
Keywords: endoglycosidase S, liquid–liquid diffusion, Streptococcus pyogenes
Abstract
Endoglycosidase S (EndoS) is an enzyme secreted by Streptococcus pyogenes that specifically hydrolyzes the β-1,4-di-N-acetylchitobiose core glycan on immunoglobulin G (IgG) antibodies. One of the most common human pathogens and the cause of group A streptococcal infections, S. pyogenes secretes EndoS in order to evade the host immune system by rendering IgG effector mechanisms dysfunctional. On account of its specificity for IgG, EndoS has also been used extensively for chemoenzymatic synthesis of homogeneous IgG glycoprotein preparations and is being developed as a novel therapeutic for a wide range of autoimmune diseases. The structural basis of its enzymatic activity and substrate specificity, however, remains unknown. Here, the purification and crystallization of EndoS are reported. Using traditional hanging-drop and sitting-drop vapor-diffusion crystallization, crystals of EndoS were grown that diffracted to a maximum of 3.5 Å resolution but suffered from severe anisotropy, the data from which could only be reasonably processed to 7.5 Å resolution. When EndoS was crystallized by liquid–liquid diffusion, it was possible to grow crystals with a different space group to those obtained by vapor diffusion. Crystals of wild-type endoglycosidase and glycosynthase constructs of EndoS grown by liquid–liquid diffusion diffracted to 2.6 and 1.9 Å resolution, respectively, with a greatly diminished anisotropy. Despite extensive efforts, the failure to reproduce these liquid–liquid diffusion-grown crystals by vapor diffusion suggests that these crystallization methods each sample a distinct crystallization space.
1. Introduction
Streptococcus pyogenes EndoS is a bacterial endoglycosidase that specifically hydrolyzes the β-1,4-di-N-acetylchitobiose core glycan on human immunoglobulin G (IgG) antibodies (Collin & Olsen, 2001a ▶). This enzyme is highly specific for, and has activity on, natively folded IgG (Collin & Olsen, 2001a ▶,b ▶), by which it contributes to increased bacterial survival, on account of reduced IgG binding to Fcγ receptors and impaired complement pathway activation (Collin et al., 2002 ▶). EndoS releases the glycan linked to residue Asn297 of the human Fc region CH2 domain, affecting the local structure of IgG (Arnold et al., 2007 ▶; Lund et al., 1996 ▶), reducing IgG binding to both complement factor C1q (Krapp et al., 2003 ▶) and to Fcγ receptors (Nimmerjahn & Ravetch, 2008 ▶). These binding events regulate two of the primary effector functions of IgG antibodies and, consequently, EndoS endows S. pyogenes with a survival advantage when infecting the host. This enzymatic property of EndoS, however, can be leveraged for the treatment of autoimmune diseases that are dependent on autoantibodies. Indeed, the administration of recombinant EndoS protein as an in vivo modulator of IgG effector functions has shown great promise in the treatment of numerous autoimmune conditions in animal models (Nandakumar et al., 2007 ▶; Albert et al., 2008 ▶; Collin et al., 2008 ▶; van Timmeren et al., 2010 ▶; Allhorn et al., 2010 ▶; Tradtrantip et al., 2013 ▶; Hirose et al., 2012 ▶; Lood et al., 2012 ▶). EndoS is also being used increasingly for in vitro glycan remodeling in order to modulate the technological and therapeutic properties of IgG (Goodfellow et al., 2012 ▶; Huang et al., 2012 ▶). Despite its role in immune evasion by a common and sometimes deadly pathogen, its potential as a treatment for numerous autoimmune diseases, and its expanding use in IgG glycosylation manipulations for biotechnological and therapeutic purposes, the structural basis of the enzymatic activity of EndoS remains poorly understood.
The primary impediment to determining the X-ray crystal structure of any protein, including EndoS, is the growth of crystals that diffract to sufficient resolution. Currently, the most widely used techniques to grow protein crystals are batch and vapor-diffusion methods (Ducruix & Giegé, 2004 ▶). In both, protein and precipitant solution are mixed immediately (i.e. batch) or equilibrated with a reservoir solution (i.e. vapor diffusion). Unfortunately, many proteins of biological interest are refractory to crystallization by these methods, or the crystals obtained using them do not diffract to sufficient resolution to allow for structure determination. Liquid–liquid diffusion, also known as counterdiffusion, represents an alternative approach to protein crystallization. In liquid–liquid diffusion, protein and precipitant solutions are initially prepared in contact with one another in a narrow geometry (e.g. in a microcapillary), such that diffusion is limiting (Otálora et al., 2009 ▶). The diffusion between the two solutions allows a broader screening of precipitant conditions than batch or vapor-diffusion methodology, which can facilitate the crystallization process (Dhouib et al., 2009 ▶; Emamzadah et al., 2009 ▶; Hansen et al., 2002 ▶; Ng et al., 2003 ▶; Ng, Clark et al., 2008 ▶). This crystallization method is being used increasingly in both the search for initial crystallization conditions and in crystal growth optimization owing to the recent development of a range of liquid–liquid-diffusion-based crystallization tools, including microfluidic devices (Hansen et al., 2002 ▶; Zheng et al., 2003 ▶, 2004 ▶; Ng, Stevens et al., 2008 ▶; Gerdts et al., 2006 ▶; Zhou et al., 2007 ▶; Du et al., 2009 ▶), microcapillary-based microbatch plastic tubes (Yadav et al., 2005 ▶; Kalinin & Thorne, 2005 ▶) and glass capillaries (Gavira et al., 2002 ▶; Ng et al., 2003 ▶; Ng, Clark et al., 2008 ▶; García-Ruiz, 2003 ▶; Garcia-Ruiz & Ng, 2007 ▶; Kurz et al., 2012 ▶). Crystal Formers (Microlytic) are commercially available microfluidic devices that have been used successfully for the initial crystallization screening of several well characterized proteins (Stojanoff et al., 2011 ▶). In this device, the mixing of precipitant and protein solutions occurs primarily by diffusive equilibration based on the concentration difference established in the crystallization channel.
Here, we report a comparative analysis of EndoS crystallization by two methods: vapor diffusion, using both hanging and sitting drops, and liquid–liquid diffusion, using Crystal Formers microfluidic devices. Although we grew crystals of EndoS using both methods, the diffraction of those grown by vapor diffusion was highly anisotropic and of insufficient resolution for structure determination. Crystals grown by liquid–liquid diffusion, conversely, diffracted to 2.6 and 1.9 Å resolution for wild-type endoglycosidase and glycosynthase constructs of EndoS, respectively, and produced high-quality native data sets. Our inability to replicate these crystals by vapor diffusion despite searching an exhaustive range of similar crystallization conditions suggests that these distinct diffusion methods for protein crystallization each sample a distinct crystallization space.
2. Materials and methods
2.1. Protein purification
Wild-type EndoS comprising amino acids 98–995, EndoSWT(98–995), was amplified via PCR from EndoS-GST and cloned into a modified form of the pCPD vector (pCPD-L) containing the C-terminal fusion protein from Vibrio cholerae MARTX toxin cysteine protease domain (CPD) (Lomino, 2011 ▶; Shen et al., 2009 ▶). The N-terminal 97 residues derive from a signal peptide and a putative coiled-coil structure, which is responsible for forming oligomers of EndoS and is likely to inhibit crystallization. It was necessary to add a leucine at the C-terminus of all EndoS constructs in order to facilitate cleavage of the CPD domain and to purify the protein. EndoS with this additional residue retains enzymatic activity. EndoSWT(98–445), EndoSWT(98–966), EndoSWT(138–995) and EndoSWT(446–995) variants were amplified by PCR from EndoSWT(98–995)-CPD-L and were introduced similarly into the pCPD-L vector. EndoSWT(98–445) and EndoSWT(446–995) are the two major degradation products of SpeB, another enzyme secreted by S. pyogenes that inactivates EndoS (Allhorn et al., 2008 ▶). The EndoSWT(98–966) and EndoSWT(138–995) constructs were selected because the N- and C-termini of the protein were predicted to be disordered using GlobPlot (Linding et al., 2003 ▶). The glycosynthase mutant EndoSD233Q(98–995), which transfers glycans to IgG as opposed to removing them as does the wild-type enzyme, was generated by site-directed mutagenesis in the pCPD-L vector.
All EndoS CPD-L variants were produced in Escherichia coli BL21(DE3) cells (Novagen) grown in 2×YT medium supplemented with 50 µg ml−1 ampicillin. Cultures were grown at 310 K to an OD600 of 0.6–0.8, at which point the temperature was lowered to 291 K for 1 h. Induction was triggered with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 291 K overnight. Cells were harvested by centrifugation and lysed by sonication using 50 mM Tris–HCl pH 7.5, 500 mM NaCl, 10% glycerol. CPD fusion proteins were purified by Ni2+-immobilized metal-affinity chromatography followed by overnight treatment with 1 mM phytic acid. EndoS constructs were further purified via size-exclusion chromatography (SEC) in 20 mM Tris–HCl pH 7.5, 50 mM NaCl. EndoSWT(98–445) and EndoSWT(138–995) expressed but degraded immediately after purification. Therefore, we attempted to crystallize the EndoSWT(98–995), EndoSD233Q(98–995), EndoSWT(98–966) and EndoSWT(446–995) proteins.
2.2. Crystallization
2.2.1. Vapor diffusion
Initial crystallization screening, using The Classics and JCSG+ Suites (Qiagen) and the PEG/Ion and SaltRx screens (Hampton Research), of EndoSWT(98–995), EndoSD233Q(98–995), EndoSWT(98–966) and EndoSWT(446–995) (each at 10–20 mg ml−1 in 20 mM Tris–HCl, 50 mM NaCl pH 7.5) was performed by sitting-drop vapor diffusion. Each drop consisted of 0.2 µl each of protein and precipitant solutions in a 1:1 ratio of protein:precipitant. These were set up at 298 K in 96-well sitting-drop iQ-plates (TTP LabTech) using a Gryphon crystallization robot (Art Robbins Instruments).
EndoSWT(98–995) and EndoSD233Q(98–995) were crystallized by sitting-drop vapor diffusion in 20% polyethylene glycol (PEG) 3350, 0.2 M potassium citrate. The crystals were improved by microseeding using a Seed Bead Kit (Hampton Research) according to the manufacturer’s instructions and the addition of 4% trimethylamine N-oxide dihydrate in hanging drops (2 µl with a 1:1 ratio of protein:precipitant) in VDX48 plates with sealant and siliconized glass cover slides (Hampton Research). The crystals measured approximately 50 × 200 × 300 µm.
EndoSWT(98–966) was crystallized by sitting-drop vapor diffusion at 298 K after 4 d in 0.2 M sodium chloride, 2 M ammonium sulfate, 0.1 M sodium cacodylate pH 7.2. The crystals were improved by microseeding in hanging drops as described above. The crystals measured approximately 50 × 15 × 70 µm.
EndoSWT(446–995) was crystallized by sitting-drop vapor diffusion at 298 K in 18% PEG 3350, 8% Tacsimate pH 6.0. Clusters of thin needles appeared after 2 d and could not be improved by microseeding. We were unable to harvest single crystals.
2.2.2. Liquid–liquid diffusion
Crystals of EndoSWT(98–995) and EndoSD233Q(98–995) (each 25 mg ml−1 in 20 mM Tris, 50 mM NaCl pH 7.5) were obtained by liquid–liquid diffusion in Crystal Formers (Microlytic). We initiated crystallization screens using the companion superSmart and PurePEGs-48 screens (Microlytic) and obtained crystals at 298 K for both constructs in three different conditions: (i) 0.2 M ammonium acetate, 0.1 M bis-tris–HCl pH 5.5, (ii) 25% PEG 3350, 0.2 M calcium chloride, 0.1 M Tris–HCl pH 8.0 and (iii) 0.2 M lithium acetate, 20% PEG 3350 using 0.7 µl each of protein and precipitant in SBS High Throughput Crystal Former plates (Microlytic). Small fragile crystals were obtained in conditions (i) and (ii), while crystals with substantially larger dimensions were obtained at 298 K in condition (iii). Crystals were reproduced using 1.4 µl of protein and precipitant solutions in Crystal Former plates (Microlytic). Crystals measuring approximately 400 × 200 × 500 µm grew within 2 weeks. Crystals of identical dimensions grew overnight when microseeding [using a Seed Bead Kit (Hampton Research) as described above] in new microcapillaries using the same crystallization condition.
2.2.3. Replicating crystals grown by liquid–liquid diffusion using vapor diffusion
We prepared a 96-condition sitting-drop vapor-diffusion crystallization screen based on liquid–liquid diffusion condition (iii). The range of precipitant concentration was 0.05–0.4 M lithium acetate and 5–30% PEG 3350. Each of the 96 conditions was tested at 298 K against the following protein concentrations: 1, 5, 10, 15, 20 and 25 mg ml−1 in 96-well sitting-drop iQ-plates (TTP Lab Tech) using a Gryphon crystallization robot (Art Robbins Instruments). All drops were either clear or precipitated; no crystals or nucleation were observed. Because crystals grew by liquid–liquid diffusion in the absence of microseeding, we did not perform cross-seeding experiments in which seeds derived from liquid–liquid diffusion crystals were used to seed vapor-diffusion experiments in our efforts to replicate the crystallization process.
2.3. Data collection
For data collection crystals were flash-cooled at 100 K. Crystals obtained by liquid–liquid diffusion were harvested by scoring the sealing film on the back of the Crystal Former with a scalpel. Once exposed, 10 µl of 20% ethylene glycol in mother liquor was added to the open channel to prevent drying of the crystals during manipulation and to protect the crystal during subsequent flash-cooling. The optimal cryoprotectant for crystals obtained by vapor diffusion was 27% PEG 3350, 0.2 M potassium citrate, 4% trimethylamine N-oxide dihydrate, 5% glycerol for wild-type and mutant EndoS(98–995) and 0.2 M sodium chloride, 2 M ammonium sulfate, 0.1 M sodium cacodylate pH 7.2, 20% glycerol for EndoSWT(98–966). X-ray diffraction data of EndoSWT(98–995), EndoSD233Q(98–995) and EndoSWT(98–966) crystals obtained by vapor diffusion were collected using a Rigaku-MSC MicroMax-007 generator equipped with an R-AXIS IV++ image-plate detector and an Oxford Instruments Cryojet. Low-resolution diffraction (>10 Å) was observed for each crystal. EndoSWT(98–995) and EndoSD233Q(98–995) crystals obtained by liquid–liquid diffusion, conversely, each diffracted to a resolution of 3.5 Å when flash-cooled in a similar fashion and using the same X-ray source and detector. Synchrotron X-ray diffraction data for EndoSWT(98–995) and EndoSD233Q(98–995) crystals obtained using liquid–liquid diffusion were collected using a MAR 300 CCD detector on the 23-ID-B beamline at the Advanced Photon Source (APS), Argonne National Laboratory, Illinois, USA and data for EndoSD233Q(98–995) crystals obtained using vapor diffusion were collected using a MAR 300 CCD detector on the 23-ID-D beamline at APS. All data-collection statistics are shown in Table 1 ▶. The data were processed and indexed with XDS (Kabsch, 2010a ▶) and scaled with XSCALE (Kabsch, 2010b ▶).
Table 1. Data-collection statistics.
Values in parentheses are for the outermost resolution shell.
| EndoSD233Q(98–995) | EndoSD233Q(98–995) | EndoSWT(98–995) | |
|---|---|---|---|
| Vapor diffusion | Liquid–liquid diffusion | Liquid–liquid diffusion | |
| Space group | I422 | P212121 | P212121 |
| Unit-cell parameters | |||
| a (Å) | 189.6 | 92.6 | 92.3 |
| b (Å) | 189.6 | 96.1 | 94.5 |
| c (Å) | 489.9 | 141.2 | 142.8 |
| α = β = γ (°) | 90 | 90 | 90 |
| Wavelength (Å) | 1.069 | 0.979 | 0.979 |
| Resolution (Å) | 30–7.55 (7.98–7.55) | 30–1.91 (2.02–1.91) | 30–2.63 (2.77–2.63) |
| R merge (%) | 10.6 (36.2) | 4.6 (47.9) | 9.4 (63.7) |
| 〈I/σ(I)〉 | 12.15 (2.42) | 10.6 (1.6) | 9.8 (1.6) |
| Completeness (%) | 93.4 (66.8) | 96.8 (95.2) | 97.8 (90.6) |
| Multiplicity | 4.8 (2.4) | 2.0 (1.8) | 2.6 (2.5) |
3. Results and discussion
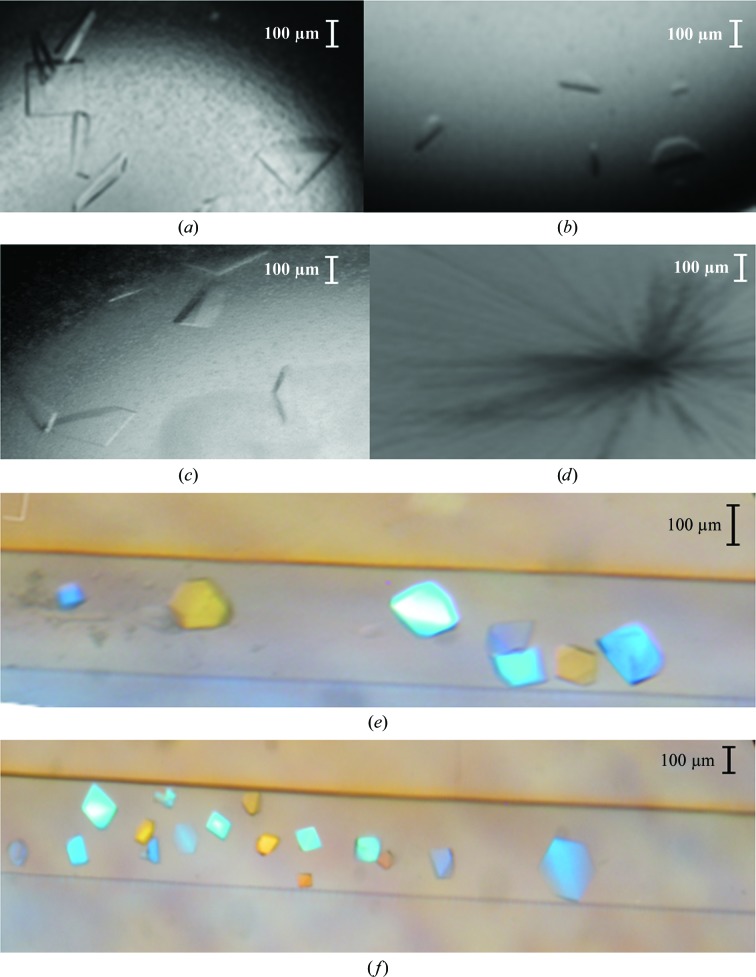
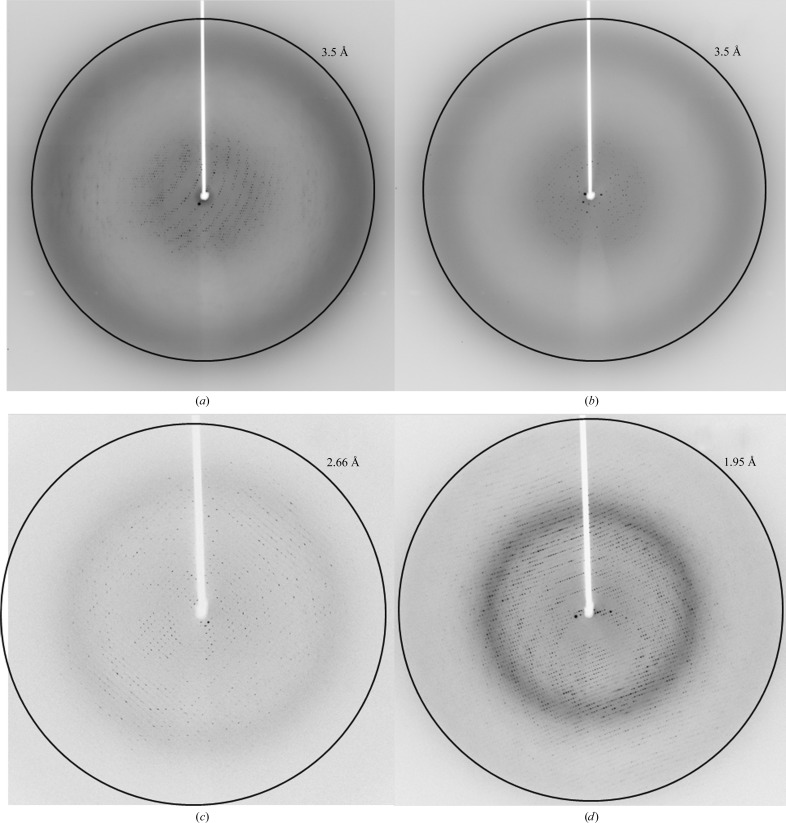
We obtained crystals of EndoSWT(98–995), EndoSWT(98–966), EndoSWT(446–995) and EndoSD233Q(98–995) by sitting-drop vapor diffusion from initial crystallization screening trials. All crystals were optimized by microseeding and reproduced by hanging-drop vapor diffusion (Figs. 1 ▶ a–1 ▶ d). However, for EndoSWT(446–995) only clusters of thin needles were obtained and we were not able to harvest any single crystals for subsequent X-ray diffraction analysis. EndoSWT(98–995) and EndoSD233Q(98–995) crystals grown by vapor diffusion achieved a size of ∼50 × 200 × 300 µm, but were extremely fragile and difficult to manipulate. Diffraction of these crystals could be observed on some images to a maximum of 3.5 Å resolution (Figs. 2 ▶ a and 2 ▶ b). The data, however, were weak or non-existent at higher resolution (Fig. 2 ▶ a) and exhibited severe diffraction anisotropy with resolution extending to 3.5 Å in one direction but only to 5 Å in other directions (Fig. 2 ▶ a). The highest quality data set collected for vapor-diffusion-grown EndoSD233Q(98–995) crystals was processed to a resolution limit of 7.5 Å along the a and b axes and to a resolution limit of 6.6 Å along the c axis (Strong et al., 2006 ▶). These crystals belong to space group I422, with unit-cell parameters a = b = 189.6, c = 489.9 Å, typical of large proteins or protein assemblies data collection and analysis often becomes challenging owing to overlaps, low intensities and high structural heterogeneity (Grimes & Stuart, 1998 ▶).
Figure 1.
Crystals of (a) EndoSWT(98–995), (b) EndoSWT(98–966), (c) EndoSD233Q(98–995) and (d) EndoSWT(446–995) obtained by vapor diffusion. Crystals of (e) EndoSWT(98–995) and (f) EndoSD233Q(98–995) obtained by liquid–liquid diffusion.
Figure 2.
Diffraction images of (a) a single EndoSD233Q(98–995) crystal obtained by vapor diffusion at ϕ = 1° and (b) at ϕ = 91°. Diffraction images of (c) single EndoSWT(98–995) and (d) EndoSD233Q(98–995) crystals obtained by liquid–liquid diffusion.
Since the quality of the X-ray diffraction data derived from all EndoS crystals grown by vapor diffusion was insufficient for structure determination, we attempted crystallization of EndoSWT(98–995) and EndoSD233Q(98–995) by liquid–liquid diffusion using commercially available microfluidic devices. We obtained crystals of both EndoSWT(98–995) and EndoSD233Q(98–995), each in three different conditions. The highest quality crystals, however, were grown in 0.2 M lithium acetate, 20% PEG 3350 (Figs. 1 ▶ e and 1 ▶ f), and the crystallization rate was increased from 2 weeks to overnight by microseeding. Although microseeding increased the speed of the crystallization process, the diffraction quality of the resulting crystals was comparable to that of crystals grown without microseeding. There was a substantial improvement in the data collected for EndoSWT(98–995) and EndoSD233Q(98–995) crystals obtained by liquid–liquid diffusion (Figs. 2 ▶ c and 2 ▶ d) relative to those obtained by vapor diffusion; they exhibited not only higher resolution, but also reduced anisotropy. The data were processed to an upper limit of 2.6 Å resolution for EndoSWT(98–995) crystals and 1.9 Å resolution for EndoSD233Q(98–995) crystals. As opposed to the crystals of the same EndoS constructs grown by vapor diffusion, these crystals belonged to space group P212121 with markedly smaller unit-cell parameters a = 92.6, b = 96.1, c = 141.2 Å.
We attempted to reproduce the EndoSD233Q(98–995) crystals that diffracted to 1.9 Å resolution obtained by liquid–liquid diffusion using vapor diffusion. To do so, we set up crystallization trials using the same crystallization reagents as in the liquid–liquid diffusion condition, but with a broader range of concentrations (i.e., 0.05–0.4 M lithium acetate and 5–30% PEG 3350), as well as numerous EndoSD233Q(98–995) concentrations (i.e., 1, 5, 10, 15, 20 and 25 mg ml−1) in 96-well sitting-drop vapor-diffusion plates. This set of conditions reproduces the entire range of possible conditions created by the liquid–liquid diffusion experiment. Surprisingly, we observed only crystallization drops that were either clear or precipitate, but no crystals of any size or shape, nor any indication of nucleation.
In protein crystallization, not only is the combination of buffer, salt and precipitant important, but the method by which the crystallization reagents are introduced to the protein solution can also have a significant impact on crystal growth and quality (García-Ruiz, 2003 ▶; Gavira et al., 2002 ▶). Our crystallization experiments using various constructs of EndoS are a case in point. Not only did liquid–liquid diffusion produce EndoSWT(98–995) and EndoSD233Q(98–995) crystals that were far superior to those obtained by vapor diffusion, we were also unable to replicate these crystals in similar conditions using the latter method. It should also be noted that none of the several conditions in which crystals grew by liquid–liquid diffusion were identical to those we previously discovered to grow crystals by vapor diffusion. Thus, it appears as if these diffusion methods each interrogate a distinct, although undoubtedly overlapping, crystallization space. The major difference between vapor and liquid–liquid diffusion crystallization methods is that in the latter the mother liquor and protein solution are mixed by diffusion limited by the geometry of a capillary (Otálora et al., 2009 ▶). This avoids convective forces prior to equilibrium caused by immediate mixing of protein and crystallization reagent solutions, such as in batch and vapor diffusion (Howard et al., 2009 ▶). Furthermore, in liquid–liquid diffusion the critical supersaturation for nucleation is reached more slowly, thus allowing a wider and denser screening of precipitation conditions than in batch or vapor-diffusion procedures. This phenomenon is most easily seen in Fig. 1 ▶(e), in which numerous small crystals have grown close to the precipitation chamber (left side of the capillary) to a point at which the concentration of protein and/or precipitant becomes optimal for the growth of the largest crystals (right side of the capillary). These larger crystals yielded our highest resolution data.
4. Conclusions
In this study, we provide a comparative analysis of liquid–liquid versus vapor-diffusion methods in the crystallization of EndoS. Although we could crystallize various constructs of EndoS using either method, we were only able to obtain crystals that diffracted to high resolution using liquid–liquid diffusion. Our failure to replicate these crystals by vapor diffusion in conditions encompassing all those produced by the liquid–liquid diffusion trials support the concept that kinetic, not just equilibrium, and physical, not just chemical, forces can play important roles in protein crystal growth and quality. These data suggest that these two crystallization methods did not necessarily interrogate an identical crystallization space and that the likelihood of successfully discovering conditions under which quality crystals of a given protein grow can be increased not only by increasing the number of crystallization reagent concentrations, but also by investigating crystal growth in the same set of conditions using both vapor- and liquid–liquid diffusion methods.
Acknowledgments
These studies were supported in part by National Institutes of Health grants R01AI090866 (to EJS) and R01GM080376 (to L-XW). We wish to thank the beamline scientists of beamlines 23-ID-D and 23-ID-B at the Advanced Photon Source. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract No. DE-AC02-06CH11357.
References
- Albert, H., Collin, M., Dudziak, D., Ravetch, J. V. & Nimmerjahn, F. (2008). Proc. Natl Acad. Sci. USA, 105, 15005–15009. [DOI] [PMC free article] [PubMed]
- Allhorn, M., Briceño, J. G., Baudino, L., Lood, C., Olsson, M. L., Izui, S. & Collin, M. (2010). Blood, 115, 5080–5088. [DOI] [PMC free article] [PubMed]
- Allhorn, M., Olsén, A. & Collin, M. (2008). BMC Microbiol. 8, 3. [DOI] [PMC free article] [PubMed]
- Arnold, J. N., Wormald, M. R., Sim, R. B., Rudd, P. M. & Dwek, R. A. (2007). Annu. Rev. Immunol. 25, 21–50. [DOI] [PubMed]
- Collin, M. & Olsen, A. (2001a). EMBO J. 20, 3046–3055. [DOI] [PMC free article] [PubMed]
- Collin, M. & Olsen, A. (2001b). Infect. Immun. 69, 7187–7189. [DOI] [PMC free article] [PubMed]
- Collin, M., Shannon, O. & Björck, L. (2008). Proc. Natl Acad. Sci. USA, 105, 4265–4270. [DOI] [PMC free article] [PubMed]
- Collin, M., Svensson, M. D., Sjöholm, A. G., Jensenius, J. C., Sjöbring, U. & Olsén, A. (2002). Infect. Immun. 70, 6646–6651. [DOI] [PMC free article] [PubMed]
- Dhouib, K., Khan Malek, C., Pfleging, W., Gauthier-Manuel, B., Duffait, R., Thuillier, G., Ferrigno, R., Jacquamet, L., Ohana, J., Ferrer, J.-L., Théobald-Dietrich, A., Giegé, R., Lorber, B. & Sauter, C. (2009). Lab Chip, 9, 1412–1421. [DOI] [PubMed]
- Du, W., Li, L., Nichols, K. P. & Ismagilov, R. F. (2009). Lab Chip, 9, 2286–2292. [DOI] [PMC free article] [PubMed]
- Ducruix, A. & Giegé, R. (2004). Crystallization of Nucleic Acids and Proteins: a Practical Approach, 2nd ed. Oxford University Press.
- Emamzadah, S., Petty, T. J., De Almeida, V., Nishimura, T., Joly, J., Ferrer, J.-L. & Halazonetis, T. D. (2009). Acta Cryst. D65, 913–920. [DOI] [PMC free article] [PubMed]
- García-Ruiz, J. M. (2003). Methods Enzymol. 368, 130–154. [DOI] [PubMed]
- Garcia-Ruiz, J. M. & Ng, J. D. (2007). Protein Crystallization Strategies for Structural Genomics, edited by N. E. Chayen, pp. 111–126. La Jolla: International University Line.
- Gavira, J. A., Toh, D., Lopéz-Jaramillo, J., García-Ruíz, J. M. & Ng, J. D. (2002). Acta Cryst. D58, 1147–1154. [DOI] [PubMed]
- Gerdts, C. J., Tereshko, V., Yadav, M. K., Dementieva, I., Collart, F., Joachimiak, A., Stevens, R. C., Kuhn, P., Kossiakoff, A. & Ismagilov, R. F. (2006). Angew. Chem. Int. Ed. Engl. 45, 8156–8160. [DOI] [PMC free article] [PubMed]
- Goodfellow, J. J., Baruah, K., Yamamoto, K., Bonomelli, C., Krishna, B., Harvey, D. J., Crispin, M., Scanlan, C. N. & Davis, B. G. (2012). J. Am. Chem. Soc. 134, 8030–8033. [DOI] [PubMed]
- Grimes, J. M. & Stuart, D. I. (1998). Nature Struct. Biol. 5, Suppl., 630–634. [DOI] [PubMed]
- Hansen, C. L., Skordalakes, E., Berger, J. M. & Quake, S. R. (2002). Proc. Natl Acad. Sci. USA, 99, 16531–16536. [DOI] [PMC free article] [PubMed]
- Hirose, M., Vafia, K., Kalies, K., Groth, S., Westermann, J., Zillikens, D., Ludwig, R. J., Collin, M. & Schmidt, E. (2012). J. Autoimmun. 39, 304–314. [DOI] [PubMed]
- Howard, E. I., Fernandez, J. M. & Garcia-Ruiz, J. M. (2009). Cryst. Growth Des. 9, 2707–2712.
- Huang, W., Giddens, J., Fan, S. Q., Toonstra, C. & Wang, L.-X. (2012). J. Am. Chem. Soc. 134, 12308–12318. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010a). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010b). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kalinin, Y. & Thorne, R. (2005). Acta Cryst. D61, 1528–1532. [DOI] [PubMed]
- Krapp, S., Mimura, Y., Jefferis, R., Huber, R. & Sondermann, P. (2003). J. Mol. Biol. 325, 979–989. [DOI] [PubMed]
- Kurz, M., Blattmann, B., Kaech, A., Briand, C., Reardon, P., Ziegler, U. & Gruetter, M. G. (2012). J. Appl. Cryst. 45, 999–1008.
- Linding, R., Russell, R. B., Neduva, V. & Gibson, T. J. (2003). Nucleic Acids Res. 31, 3701–3708. [DOI] [PMC free article] [PubMed]
- Lomino, J. V. (2011). Doctoral Dissertation thesis, University of North Carolina at Chapel Hill, USA.
- Lood, C., Allhorn, M., Lood, R., Gullstrand, B., Olin, A. I., Rönnblom, L., Truedsson, L., Collin, M. & Bengtsson, A. A. (2012). Arthritis Rheum. 64, 2698–2706. [DOI] [PubMed]
- Lund, J., Takahashi, N., Pound, J. D., Goodall, M. & Jefferis, R. (1996). J. Immunol. 157, 4963–4969. [PubMed]
- Nandakumar, K. S., Collin, M., Olsén, A., Nimmerjahn, F., Blom, A. M., Ravetch, J. V. & Holmdahl, R. (2007). Eur. J. Immunol. 37, 2973–2982. [DOI] [PubMed]
- Ng, J. D., Clark, P. J., Stevens, R. C. & Kuhn, P. (2008). Acta Cryst. D64, 189–197. [DOI] [PubMed]
- Ng, J. D., Gavira, J. A. & García-Ruíz, J. M. (2003). J. Struct. Biol. 142, 218–231. [DOI] [PubMed]
- Ng, J. D., Stevens, R. C. & Kuhn, P. (2008). Methods Mol. Biol. 426, 363–376. [DOI] [PubMed]
- Nimmerjahn, F. & Ravetch, J. V. (2008). Nature Rev. Immunol. 8, 34–47. [DOI] [PubMed]
- Otálora, F., Gavira, J. A., Ng, J. D. & García-Ruiz, J. M. (2009). Prog. Biophys. Mol. Biol. 101, 26–37. [DOI] [PubMed]
- Shen, A., Lupardus, P. J., Morell, M., Ponder, E. L., Sadaghiani, A. M., Garcia, K. C. & Bogyo, M. (2009). PLoS One, 4, e8119. [DOI] [PMC free article] [PubMed]
- Stojanoff, V., Jakoncic, J., Oren, D. A., Nagarajan, V., Navarro Poulsen, J.-C., Adams-Cioaba, M. A., Bergfors, T. & Sommer, M. O. A. (2011). Acta Cryst. F67, 971–975. [DOI] [PMC free article] [PubMed]
- Strong, M., Sawaya, M. R., Wang, S., Phillips, M., Cascio, D. & Eisenberg, D. (2006). Proc. Natl Acad. Sci. USA, 103, 8060–8065. [DOI] [PMC free article] [PubMed]
- Timmeren, M. M. van, van der Veen, B. S., Stegeman, C. A., Petersen, A. H., Hellmark, T., Collin, M. & Heeringa, P. (2010). J. Am. Soc. Nephrol. 21, 1103–1114. [DOI] [PMC free article] [PubMed]
- Tradtrantip, L., Ratelade, J., Zhang, H. & Verkman, A. S. (2013). Ann. Neurol. 73, 77–85. [DOI] [PMC free article] [PubMed]
- Yadav, M. K., Gerdts, C. J., Sanishvili, R., Smith, W. W., Roach, L. S., Ismagilov, R. F., Kuhn, P. & Stevens, R. C. (2005). J. Appl. Cryst. 38, 900–905. [DOI] [PMC free article] [PubMed]
- Zheng, B., Roach, L. S. & Ismagilov, R. F. (2003). J. Am. Chem. Soc. 125, 11170–11171. [DOI] [PubMed]
- Zheng, B., Tice, J. D. & Ismagilov, R. F. (2004). Adv. Mater. 16, 1365–1368. [DOI] [PMC free article] [PubMed]
- Zhou, X., Lau, L., Lam, W. W. L., Au, S. W. N. & Zheng, B. (2007). Anal. Chem. 79, 4924–4930. [DOI] [PubMed]