Abstract
An RNA-Seq experiment was performed using field grown well-watered and naturally rain fed cotton plants to identify differentially expressed transcripts under water-deficit stress. Our work constitutes the first application of the newly published diploid D5 Gossypium raimondii sequence in the study of tetraploid AD1 upland cotton RNA-seq transcriptome analysis. A total of 1,530 transcripts were differentially expressed between well-watered and water-deficit stressed root tissues, in patterns that confirm the accuracy of this technique for future studies in cotton genomics. Additionally, putative sequence based genome localization of differentially expressed transcripts detected A2 genome specific gene expression under water-deficit stress. These data will facilitate efforts to understand the complex responses governing transcriptomic regulatory mechanisms and to identify candidate genes that may benefit applied plant breeding programs.
Introduction
Limited water resources are one of the major environmental pressures impacting global crop production [1]. As climate change and decreases in arable land place increasing strain on available resources, it is essential to develop methods to study abiotic stress and its influence on the growth and development of the world’s major crops. Water-deficit influences a wide range of plant processes, from whole-plant growth and development to the molecular regulation of essential transcriptional pathways, and thus significantly impacts both plant physiology and metabolism. Characteristic responses of water-deficit stress can include wilting, decreased photosynthetic rate [2,3] and stomatal closure [4–6]. These responses negatively impact carbon metabolism. The production of reactive oxygen species (ROS) is also commonly found in water-deficit stressed plant cells, where they may destroy lipids and interact with major cellular signaling pathways [7].
The effects of water-deficit stress to the aerial portions of plants, including leaf, stem and flower tissues, have been well documented [8–10]. Recent research emphasized downstream effects of stresses to the integral root system, responsible for water uptake, on all plant tissues [11]. One root response is altered root architecture that may counter a change in soil properties by decreasing the development of lateral roots [12–14]. Degradation of lateral root amyloplasts is associated with increased hydrotropism in the main root [12,15]. The effect of the plant hormones abscisic acid (ABA), auxin, cytokinins, and gibberellin on root responses during water-deficit stress are also well-documented [8,16–18]. Thus, complex mechanisms contribute to root tissue responses to water-deficit stress [14,19–21]. These mechanisms are mediated by altered gene expression profiles in rice (Oryza sativa L.) [22,23], pine (Pinus pinaster Ait) [24] and maize (Zea mays L.) [25].
One crop influenced by the global reduction in available water resources is upland cotton (Gossypium hirsutum L.). Cotton is one of the world’s most valuable crops, providing much of the planet’s natural fiber for the global textile industry. Although additional economic value is captured from cottonseed and its associated products, cotton fiber represents about 90% of cotton’s total economic value [26]. China, the United States and India provide most of the world’s cotton, a combined total of more than 15.9 million metric tonnes of cotton lint and 30.4 million metric tonnes of cottonseed, a value of 22.8 billion and six billion dollars in 2011, respectively (FAOSTAT, www.faostat.fao.org). Environmental stresses such as extreme temperatures, soil salinity and water-deficit stress occur in these regions, further exacerbating population pressure as the effects of global climate change continue. Cotton is a warm-climate plant whose aerial tissues have evolved mechanisms conferring moderate tolerance to water-deficit stress [27–29]. An extensive root system also allows the plant to adjust to varying soil moisture levels. Plant breeding for water-deficit tolerance in cotton has resulted in a wide variety of adapted genotypes throughout the world [29–31].
Molecular processes in response to water-deficit stress have been studied at great length in cotton. Studies include the evaluation of global gene expression changes occurring during water-deficit in cultivated tetraploid cotton [18,22,32–34] and the diploid relatives G. arboreum L. and G. herbaceum L. [2,35–38]. Many of these experiments were conducted using microarray or cDNA-AFLP gene expression approaches. Although a number of significant changes in gene expression resulting from water-deficit stress were identified in these studies, the development of next generation sequencing technologies (NGS) offer opportunities to more accurately quantify those differences [39]. The recent publication of the whole genome sequence of the cotton diploid relative Gossypium raimondii Ulbrich [40] has expanded the use of NGS as a tool to study cotton development.
In this study, we report the first application of the diploid G. raimondii whole genome sequence and Illumina NGS technology to pursue RNA-seq analysis of global gene expression changes in field grown tetraploid cotton root tissue. Several genes and major biochemical pathways were up regulated in root tissue under water-deficit stress, confirming the success of this technique for transcriptome evaluation of tetraploid cotton species. Using NGS to assess global gene expression patterns in polyploid plant species is complicated; short reads found in several related loci can align to a single transcript or be removed from analysis, impacting accurate quantification of expression levels [41]. Gene duplication and genome reorganization events contribute to such complexity. In order to minimize the effects of genome complexity, we used the new PolyCat annotation pipeline [42] which assigns putative genome localization for many of the identified differentially expressed transcripts. Our objective was to use NGS to measure global gene expression profiles in field-grown tetraploid cotton root tissues under water-deficit stress to identify candidate genes for future research in molecular cotton breeding. Our results will provide an improved understanding of the putative transcriptional mechanisms involved in root responses to water-deficit stress in this important global crop.
Materials and Methods
Ethics Statement
The field studies did not involve any human, animal, or endangered species. The corresponding author is an Adjunct Professor with North Carolina State University and has unrestricted access to field research facilities.
Plant Materials
Root tissues from G. hirsutum cultivar ‘Siokra L-23’, selected for its previously established high level of water-deficit tolerance [27–29], were collected from field-grown plants under water-deficit and well-watered conditions at the North Carolina State University Sandhills Research Station near Jackson Springs, NC, USA according to the method described by 18. Roots were collected from three independent plants within each of the two water treatments. Samples were harvested during the third week of flowering on a single sample date, when significant differences in xylem water potential of the uppermost fully expanded leaves between treatments occurred, as determined by a pressure bomb (Model 600, PMS Instrument Company, Albany, OR). Plants were considered water-deficit stressed when leaf water potentials were -2.0 MPa or greater and well-watered when leaf water potentials were -1.9 MPa or lower [43]. Average water potential of well-watered and water-deficit stressed plants is presented in Table 1. Total RNA was isolated as previously described [18,44] using the XT buffer system with the addition of chloroform/iso-amyl alcohol extraction and LiCl precipitation steps [45].
Table 1. Leaf water potential values of upland cotton plants used in RNA-seq evaluation.
| Treatment | Plant | Leaf water potential (MPa) |
|---|---|---|
| Well-watered | 1 | -1.60 |
| 2 | -1.35 | |
| 3 | -1.45 | |
| Water-deficit | 1 | -2.20 |
| 2 | -2.70 | |
| 3 | -2.85 |
Leaf water potential values of selected upland cotton plants from both well-watered and water-deficit treatments.
RNA-Seq library construction and sequencing
Six individual barcoded libraries were created with the Illumina RNA TruSeq kit (Illumina) as per manufacturer's instructions using 2 µg of total RNA from three individual root RNA samples for each treatment. Library quality was assessed with an Agilent Bioanalyzer 2100 (Agilent) and the concentration of each individual library was calculated using qPCR. Libraries were pooled together so that each barcode was represented in equimolar amounts and sequenced in a single lane of 50bp Illumina HiSeq 2000.
Read trimming and mapping
Reads were trimmed with Sickle (https://github.com/najoshi/sickle) with a quality cutoff of 20. Genomic Short-Read Nucleotide Alignment Program (GSNAP) [46] was used to map reads to the G. raimondii 2.1 whole genome reference sequence [40], with SNP-tolerant mapping using a SNP index based on deep coverage of G. arboreum and G. raimondii, as described in [42]. The "-N 1" option was used for GSNAP to identify novel splice sites. Putative AT and DT genome localization of the differentially expressed transcripts was conducted using the PolyCat pipeline [42] which categorizes and maps DNA sequence reads of allotetraploid G. hirsutum to progenitor diploid genomes of G. arboreum (A2) and G. raimondii (D5) [47–49].
Differential expression
The total count of mapped reads for each library was converted to a CSV file and imputed into the DESeq (Version 1.9.12) package in R to test for significant differential expression between water-deficit and well-watered treatments [50] using a false discovery rate (FDR) of 5%. Data quality analysis was conducted by calculating and visualizing the Euclidean distance and principle component analysis of well-watered and water-deficit treatment samples using the DESeq (Version 1.9.12) package. Read count data was deposited in the National Center for Biotechnology Sequence Read Archive (NCBI SRA) (Accession No. PRJNA210770) and will be made available through CottonGen.
Functional annotation
Significant differentially expressed transcripts identified by DESeq analysis, and additional splice variants identified from the Gossypium raimondii v. 2.1 sequence [40] in Phytozome [51], were further evaluated for functional gene ontology annotation using default parameters in Blast2Go software [52]. Annotation was enhanced by merging the output of an additional InterProScan [53] analysis with the initial BLAST annotation so that additional transcripts without initial gene ontology association could be functionally characterized. The Blast2GO ANNEX program and an optional validation step were used to confirmed sequence annotation for each transcript [52]. Gene ontology enrichment analysis was conducted using AgriGO [54]. Differentially expressed transcripts with Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology IDs provided by Phytozome were mapped to specific pathways using the “Search and Color” Pathway tool, searching against the reference pathway (KO).
RT-qPCR RNA extraction and cDNA synthesis
Due to limited tissue amounts of the samples used for RNA-seq, root tissues were harvested from additional plants grown in the same plots and experimental conditions. Tissues were flash frozen in liquid nitrogen and stored at -80°C until being processed for RNA extraction. Individual root tissues from each plant per treatment were homogenized in liquid nitrogen and total RNA was extracted from 100 mg of homogenized tissue using the Spectrum Plant Total RNA kit with the On-Column DNase I Digest Set column DNAse (Sigma Aldrich) according to the manufacturer’s protocol. RNA was quantified using a NanoDrop spectrophotometer (ThermoFisher Scientific) and quality was examined using the Bioanalyzer 2100 (Agilent, Santa Clara, CA). A no-reverse transcription control on all RNA samples was used to determine DNA contamination using G. hirsutum alpha-tubulin (TUA11) gene (Gorai.010G125700), a reference gene identified from the RNA-seq data in this study. cDNAs were synthesized from 1 µg of total root RNA using the SuperScript® III First-Strand Synthesis SuperMix (Invitrogen) kit according to the manufacturer’s specifications. cDNA was diluted 10-fold for use in RT-qPCR reactions. Synthesized cDNAs were stored at -20°C.
RT-qPCR
Transcript sequences from G. raimondii were used in NCBI BLAST to identify the closest G. hirsutum sequence for primer design. Primers were designed using NCBI-Primer BLAST and diluted to a concentration of 5µM (Table 2). Template DNA for primer efficiencies was obtained through PCR using the primers specifically designed for RT-qPCR. PCR products were purified from agarose using the Wizard SV Gel and PCR Clean Up System (Promega) and DNA was quantified using the Qubit® dsDNA HS Assay Kit (Invitrogen). Two independent 10-fold dilutions of DNA for each dilution series were split into three reactions (12.5 µl per well). RT-qPCR was performed using Maxima SYBR Green/Rox qPCR Master Mix (2X) and the iCycler PCR Detection System with the standard two-step amp + melt protocol (Bio-Rad). Efficiencies were calculated using the protocols as described by 55,56.
Table 2. RT-qPCR target and reference gene primers for RNA-seq confirmation.
| Genename | G. raimondii transcript name | G. hirsutum sequence with greatest homology | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|---|---|
| GhPIP1;8 | Gorai.013G019300 | Gossypium hirsutum cultivar des119 aquaporin PIP1;8 | GTTTTCAGAGAGGCAACCTA | CCCAGCTCTATAAAAGGACC |
| GhAQP1 | Gorai.003G158100 | Gossypium hirsutum aquaporin 1 (AQP1) | TGGTTGTTAAGTGGGTGAAA | TAGTCCTTGTCTGTTTGAGC |
| GhNIP6;1 | Gorai.009G124500 | Gossypium hirsutum aquaporin (NIP6.1) | TCTCACTCACAAGAAAGGTG | ATCAGAGTTTCAGAGCCTTG |
| GhPIP2;8 | Gorai.009G107200 | Gossypium hirsutum PIP protein (PIP2;8) | ATTTGTGGTTGTGGGTTAGT | CAACCCAGTTCCCTTATTGA |
| GhTIP2;3 | Gorai.003G064000 | Gossypium hirsutum cultivar TM-1 aquaporin TIP2;3 | GCATCTTTTACTGGATTGCC | GATGATCTCCATCACCACTC |
| GhPOD6 | Gorai.012G141300 | Gossypium hirsutum bacterial-induced class III peroxidase (pod6) | GCTCGTGATTCTGTAGTTCT | CTGCAAATTTTTGCTTCTGC |
| GhPOD9 | Gorai.004G265900 | Gossypium hirsutum POD9 precursor (pod9) | CAAACACACTCAAACAACGA | TCTTGGTCTGTTTGAAGCAA |
| GhLea3 | Gorai.007G199900 | Gossypium hirsutum dehydrin (Lea3-D147) gene | GGACTGAAACAGAGGCTAAA | CCATCACTCCTTTCTTCTCG |
| GhCloMX | Gorai.002G078800 | Gossypium hirsutum clone MX019A11-jhj | ATCAGGCTTAGAAACACAGG | ATCTTCCTTTCCATGTTCCC |
| GhLOX1 | Gorai.006G238200 | Gossypium hirsutum bacterial-induced lipoxygenase (Lox1) | ATCCTATCAAGGCATTCGTC | TCTCTACAATCCGTTCCTCT |
| GhTHIA | Gorai.009G176400 | Gossypium hirsutum thiazole biosynthetic enzyme | ATGGACATGATCACCTATGC | AACAGACTGCTCGACAATAG |
| GhTUA11 | Gorai.010G125700 | Gossypium hirsutum alpha-tubulin (TUA11) | TTGGGATCTTTGTTGTTGGA | GTTCAAGAAGCGAATGAGTG |
Target and reference gene primers for RT-qPCR confirmation of RNA-seq results in tetraploid upland cotton. Primers were designed using NCBI-BLAST and diluted to 5 μM with annealing temperatures at 55 °C.
To confirm that each primer set used in RT-qPCR was accurately amplifying the correct G. hirsutum sequence, each purified PCR amplicon was cloned using either the TOPO Zero Blunt or TA Cloning Systems (Invitrogen), with OneShot Top10 competent cells (Invitrogen). PCR amplification using T3/T7 primers was used to confirm fragment insertion and correct orientation. Four individual colonies were bi-directionally sequenced as previously described in [44]. Sequence evaluation of inserted amplicons was conducted with Geneious software version 6.1 (Biomatters Ltd.) and homology-based BLAST search of amplicons was used to confirm sequence identity.
RT-qPCR was performed in duplicate 12.5 µl volumes with cDNAs from two independent cDNA superscript reactions and Maxima SYBR Green qPCR Master Mix (Fermentas). All reactions were analyzed with the iCycler Real Time PCR Detection System (Bio-Rad) with the default two-step amplification plus melt curve protocol for each reaction (Bio-Rad). Target transcripts included mRNA from ten genes identified as differentially expressed between water-deficit and well-watered treatments according the DESeq analysis of RNA-seq data. Transcript Gorai.012G141300 was selected internally from the RNA-seq data as a reference transcript and validated using the RefFinder program http://www.leonxie.com/referencegene.php). Relative Expression Ratios (RER) were calculated using the ΔCt method [55]. RT-qPCR protocols followed the MIQE guidelines [57].
Results
Total number of sequenced reads
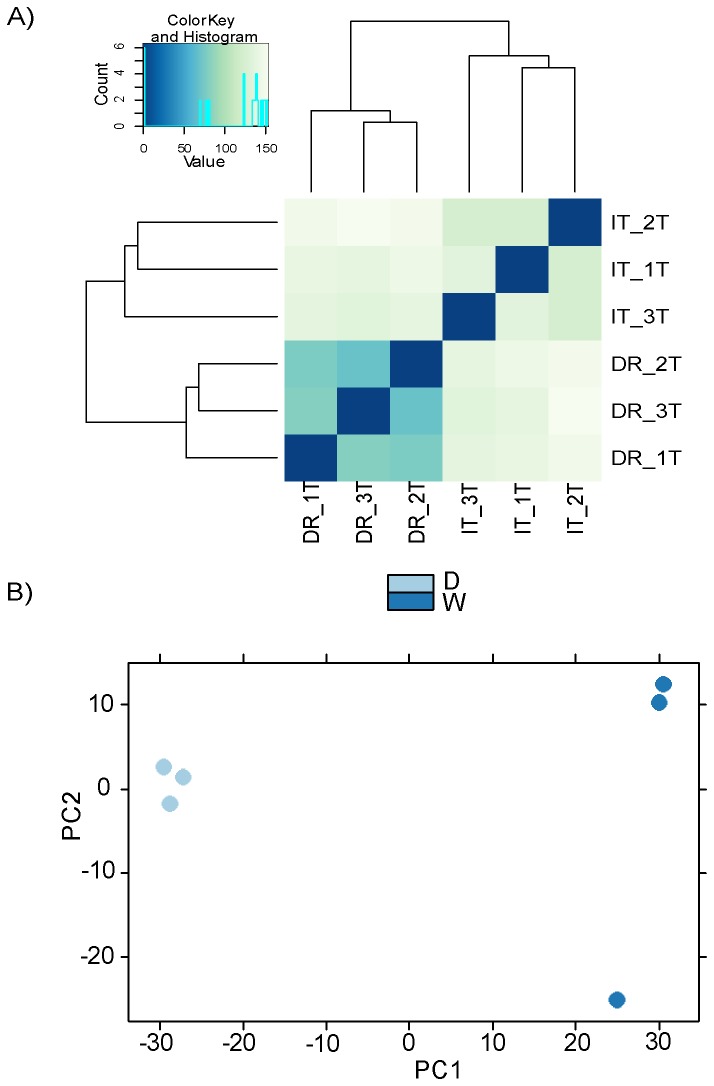
To assess global transcriptome changes occurring in the root tissue of tetraploid upland cotton during water-deficit stress, total RNA samples from three individual cotton plants of the cultivar Siokra L-23 under both water-deficit and well-watered conditions were used to create six independent libraries that were sequenced using the Illumina HiSeq 2000 sequencing platform. Approximately 109.6 million 50 bp reads from all six libraries were trimmed with Sickle and mapped to 33,930 transcripts from the G. raimondii 2.1 whole genome reference sequence [40]. Over 90% of identified transcripts had between 0 and 1000 mapped reads. Fifty percent of transcripts had fewer than 100 mapped reads, 50% of transcripts had more than 100 reads, and 7 % had more than 1000 mapped reads. Data quality was assessed using heatmap visualization of Euclidean distances and principle component analysis of all samples, conducted using DESeq (Version 1.9.12) [50] (Figure 1).
Figure 1. Data quality evaluation of mRNA-seq data.
A) Measurement of Euclidean distances and B) Principle Component Analysis (PCA) of all samples to assess data quality. Color key indicates level of similarity between libraries. Analysis was conducted using DESeq (Version 1.9.12) [50]. .
Global Transcriptome Changes during Water-Deficit Stress
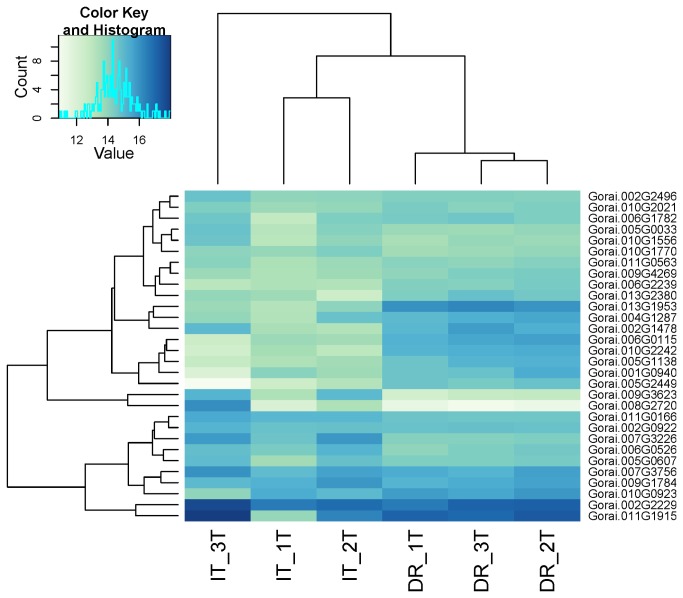
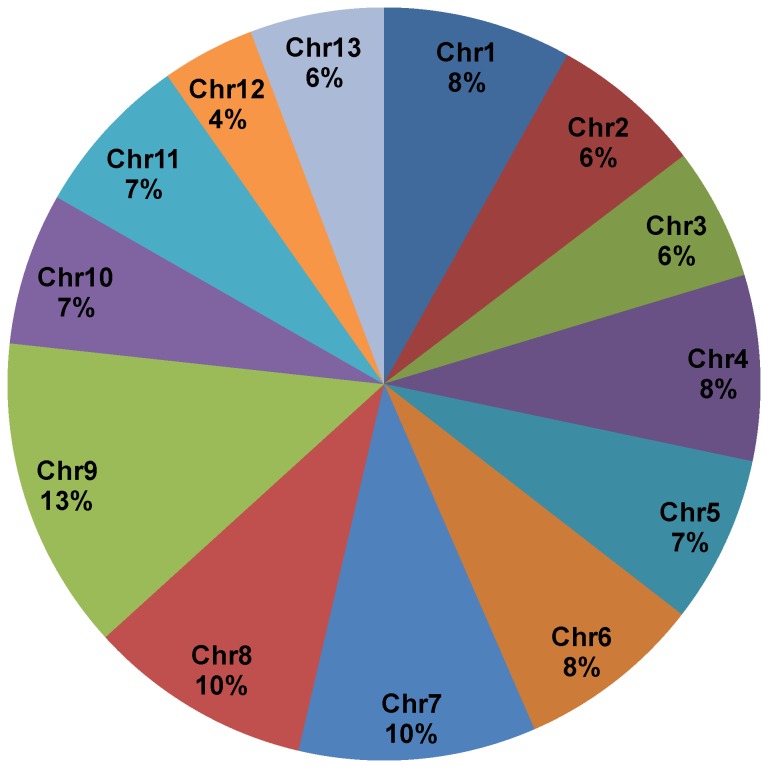
The total number of mapped sequenced reads for all identified transcripts was used for differential expression analysis in DESeq with an FDR of 0.05. A total of 1530 genes were either up or down regulated between water-deficit and well-watered upland cotton root samples. Of those 1530 genes, 913 were up-regulated under water-deficit stress and 617 down-regulated. A subset of differentially expressed genes is shown in Figure 2. Differentially expressed genes were distributed across all 13 chromosomes in the diploid progenitor genome of G. raimondii, determined by Gorai transcript IDs provided by Phytozome [40] (Figure 3). Several genes identified by a previously published gene expression study using cDNA-AFLP [18] were also found by RNA-seq. Although the total number of differentially expressed genes was different between the studies (304 in cDNA-AFLP and 1530 in RNA-seq), similar transcripts were involved in water uptake, heat stress and carbohydrate metabolism, including aquaporin water uptake protein PIP 1;3, Heat Shock Protein 26, and mannose-6-phosphate isomerase.
Figure 2. Visualization of thirty most differentially expressed genes.
Hierarchical clustering and heatmap visualization of the thirty most differentially expressed genes between well-watered and water-deficit treated upland cotton root samples, using variance stabilization with a FDR of %5 using DESeq (Version 1.9.12) [50]. Color key indicates transcript abundance for each gene.
Figure 3. Chromosomal distribution of all differentially expressed transcripts.
Distribution of differentially expressed transcripts, as determined by the alignment of G. hirsutum RNA-seq reads to the thirteen chromosomes of diploid relative G. raimondii.
Functional annotation of differentially expressed transcripts
Following differential expression analysis with DESeq, all significant transcripts and associated splice variants, a total of 2942 transcripts, were annotated using the Blast2Go application [52] . Of the 2942 total sequences, 2416 were successfully annotated; 102 genes were not analyzed because they exceeded the maximum size allowance (>8000bp) and 74 had no sequence homology in BLAST. After enhancing the annotation by including the results of an InterProScan database search and the ANNEX augmentation procedure, 112 additional annotations were added and 1821 annotations were confirmed.
Genome localization of differentially expressed transcripts
NGS and gene expression analyses are complicated in polyploid plants [58–61]. In order to putatively identify genome localization of the 1530 differentially expressed transcripts identified by RNA-seq, we used the PolyCat read mapping pipeline [42]. PolyCat uses SNPs identified between the related diploid genomes of G. arboreum (A2) and G. raimondii (D5) to map total NGS reads to either the A genome (AT) or D genome (DT) of the allopolyploid (AD1) G. hirsutum. Genes up-regulated under water-deficit predominately contained AT genome specific reads, with 407 (44.6%) of both the water-deficit and comparative well-watered transcripts having a majority of reads that mapped to the AT genome. Genes down-regulated in water-deficit were more evenly distributed, where transcripts from both treatments were comprised of reads mapping to either the AT (225, 36.5%) or DT (217, 35.4%) genomes. Only two (0.2%) of the up-regulated transcripts contained reads mapping only to the AT genome, and three (0.3%) transcripts contained reads mapping only to the DT genome. Only five (0.8%) genes down-regulated under water-deficit stress had reads that mapped to the AT genome only, and five transcripts were comprised of reads that mapped only to the DT genome. Of the total number of differentially expressed transcripts identified by RNA-seq, 101 could not be associated with a specific genome within tetraploid cotton (Table 3).
Table 3. Putative genome localization of transcripts according to the PolyCat annotation pipeline.
| Total up-regulated transcripts | AT both | DT both | AT Water-Deficit, DT Well-Watered | DT Water-Deficit, AT Well-Watered | AT only | DT only | None |
|---|---|---|---|---|---|---|---|
| 913 | 407 | 315 | 72 | 74 | 2 | 3 | 40 |
| Percentage of Total | 44.6 | 34.5 | 7.9 | 8.1 | 0.2 | 0.3 | 4.4 |
| Total down-regulated genes | AT both | DT both | AT Water-Deficit, DT Well-Watered | DT Water-Deficit, AT Well-Watered | AT only | DT only | None |
| 617 | 225 | 217 | 50 | 54 | 5 | 5 | 61 |
| Percentage of Total | 36.5 | 35.2 | 8.1 | 8.8 | 0.8 | 0.8 | 9.9 |
Putative genome localization of water-deficit stressed and well-watered associated transcripts based on SNP evaluation and comparison to diploid progenitor genomes, according to the PolyCat annotation pipeline [42]. “Both” denotes transcripts in which both water-deficit stressed and well-watered reads predominantly mapped to a specific genome, “AT Water-Deficit, DT Well-Watered” and “DT Water-Deficit, AT Well-Watered” denote those transcripts in which the predominant genome differed by treatment, “AT only and DT only” denote genes for which all reads mapped to a specific genome for both treatments. “None” denotes transcripts that could not be associated with a specific genome.
Gene ontology
Gene ontologies most highly represented for molecular function were “catalytic activity (1176)”, “binding (1196)”, “transporter activity (164)” and “nucleic acid binding transcription factor activity (189)”. Gene ontologies for cellular component were “cell (1664)”, “organelle (1142)” and “membrane (720)”. Gene ontologies for biological process were “metabolic process (1317)”, “cellular process” (1371), and “response to stimulus” (871). Not surprisingly, many of the enriched biological process ontology terms were “response to temperature stimulus (P-value: 1.40E-10), “response to high light intensity” (P-value: 1.70E-09), “response to heat” (P-value: 4.40E-09) “response to water (P-value: 3.2E-05)” and “response to water depravation (P-value: 6.2E-05)”. Enriched molecular function terms were “glycogen debranching enzyme activity” (P-value: 7.80E-08), “hydrolase activity, hydrolyzing O-glycosyl compounds” (P-value: 0.00022), “transcription regulator activity” (P-value: 0.00035) and “hydrolase activity, acting on glycosyl bonds” (P-value: 0.00064). Enriched cellular component were “protein serine/threonine phosphatase complex” (P-value: 0.0048), “stromule” (P-value: 0.0066) and “anchored to membrane” (P-value: 0.015).
KEGG pathway analysis
The primary pathways impacted in root tissue of upland cotton plants during water-deficit stress were starch and sucrose metabolism (109 sequences, 23 enzymes), glycolysis-gluconeogenesis (37 sequences, 11 enzymes), amino sugar and nucleotide sugar metabolism (35 sequences, 14 enzymes), and galactose metabolism (31 sequences, 14 enzymes). Other major plant pathways impacted included flavonoid biosynthesis (15 sequences, 6 enzymes), carotenoid biosynthesis (9 sequences, 2 enzymes), and oxidative phosphorylation (8 sequences, 2 enzymes) (Figure 4).
Figure 4. KEGG pathway visualization of starch and sucrose associated differentially expressed transcripts.
KEGG search and color pathway analysis of significant differentially expressed transcripts in starch and sucrose metabolism pathways in upland cotton root under water-deficit stress. Enzymes coded red are up regulated under water-deficit, blue genes are down regulated, and purple genes denote the reference pathway.
RT-qPCR of specific genes of interest
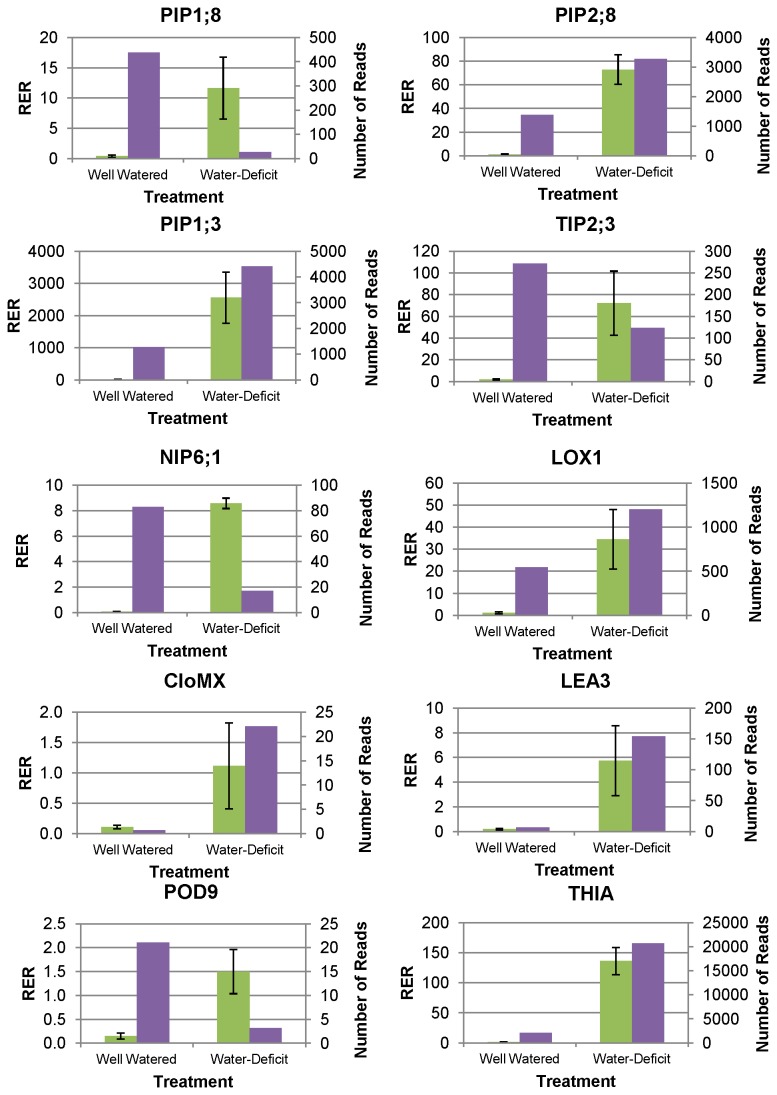
To further investigate the expression patterns of specific genes of interest from the RNA-seq experiment, we conducted RT-qPCR using 10 transcripts, representing a range in the number of reads per transcript, that were associated with specific aspects of stress physiology and water transport. Transcripts were also selected based on availability of G. hirsutum EST sequence, to simplify primer design and amplification in tetraploid cotton. Of the 10 transcripts selected, six (60%) were expressed in the pattern identified by RNA-seq. These six encoded thiazole biosynthetic enzyme (THIA), plasma membrane intrinsic aquaporin proteins PIP1;3 and PIP2.8, dehydrin (LEA3-D147), clone MX019A11-jhj (CLOMX), and bacterial-induced lipoxygenase (LOX1). In contrast, four genes (40%) had RT-qPCR gene expression patterns that differed from RNA-seq results, and were not differentially expressed between treatments. These were aquaporins PIP1;8, NIP6;1, and TIP2;3 and peroxidase precursor POD9. Overall, a majority of transcripts selected for RT-qPCR were expressed in the same manner as identified using RNA-seq (Figure 5).
Figure 5. RT-qPCR confirmation of RNA-seq results.
RT-qPCR confirmation (left Y-axis, green bars) and normalized RNA-seq read count values (right Y-axis, purple bars) of differentially expressed genes between well-watered and water-deficit treatments. RT-qPCR was calculated using the ΔCt method [55]. Error bars represent standard error of genotype means.
Discussion
Cotton is a major crop with global economic significance that requires specific environmental factors suitable for plant growth, development and production. As global climate change continues to increase demand for the world’s water resources, it is essential to identify approaches to improve our understanding of how crop plants including cotton respond to water-deficit stress. In the present study, we applied NGS technology to profile transcriptome changes in root tissue of upland cotton undergoing field water-deficit stress. A total number of 1530 differentially expressed genes were identified. In contrast from a previous study that found more genes to be down-regulated [18], most transcripts were up-regulated under water-deficit stress. To our knowledge, this is the first published use of the G. raimondii whole genome sequence and RNA-seq to measure transcriptome differences in field grown, tetraploid cotton. With this approach we have identified gene expression changes in root tissues under water-deficit stress, and many will serve as potential targets for future research and the development of molecular breeding tools for cotton breeding programs.
Many biochemical pathways are associated with root response to water-deficit
Results of the present study generally confirm previously identified biochemical mechanisms modulating the adaptation of cotton to water-deficit stress and demonstrate the utility of our methods in cotton genomics. Specifically, the induction we observed in genes associated with starch and sugar metabolism is similar to results obtained by other researchers [18,22,32–34] (Figure 4). As root tissues undergo water-deficit stress, increases in carbohydrate metabolism and other osmolyte concentrations alter the osmotic potential of the cell [9]. Water-deficit also induces several hormone responses, including cytokinins, auxin and abscisic acid [8,16–18]. Abscisic acid is an important signaling molecule in the development of root system architecture under water-deficit [12,14,62]. Genes upstream and within the abscisic acid pathway can be up-regulated under water-deficit and one gene specifically, β-carotene hydroxylase, has been shown to be induced under water-deficit stress [63]. The detection of the up regulation of the carotenoid biosynthesis ABA precursor gene 9-cis-epoxycarotenoid dioxygenase (NCED) and ABA pathway gene ABA 8'-hydroxylase in this RNA-seq data set identifies target candidate genes for further studies of water-deficit tolerance in the root system of upland cotton.
Other plant responses to water-deficit stress involve proteins responsible for cellular water uptake. Aquaporins are a large major intrinsic protein family consisting of 71 members in cotton [44] that have been shown to facilitate the movement of water and other small molecules across cell membranes [18,44,64]. Our results confirm differential aquaporin gene expression in response to water-deficit stress, as has been observed in many plant species, including cotton [18,65–68]. Specifically, we observed differential expression of aquaporin genes in both RNA-seq and RT-qPCR. These examples serve as additional evidence for the potential role of aquaporin expression in mediating water deficit stress tolerance in cotton root tissues.
Putative genome localization of water-deficit related genes in tetraploid cotton
Many agriculturally important plant species, such as wheat (Triticum aestivum L.), potato (Solanum tuberosum L.), and sugarcane (Saccharum officinarum L.) are polyploid [60]. Polyploidzation makes NGS technologies (such as RNA-seq) challenging. NGS depends on read mapping in which large amounts of nucleotide sequence are associated with genome localization; however, due to gene duplication and genomic restructuring events common in polyploids, it is difficult to accurately map reads to their genome of origin [41,42,58,69]. The development of annotation pipelines capable of assigning tetraploid transcript reads accurately to related diploid genomes is a significant improvement in the effort to assess gene expression in polyploidy plant species. In this study, we employed the use of PolyCat, a new NGS annotation pipeline capable of assigning reads from tetraploid G. hirsutum (AD1) to progenitor diploid A2 genome G. arboreum and D5 genome G. raimondii. Putative genome localization was provided by the comparison of SNP data from the sequence of the progenitor genomes to the NGS reads created by this study.
Gene expression responses to water-deficit stress have been studied extensively in a variety of cotton tissues [18,28,29,32,35,36]. While informative, to our knowledge, no previous study has reported potential genome specific responses to water-deficit. A majority of transcript reads mapped to the AT genome among genes that were up-regulated in response to water deficit; while genes that were down-regulated were represented evenly by both the A and D genomes. The up-regulation of AT genome-specific transcripts indicates the importance of the A genome diploid relative G. arboreum in water-deficit response, which has been previously identified to be a source of other stress-related genes [36–38]. Interestingly, transcripts from several genes that were either up- or down-regulated under water-deficit stress had reads that mapped to a single genome. Further investigation of these genome-specific transcripts is called for among AT and DT specific responses to water-deficit stress in tetraploid cotton.
Future considerations for transcriptome evaluation in tetraploid cotton
This study represents the first reported use of NGS technologies in combination with the recently published G. raimondii sequence to evaluate differential transcriptome profiles in upland cotton. Over 1500 genes were differentially expressed between water-deficit stress and well-watered root treatments. Expression patterns for genes associated with sugar metabolism, ABA synthesis, and water uptake were similar to those found in other published reports of gene expression analyses under water-deficit stress. Substantial up- regulation of genes associated with water-deficit, including those associated with responses to changes in temperature, high light intensity, heat, and water was detected in concert with gene ontology enrichment analysis with AgriGO [54]. Transcriptome profiling of tetraploid cotton using the G. raimondii published sequence successfully detected global gene expression changes during water-deficit stress. However, further considerations should be made when choosing genes for RT-qPCR analysis, as a majority of genes assayed by RT-qPCR had very low read count values. Very low read counts may exceed the level of accurate detection by the RT-qPCR or by other statistical methods [70–72]. Additionally, evaluating highly- conserved gene families, such as the aquaporins, may be more complicated due to gene duplication and sequence similarity, and this should be considered prior to evaluation [41]. Further comparison of RNA-seq studies using alignments to both the G. raimondii diploid genome and G. hirsutum transcriptome sequence, as it becomes more available, will be of significant interest.
Conclusions
Differentially expressed transcripts were associated with the up-regulation of important biochemical pathways needed for cellular osmotic balance, abscisic acid and cellular water uptake. Similar results from water-deficit stress research with microarray and cDNA-AFLP confirm the use and accuracy of this technique for future research in cotton genomics. An additional analysis of genome localization based on available SNP data identified AT up regulation of genes in response to water-deficit, the first discussion of a high throughput sequencing approach to quantify water-deficit responsive genome expression patterns within tetraploid cotton. Results from this study advance our current understanding of water-deficit response in the root tissue of upland cotton, opening new areas of research in cotton breeding and genomics.
Acknowledgments
Authors acknowledge assistance from Fanny Liu for Sanger DNA sequencing, Dr. Brian Dalley and the University of Utah Huntsman Cancer Institute for massively parallel DNA sequencing, and Mr. Jeff Chandler and North Carolina State University Sandhills Research Station staff for providing excellent technical field plot support. We also thank Ernie Strickland, Kendreal Wingate, and Bobby Fisher for their technical assistance. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Funding Statement
This research project was supported by funding from CRIS No. 6657-21000-006-00D of the U.S. 376 Department of Agriculture and a grant from Cotton Incorporated (Agreement No. 12-377 208). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fedoroff NV, Battisti DS, Beachy RN, Cooper PJM, Fischhoff DA et al. (2010) Radically rethinking agriculture for the 21st century. Science 327: 833–834. doi: 10.1126/science.1186834. PubMed: 20150494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deeba F, Pandey AK, Ranjan S, Mishra A, Singh R, et al. (2012) Physiological and proteomic responses of cotton (Gossypium herbaceum L.) to drought stress. Plant Physiology and Biochemistry 53: 6–18 [DOI] [PubMed]
- 3. Lawlor DW, Cornic G (2002) Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant Cell Environ 25: 275–294. doi: 10.1046/j.0016-8025.2001.00814.x. PubMed: 11841670. [DOI] [PubMed] [Google Scholar]
- 4. Ackerson RC (1980) Stomatal response of cotton to water stress and abscisic Acid as affected by water stress history. Plant Physiol 65: 455–459. doi: 10.1104/pp.65.3.455. PubMed: 16661212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103: 551–560. PubMed: 18662937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aranjuelo I, Molero G, Erice G, Avice JC, Nogués S (2011) Plant physiology and proteomics reveals the leaf response to drought in alfalfa (Medicago sativa L.). Journal of Experimental Botany 62: 111–123 [DOI] [PMC free article] [PubMed]
- 7. Jubany-Marí T, Munné-Bosch S, Alegre L (2010) Redox regulation of water stress responses in field-grown plants. Role of hydrogen peroxide and ascorbate. Plant Physiol Biochem 48: 351–358. doi: 10.1016/j.plaphy.2010.01.021. PubMed: 20199867. [DOI] [PubMed] [Google Scholar]
- 8. Zhu J-K (2011) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol, 62: 1–25. PubMed: 21314429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bray EA (1993) Responses to Water Deficit. Plant Physiology 103: 1035–1040. PubMed: 12231998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bray EA (1997) Plant responses to water deficit. Trends in Plant Science 2: 48–54. doi: 10.5363/tits.2.10_48. [DOI] [Google Scholar]
- 11. Gewin V (2010) An underground revolution. Nature 466: 552–553. doi: 10.1038/466552a. PubMed: 20671689. [DOI] [PubMed] [Google Scholar]
- 12. Sanchez-Calderon L, Ibarra-Cortes ME, Zepeda-Jazo I (2013) Root Development and Abiotic Stress Adaptation. In: Vahdati K, Leslie C. Abiotic Stress - Plant Responses and Applications in Agriculture. InTech. pp. 135–167. [Google Scholar]
- 13. Deak KI, Malamy J (2005) Osmotic regulation of root system architecture. Plant J 43: 17–28. doi: 10.1111/j.1365-313X.2005.02425.x. PubMed: 15960613. [DOI] [PubMed] [Google Scholar]
- 14. Malamy JE (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77. doi: 10.1111/j.1365-3040.2005.01306.x. PubMed: 16021787. [DOI] [PubMed] [Google Scholar]
- 15. Takahashi N, Yamazaki Y, Kobayashi A, Higashitani A, Takahashi H (2003) Hydrotropism interacts with gravitropism by degrading amyloplasts in seedling roots of Arabidopsis and Radish. Plant Physiol 132: 805–810. doi: 10.1104/pp.018853. PubMed: 12805610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiong L, Wang R-G, Mao G, Koczan JM (2006) Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic Acid. Plant Physiol 142: 1065–1074. doi: 10.1104/pp.106.084632. PubMed: 16963523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227. PubMed: 17075077. [DOI] [PubMed] [Google Scholar]
- 18. Park W, Scheffler BE, Bauer PJ, Campbell BT (2012) Genome-wide identification of differentially expressed genes under water deficit stress in upland cotton (Gossypium hirsutum L .). BMC Plant Biology 12: 90 [DOI] [PMC free article] [PubMed]
- 19. Thomas JC, Bohnert HJ (1993) Salt stress perception and plant growth regulators in the halophyte Mesembryanthemum crystallinum. Plant Physiol 103: 1299–1304. PubMed: 12232022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Talanova VV, Titov AF (1994) Endogenous abscisic acid content in cucumber leaves under the influence of unfavourable temperatures and salinity. Journal of Experimental Botany 45 : 1031–1033. doi: 10.1093/jxb/45.7.1031. [DOI] [Google Scholar]
- 21. Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I et al. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. doi: 10.1038/nature03184. PubMed: 15635403. [DOI] [PubMed] [Google Scholar]
- 22. Rabello AR, Guimarães CM, Rangel PHN, da Silva FR, Seixas D et al. (2008) Identification of drought-responsive genes in roots of upland rice (Oryza sativa L). BMC Genomics 9: 485. doi: 10.1186/1471-2164-9-485. PubMed: 18922162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moumeni A, Satoh K, Kondoh H, Asano T, Hosaka A et al. (2011) Comparative analysis of root transcriptome profiles of two pairs of drought-tolerant and susceptible rice near-isogenic lines under different drought stress. BMC Plant Biol 11: 174. doi: 10.1186/1471-2229-11-174. PubMed: 22136218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dubos C, Plomion C (2003) Identification of water-deficit responsive genes in maritime pine (Pinus pinaster Ait.) roots. Plant Mol Biol 51: 249–262. doi: 10.1023/A:1021168811590. PubMed: 12602883. [DOI] [PubMed] [Google Scholar]
- 25. Bruce WB, Edmeades GO, Barker TC (2002) Molecular and physiological approaches to maize improvement for drought tolerance. J Exp Bot 53: 13–25. doi: 10.1093/jexbot/53.366.13. PubMed: 11741036. [DOI] [PubMed] [Google Scholar]
- 26. Campbell BT, Hinze L (2010) Cotton production, processing, and uses of cotton. In: Singh B. Industrial Crops and Uses. UK: CABI; pp. 259–276. [Google Scholar]
- 27. Stiller WN, Read JJ, Constable GA, Reid PE (2005) Selection for water use efficiency traits in a cotton breeding program : cultivar differences. Crop Science 1: 1107–1113. [Google Scholar]
- 28. Nepomuceno AL, Oosterhuis DM, Stewart JM (1998) Physiological responses of cotton leaves and roots to water deficit induced by polyethylene glycol. Environmental and Experimental Botany 40: 29–41. doi: 10.1016/S0098-8472(98)00018-5. [DOI] [Google Scholar]
- 29. Voloudakis AE, Kosmas SA, Tsakas S, Eliopoulos E, Loukas M et al. (2002) Expression of selected drought-related genes and physiological response of Greek cotton varieties. Functional Plant Biology 29: 1237–1245. doi: 10.1071/PP01253. [DOI] [PubMed] [Google Scholar]
- 30. Patil D V, Deosarkar DB, Gaikwad AR (2012) Drought tolerance studies in cotton genotypes. Journal of Cotton Research and Development 26: 21–25
- 31. Sarwar M, Khan IA, Azhar FM, Ali A (2012) Generation means analysis in cotton (Gossypium hirsutum L .) for drought tolerance. Pakistan Journal of Nutrition 11: 843–847
- 32. Payton P, Rao K, Wright RJ, Allen RD (2011) Examining the drought stress transcriptome in cotton leaf and root tissue. Biotechnol Lett, 33: 821–828. PubMed: 21188619. [DOI] [PubMed] [Google Scholar]
- 33. Xie F, Sun G, Stiller JW, Zhang B (2011) Genome-wide functional analysis of the cotton transcriptome by creating an integrated EST database. PLOS ONE 6: e26980. doi: 10.1371/journal.pone.0026980. PubMed: 22087239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Venkata K, Gurusamy P, Mogilicherla D, Kumar S, Dass A et al. (2012) Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Mol Biol 78: 223–246. doi: 10.1007/s11103-011-9857-y. PubMed: 22143977. [DOI] [PubMed] [Google Scholar]
- 35. Ranjan A, Nigam D, Asif MH, Singh R, Ranjan S, et al. (2012) Genome wide expression profiling of two accession of G. herbaceum L. in response to drought. BMC Genomics 13: 94 [DOI] [PMC free article] [PubMed]
- 36. Zhang L, Li FG, Liu CL, Zhang CJ, Zhang Y (2009) Construction and analysis of cotton (Gossypium arboreum L.) drought-related cDNA library. BMC Res Notes 8: 1–8. PubMed: 19570239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X, Yao D, Wang Q, Xu W, Wei Q et al. (2013) mRNA-seq Analysis of the Gossypium arboreum transcriptome reveals tissue selective signaling in response to water stress during seedling stage. PLOS ONE 8: e54762. doi: 10.1371/journal.pone.0054762. PubMed: 23382961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barozai MY, Husnain T (2012) Identification of biotic and abiotic stress up-regulated ESTs in Gossypium arboreum . Mol Biol Rep 39: 1011–1018. doi: 10.1007/s11033-011-0826-y. PubMed: 21556756. [DOI] [PubMed] [Google Scholar]
- 39. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517. doi: 10.1101/gr.079558.108. PubMed: 18550803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J et al. (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427. doi: 10.1038/nature11798. PubMed: 23257886. [DOI] [PubMed] [Google Scholar]
- 41. Ilut DC, Coate JE, Luciano AK, Owens TG, May GD et al. (2012) A comparative transcriptomic study of an allotetraploid and its diploid progenitors illustrates the unique advantages and challenges of RNA-seq in plant species. Am J Bot 99: 383–396. doi: 10.3732/ajb.1100312. PubMed: 22301896. [DOI] [PubMed] [Google Scholar]
- 42. Page JT, Gingle AR, Udall JA (2013) PolyCat: A resource for genome categorization of sequencing reads from allopolyploid organisms. 3 517–525 [DOI] [PMC free article] [PubMed]
- 43. Guinn G, Mauney JR (1984) Fruiting of cotton. II. Effects of plant moisture status and active boll load on boll retention. Agronomy Journal 76: 94–98. doi: 10.2134/agronj1984.00021962007600010024x. [DOI] [Google Scholar]
- 44. Park W, Scheffler BE, Bauer PJ, Campbell BT (2010) Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L .). BMC Plant Biology 10: 142 [DOI] [PMC free article] [PubMed]
- 45. Wan C, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Analytical Biochemistry 223: 7–12 [DOI] [PubMed]
- 46. Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881. doi: 10.1093/bioinformatics/btq057. PubMed: 20147302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Advances in Agronomy 78: 139–186. doi: 10.1016/S0065-2113(02)78004-8. [DOI] [Google Scholar]
- 48. Senchina DS, Alvarez I, Cronn RC, Liu B, Rong J et al. (2003) Rate variation among nuclear genes and the age of polyploidy in Gossypium . Mol Biol Evol 20: 633–643. doi: 10.1093/molbev/msg065. PubMed: 12679546. [DOI] [PubMed] [Google Scholar]
- 49. Flagel LE, Wendel JF, Udall JA (2012) Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Genomics 13: 302. doi: 10.1186/1471-2164-13-302. PubMed: 22768919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. doi: 10.1186/gb-2010-11-10-r106. PubMed: 20979621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186. doi: 10.1093/nar/gkr944. PubMed: 22110026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Conesa A, Stefan G (2008) Blast2GO : A comprehensive zuite for functional analysis in plant genomics. International Journal of Plant Genomics 2008: 619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N et al. (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33: W116–W120. doi: 10.1093/nar/gni118. PubMed: 15980438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70. doi: 10.1093/nar/gkq310. PubMed: 20435677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. doi: 10.1093/nar/29.9.e45. PubMed: 11328886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rasmussen R (2001) Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K-I. Rapid Cycle Real-Time PCR SE - 3. Springer; Berlin: Heidelberg. pp. 21–34 [Google Scholar]
- 57. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622. doi: 10.1373/clinchem.2008.112797. PubMed: 19246619. [DOI] [PubMed] [Google Scholar]
- 58. Udall JA, Swanson JM, Nettleton D, Percifield RJ, Wendel JF (2006) A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics 173: 1823–1827. doi: 10.1534/genetics.106.058271. PubMed: 16702424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Osborn TC, Chris Pires J, Birchler JA, Auger DL, Chen ZJ et al. (2003) Understanding mechanisms of novel gene expression in polyploids. Trends in Genetics 19: 141–147. doi: 10.1016/S0168-9525(03)00015-5. PubMed: 12615008. [DOI] [PubMed] [Google Scholar]
- 60. Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8: 135–141. doi: 10.1016/j.pbi.2005.01.001. PubMed: 15752992. [DOI] [PubMed] [Google Scholar]
- 61. Adams KL (2007) Evolution of duplicate gene expression in polyploid and hybrid plants. J Hered 98: 136–141. doi: 10.1093/jhered/esl061. PubMed: 17208934. [DOI] [PubMed] [Google Scholar]
- 62. Munns R, Sharp RE (1993) Involvement of abscisic acid in controlling plant growth in soil of low water potential. Functional Plant Biology 20: 425–437. [Google Scholar]
- 63. Davison PA, Hunter CN, Horton P (2002) Overexpression of [beta]-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418: 203–206. doi: 10.1038/nature00861. PubMed: 12110893. [DOI] [PubMed] [Google Scholar]
- 64. Chrispeels MJ, Agre P (1994) Aquaporins: water channel proteins of plant and animal cells. Trends Biochem Sci 19: 421–425. doi: 10.1016/0968-0004(94)90091-4. PubMed: 7529436. [DOI] [PubMed] [Google Scholar]
- 65. Hayano-Kanashiro C, Calderón-Vázquez C, Ibarra-Laclette E, Herrera-Estrella L, Simpson J (2009) Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLOS ONE 4: e7531. doi: 10.1371/journal.pone.0007531. PubMed: 19888455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jain D, Chattopadhyay D (2010) Analysis of gene expression in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance. BMC Plant Biol 10: 24. doi: 10.1186/1471-2229-10-24. PubMed: 20144227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lata C, Sahu PP, Prasad M (2010) Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochem Biophys Res Commun 393: 720–727. doi: 10.1016/j.bbrc.2010.02.068. PubMed: 20171162. [DOI] [PubMed] [Google Scholar]
- 68. Jang JY, Kim DG, Kim YO, Kim JS, Kang H (2004) An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol Biol 54: 713–725. doi: 10.1023/B:PLAN.0000040900.61345.a6. PubMed: 15356390. [DOI] [PubMed] [Google Scholar]
- 69. Udall JA, Swanson JM, Haller K, Rapp RA, Sparks ME et al. (2006) A global assembly of cotton ESTs A global assembly of cotton ESTs. Genome Research: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ferrer A, Tarazona S, Garcı F, Conesa A (2011) Differential expression in RNA-seq : A matter of depth. Genome Res, 21: 2213–2223. PubMed: 21903743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bullard JH, Purdom E, Hansen KD, Dudoit S (2010) Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11: 94. doi: 10.1186/1471-2105-11-94. PubMed: 20167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Oshlack A, Wakefield MJ (2009) Transcript length bias in RNA-seq data confounds systems biology. Biol Direct 10: 1–10. PubMed: 19371405. [DOI] [PMC free article] [PubMed] [Google Scholar]