Abstract
The purpose of this study was to investigate the expression level of adiponectin and its related molecules in hypertrophied and atrophied skeletal muscle in mice. The expression was also evaluated in C2C12 myoblasts and myotubes. Both mRNA and protein expression of adiponectin, mRNA expression of adiponectin receptor (AdipoR) 1 and AdipoR2, and protein expression of adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain, and leucine zipper motif 1 (APPL1) were observed in C2C12 myoblasts. The expression levels of these molecules in myotubes were higher than those in myoblasts. The expression of adiponectin-related molecules in soleus muscle was observed at mRNA (adiponectin, AdipoR1, AdipoR2) and protein (adiponectin, APPL1) levels. The protein expression levels of adiponectin and APPL1 were up-regulated by 3 weeks of functional overloading. Down-regulation of AdipoR1 mRNA, but not AdipoR2 mRNA, was observed in atrophied soleus muscle. The expression of adiponectin protein, AdipoR1 mRNA, and APPL1 protein was up-regulated during regrowth of unloading-associated atrophied soleus muscle. Mechanical loading, which could increase skeletal muscle mass, might be a useful stimulus for the up-regulations of adiponectin and its related molecules in skeletal muscle.
Introduction
Skeletal muscle demonstrates large plasticity in response to various extracellular stimuli. Numerous studies have demonstrated that mechanical loading, which is induced by resistance exercise as well as mechanical stretching, on skeletal muscle induces an increase in muscle mass, so-called muscle hypertrophy [1], [2], [3]. On the other hand, various disease- and trauma-associated long-term inactivity, immobilization and/or unloading are well known as major causes of skeletal muscle atrophy [4], [5], [6], [7]. These extracellular stimuli also cause changes in not only muscle mass but also the metabolic properties of skeletal muscles [8], [9].
Adiponectin, one of adipokines, has been intensively studied and is now known as a molecule which plays a crucial role in the regulation of insulin sensitivity [10], [11], [12]. It has been generally considered that adiponectin is synthesized and exclusively secreted in adipocytes. However, exercise-associated improvement of insulin sensitivity is not necessarily accompanied with increase in the level of adiponectin in circulation [13], [14], [15], [16]. Therefore, the origin of the adiponectin that acts on skeletal muscle cells, which are major target cells of adiponectin, remains unclear. Recent evidences have demonstrated the expression of adiponectin in non-adipocytes such as mouse C2C12 myotubes [17] and skeletal muscles [18], [19]. It has also been reported that differentiating C2C12 myotubes have an autocrine loop of adiponectin [20], suggesting that matured skeletal muscle cells may synthesize and secrete adiponectin in an autocrine manner.
Adiponectin enhances the β-oxidation of lipids and the utilization of glucose in skeletal muscle and its actions are mediated by binding to adiponectin receptors (AdipoRs), especially AdipoR1 [21]. Although two distinct AdipoRs, AdipoR1 and AdipoR2, have been cloned, AdipoR1 is predominant in skeletal muscle [22]. It has been reported that the up-regulation of AdipoR1 enhances local adiponectin sensitivity, and is sufficient to improve skeletal muscle insulin resistance [23]. However, there is no report regarding the expression levels of adiponectin, AdipoRs, especially AdipoR2, in hypertrophied and atrophied skeletal muscles.
Recently, an adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain, and leucine zipper motif 1 (APPL1) has been characterized as a mediating molecule for signaling downstream of AdipoR [24]. APPL1 interacts with components of the insulin signalling pathway such as phosphoinositide 3-kinase and phosphatidylinositol 3-kinase [25]. Overexpression of APPL1 increases fatty acid oxidation and glucose metabolism upon adiponectin stimulation [26]. However, the expression level of APPL1 in response to changes in skeletal muscle mass also remains unclear. Therefore, in the present study, we investigated the expression levels of adiponectin, AdipoRs, and APPL1 in hypertrophied and atrophied skeletal muscle in mice. The expression levels of adiponectin-related molecules in myoblasts and myotubes were also examined.
Materials and Methods
Cultured C2C12 cells and animals
Mouse myoblast cell-line C2C12 was used in the cell culture experiment. All animal protocols were carried out in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the National Institutes of Health (Bethesda, MD) and were approved by the Animal Use Committee at Toyohashi SOZO University (A2010009, A2011001). All treatments of animals were performed under anesthesia with i.p. injection of sodium pentobarbital, and all efforts were made to prevent discomfort and suffering. Eleven-week-old male mice (C57BL/6J) were used. All mice were housed in a vivarium room with 12:12-h light:dark cycle and a maintained temperature and humidity of ∼23°C and ∼50%, respectively. Solid food and water were provided ad libitum.
Cell culture experiments
Mouse C2C12 myoblasts were routinely cultured on culture plates with a genetic type I collagen bound surface (BD BioCoat, 6 wells, BD, NJ). Cells were grown in the growth medium consisting of Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum containing high glucose (4,500 mg glucose/L, 4.0 mM L-glutamine, without sodium pyruvate) in a humidified atmosphere of 95% air and at 37°C in 5% CO2 until ∼80% confluent. The culture medium was then changed to a differentiation medium consisting of DMEM supplemented with 2% horse serum containing low glucose (1,000 mg glucose/L, 4.0 mM L-glutamine, 110 mg sodium pyruvate/L). Seven days after the exchanging of the medium, cells were collected as myotube samples for the analyses of mRNA and protein expressions. Approximately 90% of the cells had formed myotubes (data not shown). Some of the cells were collected myoblast samples immediately before the exchange of culture medium from growth to differentiation medium.
Experiment of functional overloading
Functional overloading on the soleus of left hindlimb of mice (n = 10) was performed using the methods described previously [27]. Briefly, the distal tendons of plantaris and gastrocnemius muscle were cut under anesthesia with i.p. injection of sodium pentobarbital (50 mg/kg). Sham operation was performed, and the right soleus muscle was served as a control. 1 and 3 weeks after the treatment, the soleus muscles were dissected from both hindlimbs. The right soleus muscle was served as a contralateral control. Each muscle was cross-sectionally cut into two portions at the midbelly region. Proximal and distal portions were used for the analyses of protein and mRNA expressions, respectively.
Experiment of hindlimb suspension and recovery
Twenty mice were randomly divided into 2 groups; 1) untreated pre-experimental control (n = 5) and 2) suspended groups (n = 15). Mice of the suspended group were subjected to continuous hindlimb suspension for 2 weeks. Hindlimb suspension was performed as described previously [28], [29]. Briefly, tails of the mice were cleaned, and were loosely surrounded by adhesive tapes cross-sectionally, fixing a string on the dorsal side of the tail, to keep the blood flow intact. The string was fastened to the roof of the cage at a height allowing the forelimbs to support the weight, yet preventing the hindlimbs from touching the floor and the sides of the cage. The mice could reach food and water freely by using their forelimbs. Immediately after 2 weeks of hindlimb suspension, ambulation recovery was allowed to mice (n = 10) in the suspended group.
The soleus muscles of the suspended group were dissected from bilaterally before and immediately, 2, and 4 weeks after 2 weeks of the suspension under anesthesia with i.p. injection of sodium pentobarbital (50 mg/kg). The muscles were trimmed of excess fat and connective tissues, weighed, frozen in liquid nitrogen, and stored at −80°C. The left and the right muscles were used for the analysis of protein expression and also for the analysis of mRNA expression.
Real-time Reverse Transcription-PCR
For mRNA analyses, cells and muscle tissues were incubated in RNAlater® (Qiagen GmbH, Hiden, Germany) and stored at −80°C until the extraction of total RNA. Total RNA was extracted from C2C12 cells and soleus muscles using the RNeasy Mini Kit (Qiagen) according to the manufacture's protocol. Samples (∼40 ng RNA) were reverse-transcribed into complementary DNA (cDNA) by using the first-standard cDNA Synthesis kit according to the manufacture's instructions [PrimeScript RT Master Mix (Perfect Real Time) for mRNA, Takara Bio, Otsu, Japan]. Synthesized cDNA was applied to real-time reverse transcription-PCR (Thermal Cycler Dice® Real Time System II MRQ, Takara Bio) using Takara SYBR Premix Ex Taq II for mRNA, and analyzed with Takara Thermal Cycler Dice® Real Time System Software Ver. 4.00 according to the manufacturer's instructions. The real-time cycle conditions were 95°C for 30 s followed by 40 cycles at 95°C for 5 s and at 60°C for 30 s for mRNA. Relative gene expression was quantified by the 2−ΔΔCT method. The quality of extracted RNA was evaluated by O.D. 260/280 ratio. In the present study, the values of all samples were more than 1.9. Specificity was confirmed by electrophoretic analysis of the reaction products and by inclusion of template- or reverse transcriptase-free controls. To normalize the amount of total RNA present in each reaction, we determined the best internal control for mRNA expressions by using NormFinder that is an algorithm for identifying the optimal normalization gene among a set of candidates [30], glyceraldehyde-3-phosphase dehydrogenase (GAPDH), β-actin, and S18 ribosomal RNA (18 S rRNA). Judging from these comparisons (Figs S1–S3), GAPDH was the most stable mRNA. Therefore, GAPDH mRNA was used for an internal control for mRNA expressions in each experiment.
The primers were designed by using the Takara Bio Perfect Real Time Support System (Takara Bio). Primers used for detection mouse cDNA were as follows: adiponectin, 3′-GTCAGTGGATCTGACGACACCAA-5′ (forward) and 3′-ATGCCTGCCATCCAACCTG-5′ (reverse); AdipoR1, 3′-CTGGGCATCTCTGCCATCA-5′ (forward) and 3′-CTTGACAAAGCCCTCAGCGATA-5′ (reverse); AdipoR2, 3′-ATCAGCAGCCAGACGCACTC-5′ (forward) and 3′-TGACCAGTCCCAAAGACCTCTACTC-5′ (reverse); GAPDH, 3′-TGTGTCCGTCGTGGATCTGA-5′ (forward) and 3′-TTGCTGTTGAAGTCGCAGGAG-5′ (reverse); β-actin, 3′- CATCCGTAAAGACCTCTATGCCAAC-5′ (forward) and 3′- ATGGAGCCACCGATCCACA-5′ (reverse) and 18 S rRNA, 3′-ACTCAACACGGGAAACCTCA-5′ (forward) and 3′- AACCAGACAAATCGCTCCAC-5′ (reverse).
Immunoblotting analyses
For the analyses of protein expressions, collected cells and muscles were homogenized in an isolation buffer (CelLytic MT, Sigma-Aldrich, St. Louis, MO) with 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 µg/ml leupeptin with glass homogenizer. The homogenates were then sonicated and centrifuged at 12,000 rpm (4°C for 10 min), the supernatant was collected. A part of the supernatant was solubilized in sodium-dodecyl sulfate (SDS) sample buffer {30% (vol/vol) glycerol, 5% (vol/vol) 2-mercaptoethanol, 2.3% (wt/vol) SDS, 62.5 mM Tris·HCl, 0.05% (wt/vol) bromophenol blue, and pH 6.8} at a concentration of 0.5 mg of protein per milliliter and boiled for 5 min. The SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out on 10% polyacrylamide [bisacrylamide/acrylamide, 1∶20 (wt/wt)] slab gel (60×85×1 mm) containing 0.5% SDS at a constant current of 20 mA for 120 min. Equal amounts of protein (10 µg) were loaded on each gel. Molecular weight markers (ECL DualVue Western Blotting Markers, GE Healthcare, Buckinghamshire, UK) were applied to both sides of 14 lanes as the internal controls for transfer process or electrophoresis.
Following SDS-PAGE, proteins were transferred to polyvinylidene difluoride (PVDF) membranes (0.2 µm pore size, Bio-Rad) at a constant voltage of 100 V for 60 min at 4°C. The membranes were blocked for 1 h using a blocking buffer: 5% skim milk with 0.1% Tween 20 in Tris-buffered saline (TTBS) with pH 7.5. The membranes were incubated for overnight at 4°C with a polyclonal antibody for adiponectin (R&D Systems, Minneapolis, MN), APPL1 (Cell Signaling Technology, Beverly, MA), GAPDH (Sigma-Aldrich), and β-actin (Sigma-Aldrich) and then reacted with a secondary antibody conjugated to horseradish peroxidase (anti-rabbit IgG: Cell Signaling Technology, anti-goat IgG: Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 or 2 h. After the final wash, protein bands were visualized using chemiluminescence (ECL Advance Western blotting kit, GE Healthcare), and signal density was measured using Light-Capture (AE-6971) with CS Analyzer Ver. 2.08b (ATTO corporation, Tokyo, Japan).
In the present study, we investigated which was the best internal control for protein expression, GAPDH or β-actin (Figs. S4–S6). As the most stable protein expression of GAPDH per a unit of muscular soluble protein was observed in the cell culture and the functional overloading experiment, the protein expression level of GAPDH in each sample of these experiments was used for an internal control. The protein expression levels of adiponectin and APPL1 from these experiments were normalized by using the expression level of GAPDH protein. On the other hand, in the hindlimb suspension and recovery experiment, the protein expression levels of adiponectin and APPL1 were normalized using the protein expression level of β-actin. Each sample was investigated in, at least, duplicate to ensure that results were not influenced by loading errors.
Statistical analyses
All values were expressed as means ± SEM. The statistical significance of the values from the cell culture experiments was analyzed by F-test followed by unpaired Student's t-test. For the statistical analyses of the values from the experiments of functional overloading and hindlimb suspension and recovery, the normal distribution of the data was evaluated by using Kolmogorov-Smirnov test. Statistical significance of the values from the functional overloading experiment was analyzed by using 2-way (treatment x time) analysis of variance (ANOVA) followed by Turkey-Kramer post hoc test (relative muscle wet weight and AdipoR1 mRNA) or, if normality failed, by Kruskal-Wallis test followed by Steel-Dwass test (adiponectin mRNA and protein, AdipoR2 mRNA, and APPL1 protein). Since all data from the experiment in which hind-limb suspension followed by ambulation recovery was carried out, was confirmed to fit the normal distribution, the statistical significance of the values was analyzed by using one-way ANOVA followed by Turkey-Kramer post hoc test. Spearman's rank correlation coefficients were calculated to assess the interrelationships between the relative muscle wet weight and the protein expression levels of adiponectin or APPL1, or the mRNA expression levels of AdipoR1 and AdipoR2. The significance level was accepted at p<0.05.
Results
Myoblasts and myotubes
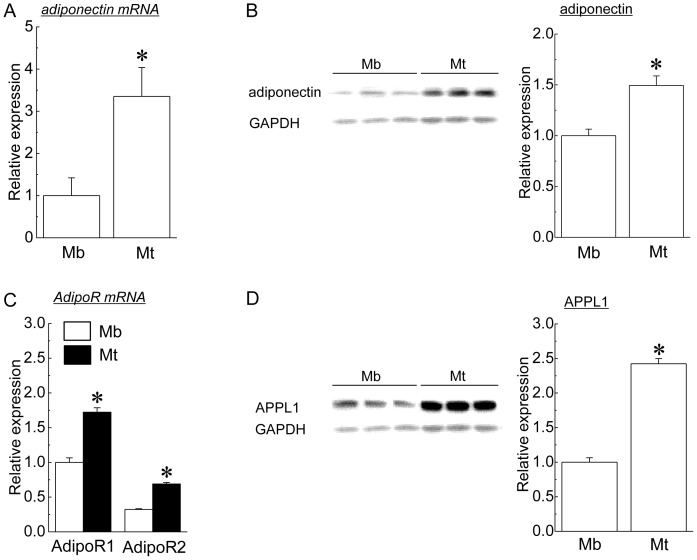
In the present study, we compared the expression levels of adiponectin and its related molecules between C2C12 myoblasts and myotubes. Differentiation of myoblasts to myotubes was judged by using the microscopic images of the cells (Fig. S7). Figure 1 shows the mean expression levels of adiponectin and its related molecules in myoblasts and myotubes of C2C12. Both mRNA and protein expressions of adiponectin were observed in not only myotubes but also myoblasts (Figs. 1A and 1B). Expression of adiponectin mRNA and protein in myotubes was significantly higher than for those in myoblasts (p<0.05). In addition, mRNA expression of AdipoR1 and AdipoR2 was also observed in both myoblasts and myotubes (Fig. 1C). The mRNA expression levels of both AdipoR1 and AdipoR2 in myotubes were also significantly higher than for those in myoblasts (p<0.05). The expression of APPL1 protein was observed in both myoblasts and myotubes. The mean expression level of APPL1 protein in myotubes was significantly higher than that in myoblasts (p<0.05, Fig. 1D).
Figure 1. Expression levels of adiponectin and its related molecules in myoblasts (Mb) and myotubes (Mt).
A: adiponectin mRNA, B: adiponectin protein, C: adiponectin receptor 1 (AdipoR1) mRNA and adiponectin receptor 2 (AdipoR2) mRNA, D: adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain, and leucine zipper motif 1 (APPL1) protein. The protein expression levels of adiponectin and APPL1 were shown as relative values to the expression level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein. Mean ± SEM. n = 6. *: significantly different from the value of myoblasts (Mb) (p<0.05).
Effects of functional overloading
The wet weights of soleus muscle relative to body weight in both untreated contralateral control and overloaded groups are shown in Fig. 2. Two-way ANOVA (treatment x time) for the changes in the relative soleus weight showed a significant effect of treatment (p<0.05), but not of time. Muscle wet weight was increased by ∼1.45-fold following 1 and 3 weeks of overloading, compared with the values of contralateral control.
Figure 2. Changes in soleus muscle wet weight in response to functional overloading.
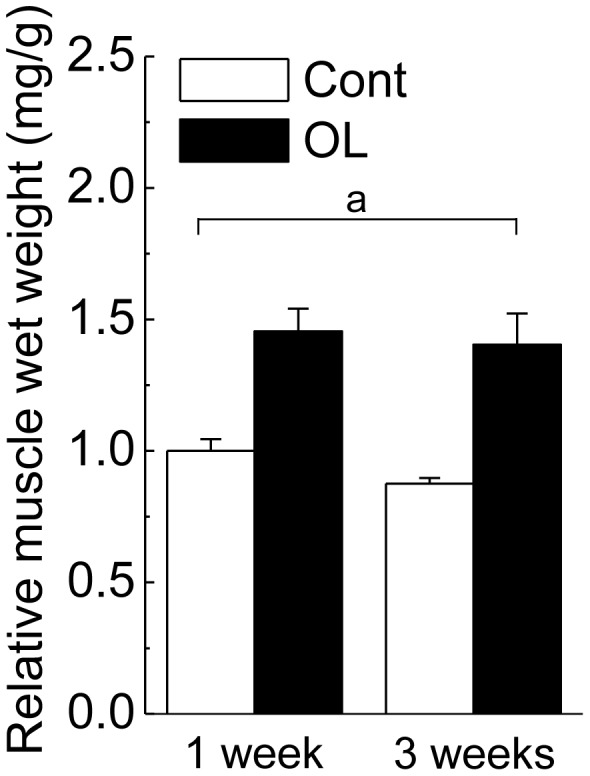
The values show the muscle wet weight relative to body weight. Cont: untreated control group, OL: functional overloaded group, 1 week and 3 weeks: 1- and 3-week of functional overloading. Mean ± SEM. n = 5/group at each time point. pone.0081929.a.tif: a significant effect of treatment (p<0.05) revealed by two-way ANOVA (treatment and time).
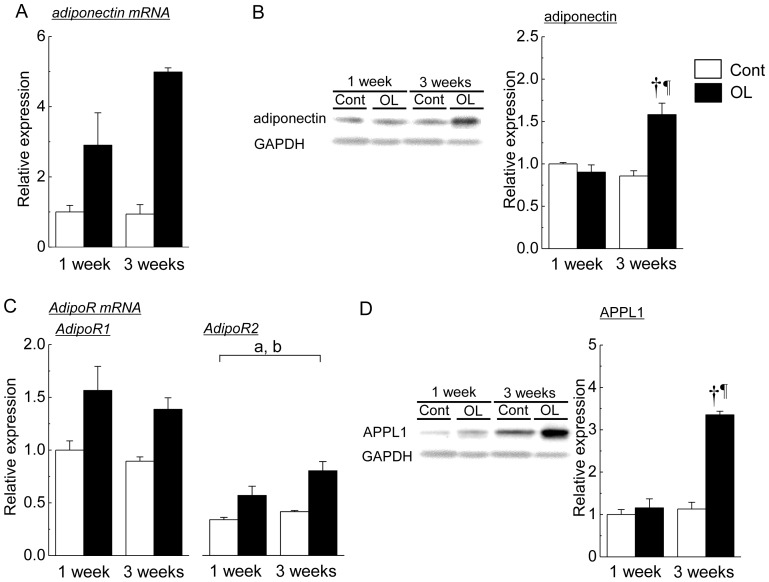
Figure 3 shows the changes in adiponectin and its related molecules in the soleus muscle of both the contralateral control and overloaded soleus muscles. Although there was no significant difference in mRNA expression of adiponectin (Fig. 3A), the protein expression level was significantly increased following 3 weeks of functional overloading, compared with the levels of contralateral control and following 1 week of overloading (Fig. 3 B, p<0.05). Two-way ANOVA revealed that significant effects of treatment and time in changes in mRNA expression levels of AdipoR2 were observed (p<0.05, Fig. 3C). Figure 3D shows the changes in the protein expression level of APPL1 in the soleus muscle of both groups. A significant up-regulation of APPL1 protein expression was observed following 3 weeks of functional overloading, compared with the levels of contralateral control and following 1 week of overloading (p<0.05).
Figure 3. Expression levels of adiponectin and its related molecules in soleus muscle in response to functional overloading.
A: adiponectin mRNA, B: adiponectin protein, C: adiponectin receptor 1 (AdipoR1) mRNA and adiponectin receptor 2 (AdipoR2) mRNA, D: adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain, and leucine zipper motif 1 (APPL1) protein. The protein expression levels of adiponectin and APPL1 were shown as the relative values to the expression level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein. Mean ± SEM. n = 5. Cont: untreated control group, overloaded: functional overloaded group. See figures 1 and 2 for other abbreviations. a and b: a significant effect of treatment (p<0.05) and a significant effect of time revealed by two-way ANOVA (treatment and time). *, †, and ¶: p<0.05 vs. 1 week of control group, 1 week of overloaded group, and 3 weeks of control group, respectively.
Effects of unloading and reloading
Changes in the relative soleus weight during 2-week of hindlimb suspension followed by 4-week of ambulation recovery are shown in Fig. 4. Immediately after 2 weeks of the suspension (R0), the relative soleus weight was significantly decreased (p<0.05). The relative weights at 2 (R2) and 4 weeks (R4) of recovery after the suspension were significantly higher than that at R0 (p<0.05).
Figure 4. Muscle wet weight in response to 2 weeks of hindlimb suspension followed by 4 weeks of recovery.
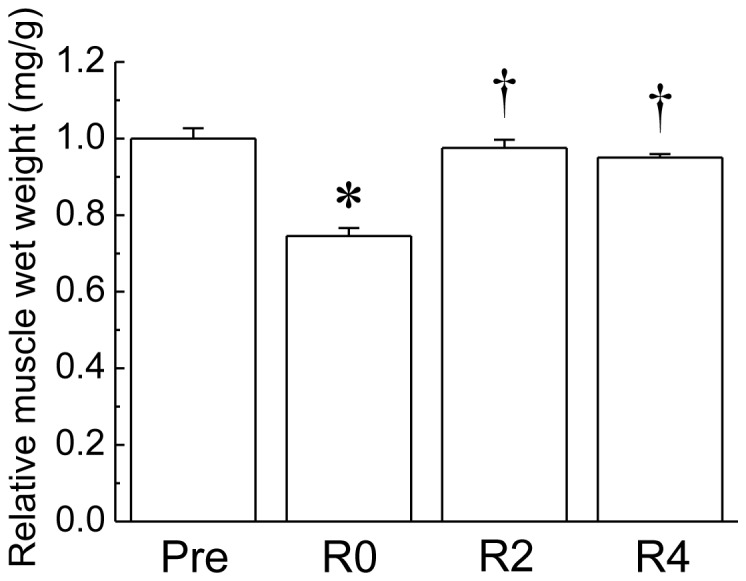
Pre: before hindlimb suspension. R0, R2, and R2: immediately, 2 and 4 weeks of respectively recovery period after the suspension. The values show the muscle wet weight relative to body weight. Mean ± SEM. n = 5. * and †: p<0.05 vs. Pre and R0, respectively.
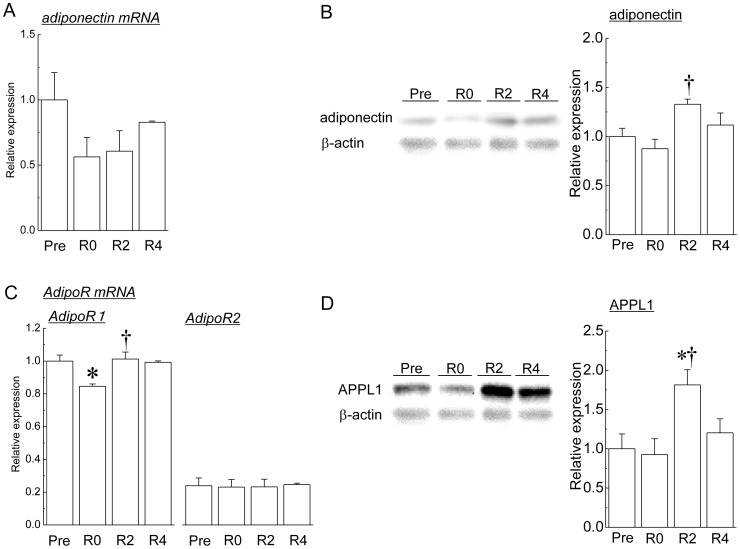
Figure 5 shows the changes in adiponectin and its related molecules in soleus muscle during the suspension followed by the recovery period. Although there were no significant differences in the mRNA expression levels of adiponectin during the experimental period (Fig. 5A), the protein expression level of adiponectin tended to be decreased by 2 weeks of the suspension, and was significantly increased by 2 weeks of the recovery (p<0.05, Fig. 5B). The mean expression level of AdipoR1 mRNA was significantly decreased by 2 weeks of the suspension, and was significantly increased during 2 weeks of the recovery (p<0.05, Fig. 5C). Conversely, no significant changes were observed in the expression level of AdipoR2 mRNA during the suspension and the recovery (Fig. 5C). Significant increase in the mean expression of APPL1 protein was observed 2 weeks after the recovery, compared with the expression levels before and immediately after the suspension (p<0.05, Fig. 5D).
Figure 5. Expression levels of adiponectin and its molecules in muscle in response to hindlimb unloading followed by reloading.
A: adiponectin mRNA, B: adiponectin protein, C: adiponectin receptor 1 (AdipoR1) mRNA and adiponectin receptor 2 (AdipoR2) mRNA, D: adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain, and leucine zipper motif 1 (APPL1) protein. The protein expression levels of adiponectin and APPL1 were shown as the relative values to the expression level of β-actin protein. Mean ± SEM. n = 5. See figures 1, 2, and 3 for abbreviations. * and †: p<0.05 vs. Pre and R0, respectively.
Interrelationship between muscle wet weight and adiponectin, AdipoRs or APPL1
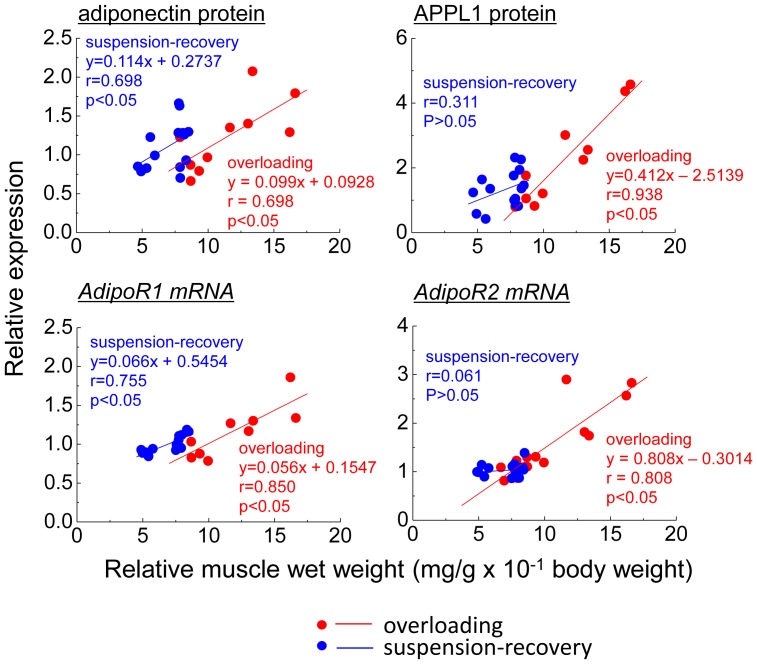
Figure 6 shows the interrelationships between soleus muscle wet weight relative to body weight and the expression levels of adiponectin-related molecules. The values were plotted from the contralateral control and 3 weeks of functional overloading in the functional overloading experiment both before and immediately, 2, and 4 weeks respectively after 2 weeks of the suspension in the suspension-recovery experiment, since there was no significant increase in the relative soleus muscle weight following 1 week of the overloading. In the overloading experiment, there were positive interrelationships between the soleus muscle wet weight relative to body weight and the expression levels of adiponectin protein, APPL1 protein, AdipoR1 mRNA, or AdipoR2 mRNA (p<0.05). Positive relation between the muscle weight and the expression levels of adiponectin protein or AdipoR1 mRNA were also observed in the suspension-recovery experiment (p<0.05). However, there was no significant relationship between soleus muscle weight and APPL1 protein or AdipoR2 mRNA in the suspension-recovery experiment.
Figure 6. Interrelationship between soleus muscle wet weight relative to body weight and the expression levels of the expression levels of adiponectin protein, APPL1 protein, AdipoR1 mRNA or AdipoR2 mRNA.
Red circle: data from the experiment of functional overloading experiment (contralateral control and following 3 weeks of functional overloading). Blue circle: data from the experiment of hindlimb suspension and recovery (before, 2 weeks of the suspension, and 2 and 4 weeks of recovery following the suspension). A linear regression line was calculated by using of data each experiment in each graph.
Discussion
The present study showed both mRNA and protein expressions of adiponectin, mRNA expressions of AdipoR1 and AdipoR2, and protein expression of APPL1 in C2C12 myoblasts. In C2C12 myotubes, the expression levels of adiponectin-related molecules were higher than those in myoblasts. The expression of adiponectin-related molecules in antigravitational soleus muscle was observed at mRNA (adiponectin, AdipoR1, AdipoR2) and protein (adiponectin, APPL1) levels. The protein expression levels of adiponectin and APPL1 in soleus muscle were up-regulated following 3 weeks of functional overloading. On the other hand, down-regulation of AdipoR1 mRNA, but not AdipoR2 mRNA, was observed in atrophied soleus muscle. The expression levels of adiponectin protein, AdipoR1 mRNA, and APPL1 protein were up-regulated during the regrowth of unloading-associated atrophied soleus muscle. There were significant positive interrelationships between the muscle wet weight and the expression levels of adiponectin-related molecules in overloading-associated hypertrophied skeletal muscle. However, there were significant interrelationships between the muscle wet weight and the expression levels of adiponectin protein or AdipoR1 mRNA, but not APPL1 protein and AdipoR2 mRNA, in atrophied and regrowing skeletal muscle.
Myogenic differentiation and adiponectin expression
In the present study, the expression of adiponectin and its related molecules was observed in both myoblasts and myotubes of cultured C2C12 cells. Although the expression of AdipoR1, AdipoR2, and APPL1 in myoblasts was also confirmed in the previous study [20], [31], [32], this is the first study showing the mRNA and protein expression of adiponectin in C2C12 myoblasts. A previous study [20] has reported that the protein expression of adiponectin had been observed in C2C12 myotubes, but not in myoblasts. Although we have no clear explanation for the discrepancy between the results obtained in the previous investigation [20] and the present study, the mRNA expression of adiponectin in myoblasts may support our result that myoblasts have endogenous expression of adiponectin protein.
We also found that the up-regulation of adiponectin and its related molecules was observed in myotubes, rather than myoblasts. Up-regulation of APPL1 protein during myogenic differentiation was consistent with the results from the previous study [32]. However, it has been reported that the mRNA expression levels of AdipoR1 and AdipoR2 were constant during the differentiation [20], but this result was inconsistent with the present study. In the previous study, however, the mRNA expression levels of AdipoRs were evaluated using 3-day-differentiating myotubes [20]. On the other hand, we investigated the mRNA expression levels of 7-day-differentiating myotubes. Since the number of undifferentiated C2C12 cells on the ∼4th day of the differentiation were higher than that on the 7th day of the differentiation (Fig. S7), the discrepancy in the changes in AdipoR mRNAs expression between the results of the previous study [20] and the present study may be attributed to the differentiation stages of C2C12 myotubes.
Overloading and unloading followed by reloading
In the present study, soleus muscle wet weight was increased by ∼1.45-fold following 3 weeks of overloading, compared with the values of contralateral control. A similar increment of overloading-associated soleus muscle was observed in the previous studies [27], [33], [34]. Protein expression levels of adiponectin in skeletal muscle were up-regulated by 3 weeks of functional overloading. In addition, the up-regulation of adiponectin in skeletal muscle was also observed by reloading on unloading-associated atrophied muscle. This is the first study showing the expression levels of adiponectin in skeletal muscle in response to overloading and unloading followed by reloading.
It has been suggested that some of the overloading- and reloading-associated increase in antigravitational soleus muscle wet weight may be attributed to some damage, inflammation and swelling in the loaded muscle during the early stage of adaptation (∼5–7 days of overloading and ∼4–5 days of reloading, respectively) [35], [36], [37]. However, it is unlikely that the increase of soleus muscle in the present study (except for 1 week of the overloading) is affected by such acute affects because the muscle was sampled after 3 weeks of overloading, and after 2 and 4 weeks of reloading following unloading. In addition, the previous study reported that muscle protein content increased following 3 weeks of overloading [27] and 2 weeks of reloading following 2 weeks of hindlimb suspension [38].
In the present study, up-regulation of AdipoR1 mRNA was observed in reloaded soleus muscle. Up-regulation of AdipoR2 was also observed in overloaded muscle. On the other hand, the down-regulation of AdipoR1 mRNA, but not AdipoR2 mRNA, was observed in atrophied soleus muscle. Several reports have shown that AdipoR1 mRNA level in skeletal muscle was up-regulated by running exercise in rats [39] and mice [40], and by cycling, running or swimming exercise in healthy and type 2 diabetes human [16]. Since stress proteins, namely heat shock proteins, in skeletal muscle are up-regulated in functionally overloading [41] as well as reloading following hindlimb unloading [29], AdipoR1 in skeletal muscle may be up-regulated by loading-related cellular stress.
It has been reported that the expression level of AdipoR1in skeletal muscle correlates with glucose and lipid metabolism, and insulin sensitivity [42], [43], [44]. Although insulin sensitivity was not evaluated in the present study, loading-associated up-regulation of AdipoR1 in skeletal muscle could improve insulin sensitivity. However, a physiological significance of AdipoR2 expression in response to overloading and reloading following unloading remains unclear.
In the present study, APPL1 protein was also up-regulated in overloaded and reloaded soleus muscle following unloading. This is the first study showing the responses of APPL1 expression in skeletal muscle in response to overloading, unloading, and reloading. Although we have no clear explanation regarding the up-regulation of APPL1in hypertrophied and reloaded skeletal muscle, the up-regulation of APPL1 may be associated with the up-regulations of adiponectin and AdipoRs in skeletal muscle. Collectively, all the aforementioned results suggest that the expressions of adiponectin and its related molecules including APPL1 could be up-regulated by increasing load on skeletal muscle, suggesting that these phenomena could enhance adiponectin action.
Numerous studies have shown the transition of type II to type I fibers in overloading-associated soleus muscle hypertrophy in mice [34] and in rats [45], [46]. In addition, unloading-associated decrease in type I myosin heavy chain (MHC) [47], [48], [49], [50] and reloading-associated recovery of type I MHC [48], [51], [52] in rats and mice are also well known. Muscle hypertrophy- and atrophy-associated transition of fiber types may influence the expression levels of adiponectin-related molecules since type IIA and IID fibers contain high adiponectin in comparison to type IIB and type I fibers [18]. In the present study, the expression levels of adiponectin-related molecules were up-regulated in overloading-associated hypertrophied mouse soleus muscle. On the contrary, protein expression level of adiponectin tended to be decreased by 2 weeks of the suspension, and was significantly increased by 2 weeks of the recovery (Fig. 5). Although we did not investigate the changes in fiber type in soleus muscle following overloading, hindlimb unloading, and reloading, it is unlikely that a shift in fiber type distribution affected adiponectin regulation in soleus muscle.
In the present study, there were positive interrelationships between soleus muscle wet weight relative to body weight and the expression levels of adiponectin protein or AdipoR1 mRNA in the experiments of functional overloading and hindlimb suspension-recovery experiments, suggesting that muscle mass might implicate adiponectin content. However, positive relationships between the muscle weight and APPL1 protein or AdipoR2 mRNA were observed in the overloading experiment, but not in the suspension-recovery experiment. Therefore, there is a possibility that the regulation mechanism(s) of adiponectin-related molecules expressions during hypertrophy of normal skeletal muscle might be different from that during the regrowth of atrophied skeletal muscle. Additional studies will be needed to elucidate this issue.
In conclusion, both mRNA and protein expressions of adiponectin, mRNA expressions of AdipoR1 and AdipoR2, and protein expression of APPL1 in myoblasts was observed. These expressions were up-regulated by myogenic differentiation. Up-regulation of adiponectin-related molecules was observed during functional overloading-associated muscle hypertrophy and during the regrowth of unloading-associated atrophied soleus muscle although the physiological role(s) of up-regulation of adiponectin in hypertrophied and regrowing skeletal muscle remains unclear. Therefore, mechanical loading, which is a type of hypertrophic stimuli, might up-regulate adiponectin and its related molecules in skeletal muscle.
Supporting Information
Relative mRNA expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and 18 S rRNA in myoblasts (Mb) and myotubes (Mt). Mean ± SEM. n = 6.
(TIF)
Relative mRNA expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and 18 S rRNA in response to functional overloading. Cont: untreated control group, OL: functional overloaded group, 1 week and 3 weeks: 1- and 3-week of functional overloading. Mean ± SEM. n = 5/group at each time point.
(TIF)
Relative mRNA expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and 18 S rRNA in response to 2 weeks of hindlimb suspension followed by 4 weeks of recovery. Pre: before hindlimb suspension. R0, R2, and R2: immediately, 2 and 4 weeks of recovery after the suspension, respectively. Mean ± SEM. n = 5/group at each time point.
(TIF)
Relative protein expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin in myoblasts (Mb) and myotubes (Mt). Mean ± SEM. n = 6.
(TIF)
Relative protein expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin in response to functional overloading. Cont: untreated control group, OL: functional overloaded group, 1 week and 3 weeks: 1- and 3-week of functional overloading. Mean ± SEM. n = 5/group at each time point.
(TIF)
Relative protein expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin in response to 2 weeks of hindlimb suspension followed by 4 weeks of recovery. Pre: before hindlimb suspension. R0, R2, and R2: immediately, 2 and 4 weeks of recovery after the suspension, respectively. Mean ± SEM. n = 5/group at each time point.
(TIF)
Representative images of undifferentiated myoblasts and differentiating myotubes 2, 4, and 7 days after the initiation of differentiation. Day 2, 4, and 7: 2, 4, and 7 days after the initiation of differentiation
(TIF)
Acknowledgments
The authors greatly appreciate Dr. L. L. Tang for his technical assistance. We also thank Dr. Hideo Miyahara for his valuable advice regarding statistical analyses.
Funding Statement
This study was supported, in part, by Grants-in-Aid for Scientific Research (B, 20300218, K. Goto; A, 22240071, T. Yoshioka; S, 19100009, Y. Ohira) and Grants-in-Aid for challenging Exploratory Research (24650411, K. Goto; 24650407, Y. Ohira) from Japan Society for the Promotion of Science and the Science Research Promotion Fund from The Promotion and Mutual Aid Corporation for Private Schools of Japan (K. Goto). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Carson JA, Booth FW (1998) Effect of serum and mechanical stretch on skeletal alpha-actin gene regulation in cultured primary muscle cells. Am J Physiol 275: C1438–1448. [DOI] [PubMed] [Google Scholar]
- 2. Goldspink G (1999) Changes in muscle mass and phenotype and the expression of autocrine and systemic growth factors by muscle in response to stretch and overload. J Anat 194: 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perrone CE, Fenwick-Smith D, Vandenburgh HH (1995) Collagen and stretch modulate autocrine secretion of insulin-like growth factor-1 and insulin-like growth factor binding proteins from differentiated skeletal muscle cells. J Biol Chem 270: 2099–2106. [DOI] [PubMed] [Google Scholar]
- 4. Goto K, Okuyama R, Honda M, Uchida H, Akema T, et al. (2003) Profiles of connectin (titin) in atrophied soleus muscle induced by unloading of rats. J Appl Physiol 94: 897–902. [DOI] [PubMed] [Google Scholar]
- 5. Kim SJ, Roy RR, Kim JA, Zhong H, Haddad F, et al. (2008) Gene expression during inactivity-induced muscle atrophy: effects of brief bouts of a forceful contraction countermeasure. J Appl Physiol 105: 1246–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Min K, Smuder AJ, Kwon OS, Kavazis AN, Szeto HH, et al. (2011) Mitochondrial-targeted antioxidants protect skeletal muscle against immobilization-induced muscle atrophy. J Appl Physiol 111: 1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vazeille E, Codran A, Claustre A, Averous J, Listrat A, et al. (2008) The ubiquitin-proteasome and the mitochondria-associated apoptotic pathways are sequentially downregulated during recovery after immobilization-induced muscle atrophy. Am J Physiol Endocrinol Metab 295: E1181–1190. [DOI] [PubMed] [Google Scholar]
- 8. Yamashita K, Yoshioka T (1992) Activities of creatine kinase isoenzymes in single skeletal muscle fibres of trained and untrained rats. Pflügers Arch 421: 270–273. [DOI] [PubMed] [Google Scholar]
- 9. Yoshioka T, Takekura H, Yamashita K (1992) Effect of endurance training on disuse muscle atrophy induced by body suspension in rats. Med Sport Sci 37: 150–161. [Google Scholar]
- 10. Hotamisligil GS, Shargill NS, Spiegelman BM (1993) Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 259: 87–91. [DOI] [PubMed] [Google Scholar]
- 11. Yamauchi T, Kamon J, Waki H, Murakami K, Motojima K, et al. (2001) The mechanisms by which both heterozygous peroxisome proliferator-activated receptor gamma (PPARgamma) deficiency and PPARgamma agonist improve insulin resistance. J Biol Chem 276: 41245–41254. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, et al. (1994) Positional cloning of the mouse obese gene and its human homologue. Nature 372: 425–432. [DOI] [PubMed] [Google Scholar]
- 13. Hulver MW, Zheng D, Tanner CJ, Houmard JA, Kraus WE, et al. (2002) Adiponectin is not altered with exercise training despite enhanced insulin action. Am J Physiol Endocrinol Metab 283: E861–865. [DOI] [PubMed] [Google Scholar]
- 14. Boudou P, Sobngwi E, Mauvais-Jarvis F, Vexiau P, Gautier JF (2003) Absence of exercise-induced variations in adiponectin levels despite decreased abdominal adiposity and improved insulin sensitivity in type 2 diabetic men. Eur J Endocrinol 149: 421–424. [DOI] [PubMed] [Google Scholar]
- 15. Oberbach A, Tönjes A, Klöting N, Fasshauer M, Kratzsch J, et al. (2006) Effect of a 4 week physical training program on plasma concentrations of inflammatory markers in patients with abnormal glucose tolerance. Eur J Endocrinol 154: 577–585. [DOI] [PubMed] [Google Scholar]
- 16. Blüher M, Bullen JW Jr, Lee JH, Kralisch S, Fasshauer M, et al. (2006) Circulating adiponectin and expression of adiponectin receptors in human skeletal muscle: associations with metabolic parameters and insulin resistance and regulation by physical training. J Clin Endocrinol Metab 91: 2310–2316. [DOI] [PubMed] [Google Scholar]
- 17. Delaigle AM, Jonas JC, Bauche IB, Cornu O, Brichard SM (2004) Induction of adiponectin in skeletal muscle by inflammatory cytokines: in vivo and in vitro studies. Endocrinology 145: 5589–5597. [DOI] [PubMed] [Google Scholar]
- 18. Krause MP, Liu Y, Vu V, Chan L, Xu A, et al. (2008) Adiponectin is expressed by skeletal muscle fibers and influences muscle phenotype and function. Am J Physiol Cell Physiol 295: C203–C212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang B, Chen L, Qian Y, Triantafillou JA, McNulty JA, et al. (2006) Changes of skeletal muscle adiponectin content in diet-induced insulin resistant rats. Biochem Biophys Res Commun 341: 209–217. [DOI] [PubMed] [Google Scholar]
- 20.Fiaschi T, Cirelli D, Comito G, Gelmini S, Ramponi G, et al. (2009) Globular adiponectin induces differentiation and fusion of skeletal muscle cells. Cell Res 19: : 584–597, 2009. [DOI] [PubMed] [Google Scholar]
- 21. Yamauchi T, Nio Y, Maki T, Kobayashi M, Takazawa T, et al. (2007) Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat Med 13: 332–339. [DOI] [PubMed] [Google Scholar]
- 22. Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, et al. (2003) Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423: 762–769. [DOI] [PubMed] [Google Scholar]
- 23. Patel SA, Hoehn KL, Lawrence RT, Sawbridge L, Talbot NA, et al. (2012) Overexpression of the adiponectin receptor AdipoR1 in rat skeletal muscle amplifies local insulin sensitivity. Endocrinology 153: 5231–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deepa SS, Dong LQ (2009) APPL1: role in adiponectin signaling and beyond. Am J Physiol Endocrinol Metab 296: E22–E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cleasby ME, Lau Q, Polkinghorne E, Patel SA, Leslie SJ, et al. (2011) The adaptor protein APPL1 increases glycogen accumulation in rat skeletal muscle through activation of the PI3-kinase signalling pathway. J Endocrinol 210: 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mao X, Kikani CK, Riojas RA, Langlais P, Wang L, et al. (2006) APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol 8: 516–523. [DOI] [PubMed] [Google Scholar]
- 27. Morioka S, Goto K, Kojima A, Naito T, Matsuba Y, et al. (2008) Functional overloading facilitates the regeneration of injured soleus muscles in mice. J Physiol Sci 58: 397–404. [DOI] [PubMed] [Google Scholar]
- 28. Matsuba Y, Goto K, Morioka S, Naito T, Akema T, et al. (2009) Gravitational unloading inhibits the regenerative potential of atrophied soleus muscle in mice. Acta Physiol (Oxf) 196: 329–339. [DOI] [PubMed] [Google Scholar]
- 29. Yasuhara K, Ohno Y, Kojima A, Uehara K, Beppu M, et al. (2011) Absence of heat shock transcription factor 1 retards the regrowth of atrophied soleus muscle in mice. J Appl Physiol 111: 1142–1149. [DOI] [PubMed] [Google Scholar]
- 30. Andersen CL, Leder-Jensen J, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 31. Fiaschi T, Tedesco FS, Giannoni E, Diaz-Manera J, Parri M, et al. (2010) Globular adiponectin as a complete mesoangioblast regulator: role in proliferation, survival, motility, and skeletal muscle differentiation. Mol Biol Cell 21: 848–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bae GU, Lee JR, Kim BG, Han JW, Leem YE, et al. (2010) Cdo interacts with APPL1 and activates Akt in myoblast differentiation. Mol Biol Cell 21: 2399–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koya T, Nishizawa S, Ohno Y, Goto A, Ikuta A, et al. (in press) Heat shock transcription factor-1 deficiency attenuates overloading-associated hypertrophy of mouse soleus muscle. PLoS ONE. [DOI] [PMC free article] [PubMed]
- 34. Timson BF, Bowlin BK, Dudenhoeffer GA, George JB (1985) Fiber number, area, and composition of mouse soleus muscle following enlargement. J Appl Physiol 58: 619–624. [DOI] [PubMed] [Google Scholar]
- 35. Adams GR, Haddad F, Baldwin KM (1999) Time course of changes in markers of myogenesis in overloaded rat skeletal muscles. J Appl Physiol 87: 1705–1712. [DOI] [PubMed] [Google Scholar]
- 36. Armstrong RB, Marum P, Tullson P, Saubert CW (1979) Acute hypertrophic response of skeletal muscle to removal of synergists. J Appl Physiol 46: 835–842. [DOI] [PubMed] [Google Scholar]
- 37.Tidball JG, Wehling-Henricks M (2007) Macrophages promote muscle membrane repair and muscle fibre growth and regeneration during modified muscle loading in mice in vivo. J Physiol 578 (Pt 1) 327–336. [DOI] [PMC free article] [PubMed]
- 38. Goto K, Honda M, Kobayashi T, Uehara K, Kojima A, et al. (2004) Heat stress facilitate the recovery of atrophied soleus muscle in rats. J Physiol Sci 54: 285–293. [DOI] [PubMed] [Google Scholar]
- 39. Zeng Q, Isobe K, Fu L, Ohkoshi N, Ohmori H, et al. (2007) Effects of exercise on adiponectin and adiponectin receptor levels in rats. Life Sci 80: 454–459. [DOI] [PubMed] [Google Scholar]
- 40. Huang H, Iida KT, Sone H, Yokoo T, Yamada N, et al. (2006) The effect of exercise training on adiponectin receptor expression in KKAy obese/diabetic mice. J Endocrinol 189: 643–653. [DOI] [PubMed] [Google Scholar]
- 41. Locke L (2008) Heat shock protein accumulation and heat shock transcription factor activation in rat skeletal muscle during compensatory hypertrophy. Acta Physiol (Oxf) 192: 403–411. [DOI] [PubMed] [Google Scholar]
- 42. Debard C, Laville M, Berbe V, Loizon E, Guillet C, et al. (2004) Expression of key genes of fatty acid oxidation, including adiponectin receptors, in skeletal muscle of Type 2 diabetic patients. Diabetologia 47: 917–925. [DOI] [PubMed] [Google Scholar]
- 43. Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, et al. (2010) Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca2+ and AMPK/SIRT1. Nature 464: 1313–1319. [DOI] [PubMed] [Google Scholar]
- 44. Staiger H, Kaltenbach S, Staiger K, Stefan N, Fritsche A, et al. (2004) Expression of adiponectin receptor mRNA in human skeletal muscle cells is related to in vivo parameters of glucose and lipid metabolism. Diabetes 53: 2195–2201. [DOI] [PubMed] [Google Scholar]
- 45. Sakuma K, Yamaguchi A, Katsuta S (1995) Are region-specific changes in fibre types attributable to nonuniform muscle hypertrophy by overloading? Eur J Appl Physiol 71: 499–504. [DOI] [PubMed] [Google Scholar]
- 46. Yamaguchi A, Sakuma K, Morita I, Soya H, Takeda G, et al. (1996) Changes in fibre types in rat soleus and plantaris muscles following hypophysectomy and compensatory overload. Acta Physiol Scand 158: 89–95. [DOI] [PubMed] [Google Scholar]
- 47. Stelzer JE, Widrick JJ (2003) Effect of hindlimb suspension on the functional properties of slow and fast soleus fibers from three strains of mice. J Appl Physiol 95: 2425–2433. [DOI] [PubMed] [Google Scholar]
- 48. Däpp C, Schumutz S, Hoppeler H, Flück M (2004) Transcriptional reprogramming and ultrastructure during atrophy and recovery of mouse soleus muscle. Physiol Genomics 20: 97–107. [DOI] [PubMed] [Google Scholar]
- 49. Desaphy JF, Pierno S, Liantonio A, Giannuzzi V, Dogennaro C, et al. (2010) Antioxidant treatment of hindlimb-unloading mouse counteracts fiber type transition but not atrophy of disused muscles. Pharmacol Res 61: 553–563. [DOI] [PubMed] [Google Scholar]
- 50. Sandonà D, Desaphy J-F, Camerino GM, Bianchini E, Ciciliot S, et al. (2012) Adaptation of mouse skeletal muscle to long-term microgravity in the MDS mission. PLoS ONE 7: e33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pandorf CE, Jiang WH, Qin AX, Bodell PW, Baldwin KM, et al. (2009) Calcineurin plays a modulatory role in loading-induced regulation of type I myosin heavy chain gene expression in slow skeletal muscle. Am J Physiol Regul Integr Comp Physiol 297: R1037–R1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miyazaki M, Hitomi Y, Kizaki T, Ohno H, Katsumura T, et al. (2006) Calcineurin-mediated slow-type fiber expression and growth in reloading condition. Med Sci Sports Exerc 38: 1065–1072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative mRNA expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and 18 S rRNA in myoblasts (Mb) and myotubes (Mt). Mean ± SEM. n = 6.
(TIF)
Relative mRNA expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and 18 S rRNA in response to functional overloading. Cont: untreated control group, OL: functional overloaded group, 1 week and 3 weeks: 1- and 3-week of functional overloading. Mean ± SEM. n = 5/group at each time point.
(TIF)
Relative mRNA expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and 18 S rRNA in response to 2 weeks of hindlimb suspension followed by 4 weeks of recovery. Pre: before hindlimb suspension. R0, R2, and R2: immediately, 2 and 4 weeks of recovery after the suspension, respectively. Mean ± SEM. n = 5/group at each time point.
(TIF)
Relative protein expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin in myoblasts (Mb) and myotubes (Mt). Mean ± SEM. n = 6.
(TIF)
Relative protein expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin in response to functional overloading. Cont: untreated control group, OL: functional overloaded group, 1 week and 3 weeks: 1- and 3-week of functional overloading. Mean ± SEM. n = 5/group at each time point.
(TIF)
Relative protein expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin in response to 2 weeks of hindlimb suspension followed by 4 weeks of recovery. Pre: before hindlimb suspension. R0, R2, and R2: immediately, 2 and 4 weeks of recovery after the suspension, respectively. Mean ± SEM. n = 5/group at each time point.
(TIF)
Representative images of undifferentiated myoblasts and differentiating myotubes 2, 4, and 7 days after the initiation of differentiation. Day 2, 4, and 7: 2, 4, and 7 days after the initiation of differentiation
(TIF)