Abstract
RhoA GTPase plays a crucial role in numerous biological functions and is linked to cancer metastasis. However, the understanding of the molecular mechanism responsible for RhoA transcription is still very limited. Here we show that RhoA transcription is orchestrated by the Myc/Skp2/Miz1/p300 transcription complex. Skp2 cooperates with Myc to induce RhoA transcription by recruiting Miz1 and p300 to the RhoA promoter independently of SCF-Skp2 E3 ligase activity. Deficiency of this complex results in impairment in RhoA expression, cell migration, invasion, and breast cancer metastasis, recapitulating the phenotypes observed in RhoA knockdown, and RhoA restoration rescues the defect in cell invasion. Strikingly, the overexpression of Myc/Skp2/Miz1 complex is found in metastatic human cancers and correlated with RhoA expression. Our study provides great insight into how oncogenic Skp2 and Myc coordinate to induce RhoA transcription and establishes a novel SCF-Skp2 E3 ligase-independent function for oncogenic Skp2 in transcription and cancer metastasis.
Cancer metastasis is a complex process involving several key steps that allow disseminating primary cancer cells to colonize into distant site1,2. It causes the majority of deaths of patients with cancer, and the treatment of this devastating form of disease remains a big challenge.
RhoA, a well-known member of the Rho family of GTPases, regulates numerous biological functions and is implicated in cancer metastasis3,4. Although mutation of RhoA has not been found in human cancers4-6, upregulation of RhoA mRNA and protein levels have been well documented in various human cancers7-9. Moreover, RhoA overexpression overcomes senescence checkpoints and induces preneoplastic transformation of human mammary epithelial cells10, whereas RhoA deficiency suppresses the invasiveness of breast cancer cells11. Importantly, recent study also demonstrates that microRNA-31 (miR-31) suppresses breast cancer metastasis partly through downregulation of RhoA gene expression12. Although accumulating evidence unequivocally delineates that RhoA overexpression is positively correlated with cancer metastasis13,14, the transcription machinery responsible for RhoA gene expression has been so far a mystery.
The Myc transcription factor heterodimerizes with Max and binds to the E-box motif CACGTG to activate transcription by cooperating with multiple coactivator complexes15-17. It serves a master regulator in various biological functions and is linked to metastasis. Recent studies demonstrate that Myc ubiquitination plays a critical role in Myc transcriptional activation in a subset of target genes important for cell proliferation18-20. Although a handful of Myc target genes involved in Myc-driven cell cycle progression and apoptosis have been identified, its target genes responsible for Myc-mediated cell migration and metastasis still remain elusive.
In this study, we aimed to identify the transcription machinery critical for RhoA transcription and cancer metastasis. Our study reveals that the Myc/Skp2/Miz1 transcription complex is critical for RhoA gene transcription, cell invasion, and cancer metastasis.
Results
Myc serves a transcription factor for RhoA
To explore the transcriptional machinery for RhoA, we analyzed the 5’-sequence upstream of the transcription start site up to 9 kilobase-pairs (Kb) using the computer program TFSEARCH (http://www.cbrc.jp/reSEARCH/db/TFSEARCH.html). We found that there are one canonical (CACGTG) and five noncanonical E-boxes (CACGCG) present within 2 Kb upstream of transcription start site (Fig. 1a), reminiscent of the binding sites for Myc and Twist17,21,22. Myc and Twist contribute to multiple steps of tumourigenesis and are linked to cancer metastasis16,21-24. To determine the role of Myc and Twist in RhoA transcription, we cloned this human RhoA promoter fragment (nucleotides −1850 to +1) into the pGL3 luciferase vector for a luciferase activity assay. Notably, RhoA transcriptional activity and protein expression were induced by Myc overexpression, but not by Twist overexpression (Fig. 1a and Supplementary information, Fig. S1a, b). This result suggests that RhoA transcription is selectively induced by Myc.
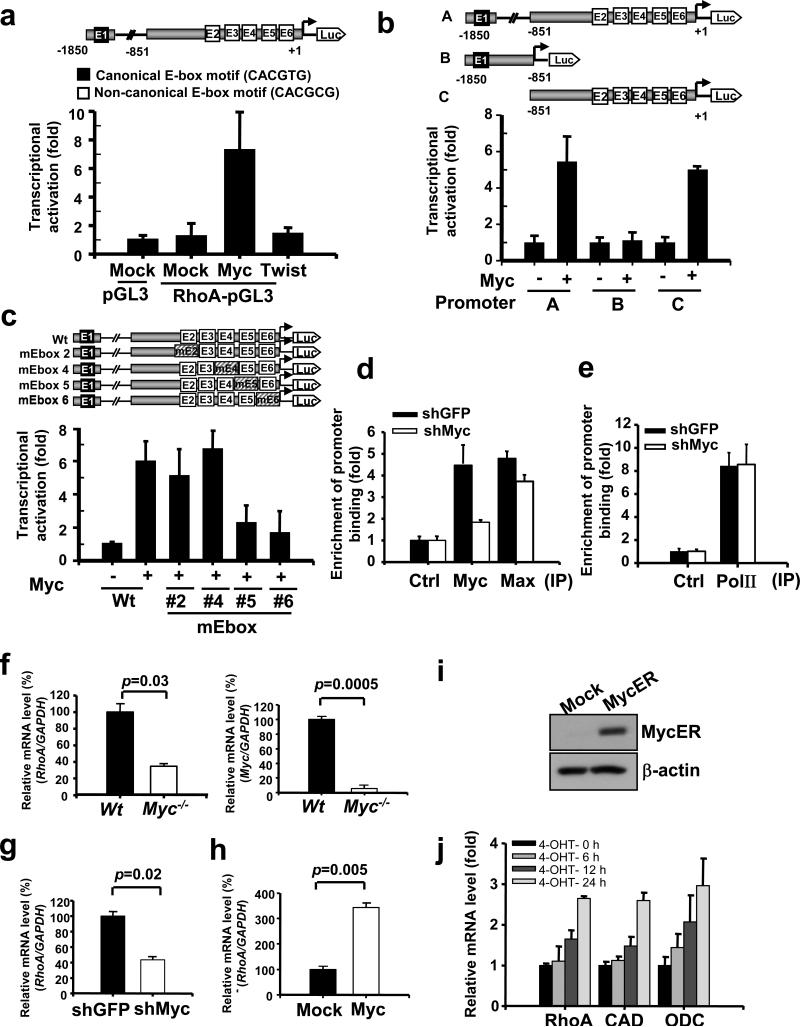
Figure 1. Myc is a transcription factor for RhoA.
(a) 293T cells were transfected with the indicated plasmids and harvested for a RhoA reporter assay. (b) Various RhoA promoter fragment constructs used for the reporter assay. (c) Reporter assay in 293T cells transfected with various RhoA promoter constructs with mutations in different E-box regions. (d, e) ChIP assay in PC-3 cells with GFP- or Myc-knockdown. Myc knockdown efficiency was shown in the left panel of Fig. 2b. (f) Analysis of RhoA and Myc mRNA levels in Wt and Myc-null Rat1 cells by real-time PCR. The quantified results were presented as mean ± s.d. (n=3) (g) Analysis of RhoA mRNA levels in MDA-MB-231 cell lines infected with GFP or Myc knockdown. (h) Analysis of RhoA mRNA levels in MDA-MB-231 cell lines infected with MSCV or MSCV-Myc. (i) Western blot analysis of Myc protein expression in MDA-MB-231 cells infected with pBabe or pBabe-MycER. (j) Analysis of RhoA , CAD, and ODC mRNA levels in MDA-MB-231 cell lines following the time course of MycER activation induced by 4-OHT (4-hydoxytamoxifen). Results in panel a-c were shown as mean ± s.d. of one representative experiment (a, c from 4 independent experiments; b from 3 independent experiments) performed in triplicate. Results in panel d-h, j were presented as mean ± s.d. (n=3).
To dissect the promoter region required for RhoA transcription by Myc, we generated two fragments from the full-length RhoA promoter. To our surprise, the transcriptional activity of the distal region (fragment B) containing the canonical E-box motif was not induced by Myc, whereas the transcriptional activity of fragment C containing 5 non-canonical E-box motifs was readily induced by Myc (Fig. 1b), suggesting that Myc may affect RhoA transcription by recognizing any one of 5 non-canonical E-box motifs within fragment C. To validate this notion, we mutated these non-canonical E-box motifs individually and used them in a reporter assay. We successfully generated full-length RhoA promoters with mutations in E-box 2, 4, 5, or 6 located within fragment C; a promoter with mutations in E-box 3 was not obtained because of its high GC-rich content. The basal transcriptional activity of these promoters was not affected by the mutations (Supplementary information, Fig. S1c). Although mutations in E-boxes 2 and 4 did not significantly affect the ability of Myc to induce RhoA transcription activation, mutations in either E-box 5 or E-box 6 impaired the effect of Myc on RhoA transcription activation (Fig. 1c), suggesting that Myc may bind to E-box 5 and 6 to regulate RhoA transcription.
To further corroborate this notion, we performed in vivo chromatin immunoprecipitation (ChIP) assays to address whether Myc binds to the RhoA promoter region surrounding E-boxes 5 and 6. The ChIP assay revealed that endogenous Myc bind to the E-box 5 and 6, whereas Myc knockdown attenuated this effect (Fig. 1d, 2b). We found that Max was also present in this RhoA promoter, but the recruitment of Max to this RhoA promoter was only slightly affected by Myc knockdown (Fig. 1d). Control experiments showed that RNA polymerase II bound to the GAPDH promoter irrespectively of Myc expression (Fig. 1e). Real-time PCR revealed that RhoA mRNA in Myc-null cells was much lower than in Wild-type (Wt) cells (Fig. 1f). Similarly, Myc knockdown reduced RhoA mRNA expression, whereas Myc overexpression induced it (Fig. 1g, h).
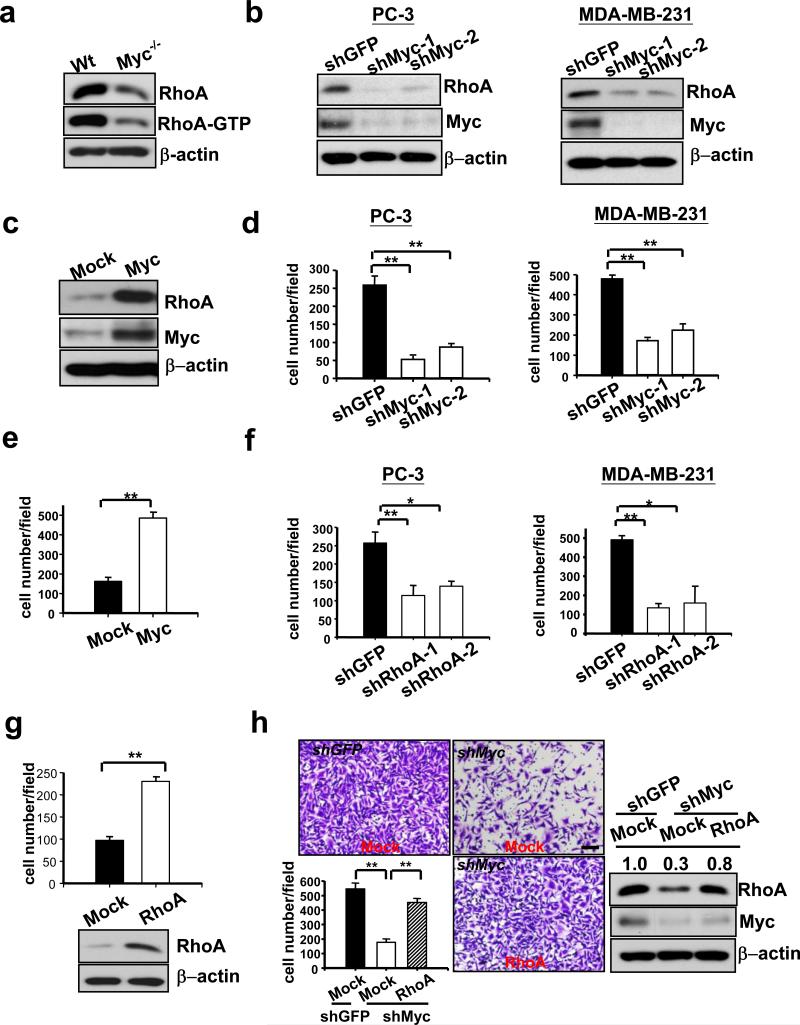
Figure 2. Myc regulates cell invasion through RhoA.
(a) Western blot analysis of active RhoA (RhoA-GTP) and total RhoA expression in Wt Myc and Myc-null Rat1 cells. (b) Western blot analysis of RhoA expression in two cancer lines with GFP or Myc knockdown. Two Myc lentiviral shRNAs were used in this assay. (c) Western blot analysis of RhoA expression in MDA-MB-231 cells infected with MSCV or MSCV-Myc. (d) Matrigel cell invasion assay in two cancer cell lines with GFP or Myc knockdown. (e) Matrigel cell invasion assay in MDA-MB-231 cells infected with MSCV or MSCV-Myc. (f) Matrigel cell invasion assay in two cancer cell lines with GFP or RhoA knockdown. Two RhoA lentiviral shRNAs were used in this assay. (g) Matrigel cell invasion assay and Western blot analysis in MDA-MB-231 cells infected with MSCV or MSCV-RhoA. (h) Matrigel cell invasion assay and Western blot analysis in MDA-MB-231 cells with GFP, Myc knockdown, or Myc knockdown plus RhoA overexpression. The relative intensity of RhoA was quantified using ImageQuant software and normalized with β-actin. The quantified results were presented as mean ± s.d. (n=4). * indicates p<0.05; ** indicates p<0.01. Scale bar = 100 μm.
We examined the kinetics of RhoA transcription by using inducible MycER system20,25. MDA-MB-231 cells with stable MycER expression were generated, treated with 4-OHT (4-hydoxytamoxifen) to activate MycER proteins, and harvested for RhoA expression (Fig.1i). RhoA mRNA levels were induced following the MycER activation by 4-OHT in a time-dependent manner, similar to that of CAD and ODC (ornithine decarboxylase), two known Myc target genes26,27 (Fig. 1j). These results suggest that Myc serves as a transcriptional factor to activate RhoA transcription.
RhoA mediates Myc-regulated cell migration and invasion
We next determined whether Myc levels are critical for RhoA protein expression and activity. Both RhoA protein expression and RhoA activity were greatly reduced in Myc-null cells, indicating that Myc regulates RhoA protein expression and in turn affects RhoA activity (Fig. 2a). Similarly, knockdown of Myc expression profoundly reduced RhoA protein expression in cancer cells, while Myc overexpression promoted it (Fig. 2b, c). RhoA protein expression was also induced following the MycER activation in a time-dependent manner (Supplementary information, Fig. S1d). Since RhoA plays a critical role in cell migration and invasion3,4, we determined the role of Myc in these processes. Notably, Myc knockdown in cancer cells robustly attenuated cell migration and invasion, whereas Myc overexpression promoted these processes (Fig. 2d, e and Supplementary information, Fig. S1e, h). Importantly, RhoA knockdown or overexpression phenocopied the effects of Myc on cell migration and invasion (Fig. 2f, g and Supplementary information, Fig. S1f, g, i), raising the possibility that Myc may regulate these processes by affecting RhoA expression. In support of this notion, we found that restoration of RhoA expression rescued cell invasion defect in Myc-knockdown cells (Fig, 2h). The effect of Myc and RhoA deficiency on cell migration and invasion was not due to their ability to regulate cell proliferation and apoptosis, since Myc and RhoA knockdown did not significant affect either cell proliferation or apoptosis within 2 days given that all our migration and invasion assays were performed within 24 hours (Supplementary information, Fig. S2a-c). However, Myc knockdown significantly inhibited cell proliferation at later time points and primary tumour formation (Supplementary information, Fig. S2a, d), supporting its oncogenic functions16,17. Our results identify RhoA as a critical effector for Myc-mediated cell migration and invasion.
Skp2 cooperates with Myc to promote RhoA transcription independently of SCF-Skp2 E3 ligase activity
Although ubiquitination is thought to regulate protein degradation, recent studies suggest that ubiquitination also has a nonproteolytic role in the control of transcription28,29. HectH9 and Skp2 E3 ligases have been demonstrated to induce Myc ubiquitination and to cooperate with Myc to induce a subset of target genes important for cell cycle progression and proliferation18-20. To investigate whether Myc ubiquitination is also important for Myc-mediated RhoA transcription, we examined the impact of HectH9 and Skp2 on RhoA transcription. HectH9 overexpression failed to promote RhoA transcriptional activation, protein expression, and Myc-mediated RhoA transcription, although it induced Myc ubiquitination (Fig. 3a and Supplementary information, Fig. S3a, b)18. Moreover, we found that HectH9 was not required for RhoA protein expression and the ability of Myc to induce RhoA transcription (Supplementary information, Fig. S3c, d). These results suggest that a subset of Myc target genes such as RhoA is not regulated by HectH9 and HectH9-mediated Myc ubiquitination. We found; however, that RhoA transcription was induced by Skp2 overexpression, and a synergistic effect on RhoA transcription was observed upon Skp2 and Myc overexpression (Fig. 3a). Skp2 is an F-box protein that forms a Skp2-SCF complex with Skp1, Cullin-1, and Rbx1 to constitute an E3 ligase activity, and its F-box domain is required for SCF-Skp2 E3 ligase activity and Skp2 oncogenic functions30,31. Since Skp2 regulated Myc ubiquitination (Supplementary information, Fig. S3e), we wished to determine whether SCF-Skp2 E3 ligase activity is required for Skp2's effect on RhoA transcription. Consistent with the previous reports19,32, we found that the Skp2-LRR mutant devoid of the N-terminal and F-box domain failed to form a Skp2-SCF complex and lost its E3 ligase activity towards promoting ubiquitination of p27 and Myc (Supplementary information, Fig. S4a-c), two well-known Skp2 substrates19,20,30,31. Surprisingly, Skp2-LRR synergized with Myc to activate RhoA transcription as Wt Skp2 did (Fig. 3b). This result suggests that SCF-Skp2 E3 ligase activity and Myc ubiquitination induced by Skp2 are not critical for RhoA transcription.
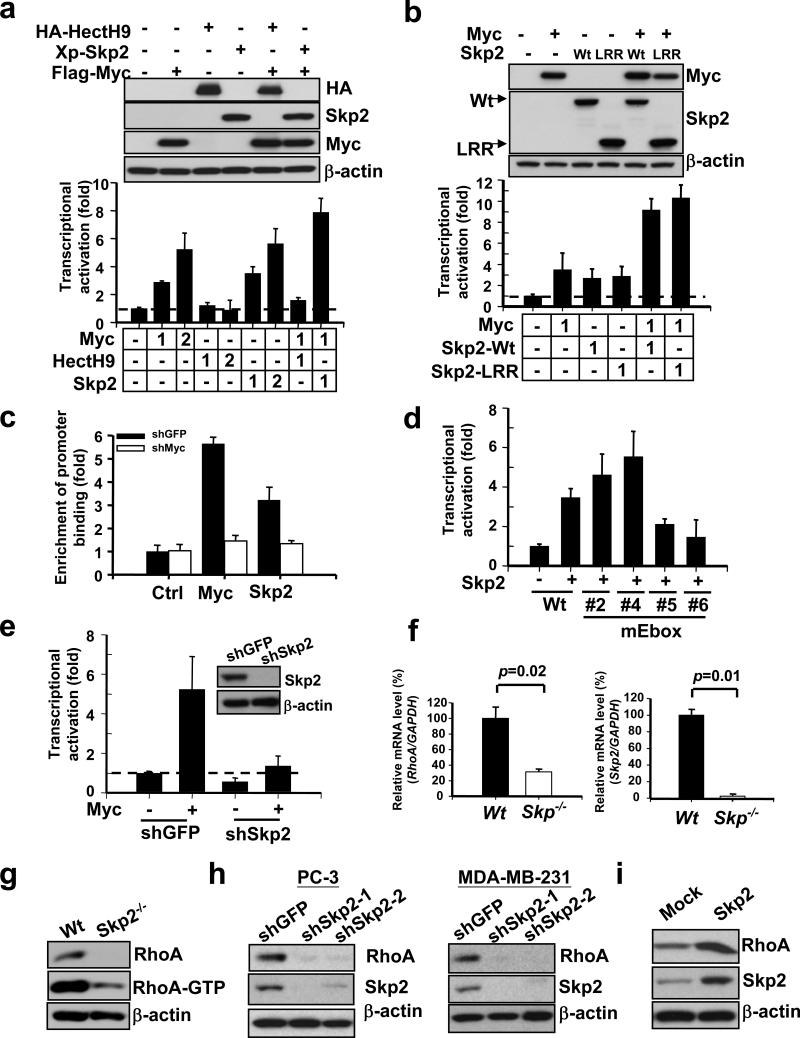
Figure 3. Skp2 cooperates with Myc to regulate RhoA transcription independent of Myc ubiquitination and SCF-Skp2 E3 ligase activity.
(a, b) RhoA reporter assay in 293T cells transfected with the indicated plasmids. The number indicates the amount (μg) of the plasmids used in this assay (lower panel). Western blot analysis showing the expression of constructs as indicated (upper panel). (c) ChIP assay in PC-3 cells with GFP or Myc knockdown. The quantified results were presented as mean ± s.d. (n=3) (d) RhoA reporter assay in 293T cells transfected with various RhoA promoter constructs with mutations in different E-box regions. (e) RhoA reporter assay in 293T cells with GFP or Skp2 knockdown (left panel). Western blot analysis of Skp2 expression in 293T cells with GFP or Skp2 knockdown (right panel). (f) Analysis of RhoA and Skp2 mRNA levels in Wt and Skp2−/− primary MEFs by real-time PCR. The quantified results were presented as mean ± s.d. (n=3). (g) Western blot analysis of active RhoA (RhoA-GTP) and total RhoA expression in Wt and Skp2−/− primary MEFs. (h) Western blot analysis of RhoA expression in various cancer cell lines infected with GFP shRNA, Skp2 shRNA. Two Skp2 lentiviral shRNAs were used in this assay. (i) Western blot analysis in PC-3 cells infected with pBabe or pBabe-Skp2. Results in panel a, b, d, e were shown as mean ± s.d. of one representative experiment (from 3 independent experiments) performed in triplicate.
We next determined whether Skp2-induced RhoA transcription requires Myc. The ChIP assay demonstrated that both endogenous Skp2 and Myc bind to the RhoA promoter, and Myc knockdown prevented Skp2 from binding to this promoter (Fig. 3c), suggesting that Myc recruits Skp2 to the RhoA promoter to activate RhoA transcription. This notion was further supported by the fact that Skp2 failed to activate RhoA promoters with mutations in either E-box 5 or E-box 6, the region critical for Myc binding (Fig. 1c, 3d).
Interestingly, while Myc induced RhoA transcriptional activation in GFP-knockdown cells, it failed to activate RhoA transcription in Skp2-knockdown cells (Fig. 3e), suggesting that Skp2 is required for Myc-mediated RhoA transcription. Consistent with the role of Skp2 in RhoA transcription, we found that Skp2−/− MEFs displayed a profound reduction in RhoA mRNA levels, protein levels, and activity (Fig. 3f, g). Similarly, Skp2 knockdown markedly reduced RhoA levels, while Skp2 overexpression induced RhoA protein expression (Fig. 3h, i, and Supplementary information, Fig. S5a, b). Our results therefore underscore the critical role of the Myc and Skp2 network in RhoA transcription and suggest that SCF-Skp2 E3 ligase activity is not required for this function.
Skp2 regulates cell migration and invasion by affecting RhoA expression
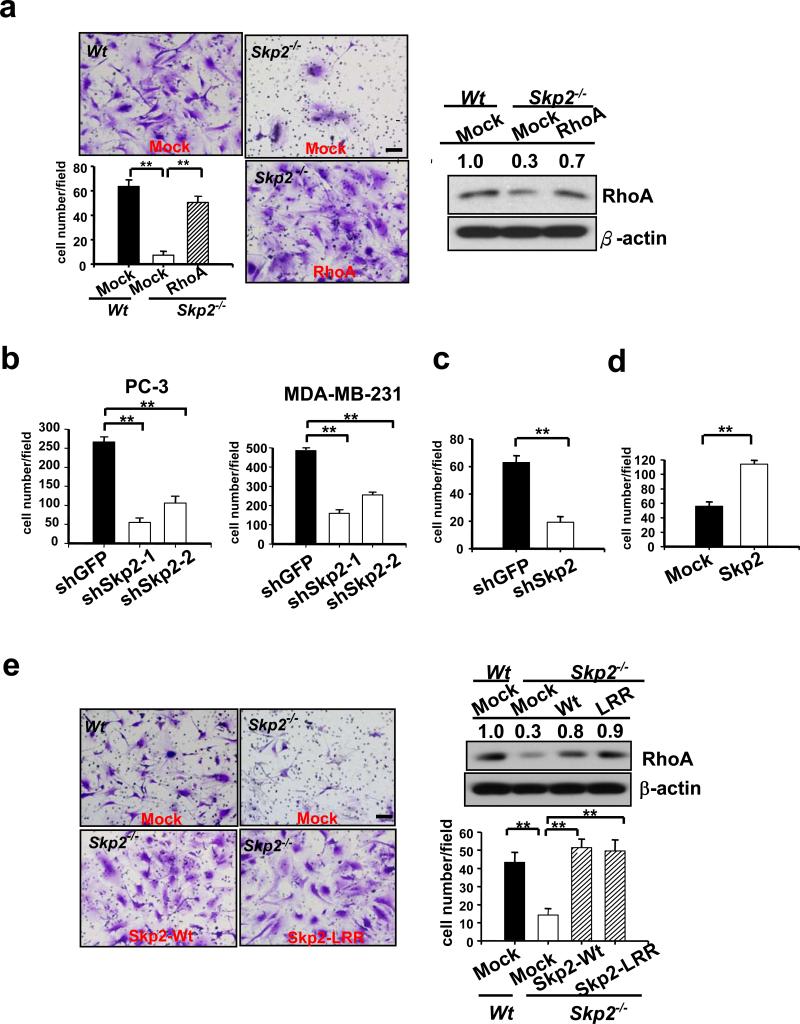
Skp2 overexpression is associated with cancer progression and metastasis33,34 suggesting that Skp2 may have an important role in cancer cell migration, invasion, and metastasis. Indeed, we observed that Skp2−/− MEFs or Skp2-knockdown cells displayed a marked reduction in cell migration and invasion (Fig. 4, a-c and Supplementary information, Fig. S5c, d), recapitulating the phenotypes in Myc- and RhoA- knockdown cells (Fig. 2d, f and Supplementary information, Fig. S1e, g). The effect of Skp2 deficiency on cell migration and invasion was not due to its ability to regulate cell proliferation and apoptosis, since Skp2 knockdown did not significant affect either cell proliferation or apoptosis within 2 days (Supplementary information, Fig. S2a, c). However, Skp2 knockdown inhibited cell proliferation at later time points and primary tumour formation (Supplementary information, Fig. S2a, d), supporting its oncogenic activity30,31,34. Likewise, Skp2 overexpression markedly promoted cell migration as Myc and RhoA overexpression did (Fig. 4d and Supplementary information, Fig. S1h, i). Notably, restoration of RhoA expression rescued both cell migration and invasion defects in Skp2−/− MEFs and Skp2-knockdown cells (Fig. 4a and Supplementary information, Fig. S5c, e), suggesting that RhoA serves as a critical downstream effector for Myc and Skp2 in cell migration and invasion. Notably, the restoration of Skp2-LRR mutant also rescued RhoA protein expression and cell invasion defect in Skp2−/− MEFs as Wt Skp2 did (Fig. 4e). Our results therefore suggest a novel E3 ligase-independent function for Skp2 in RhoA transcription and cell invasion.
Figure 4. Skp2 regulates cell invasion independent of SCF-Skp2 E3 ligase activity.
(a) Matrigel cell invasion assay and Western blot analysis of RhoA expression in Wt and Skp2−/− primary MEFs infected with MSCV or MSCV-RhoA. (b) Matrigel cell invasion assay in two cancer cell lines infected with GFP or Skp2 knockdown. (c) Transwell cell migration assay in PC-3 cells infected with GFP or Skp2 knockdown. (d) Transwell cell migration assay in PC-3 cells infected with pBabe or pBabe-Skp2. (e) Matrigel cell invasion assay and Western blot analysis of RhoA expression in Wt and Skp2−/− primary MEFs infected with pBabe, pBabe-Skp2, or pBabe-Skp2-LRR as indicated. The relative intensity of RhoA was quantified using ImageQuant software and normalized with β-actin. Results in panel b-d were shown as mean ± s.d. (n=4). * indicates p<0.05; ** indicates p<0.01. Scale bar = 100 μm
Skp2 recruits p300 and Miz1 to the Myc transcription complex to induce RhoA transcription
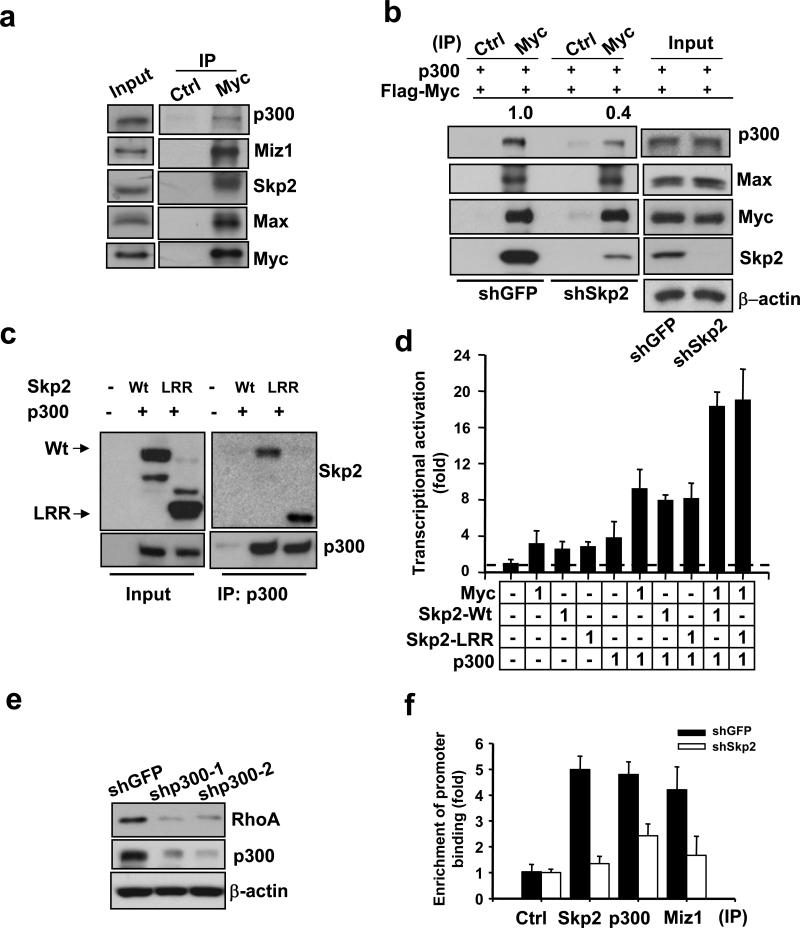
Since Skp2 is required for Myc-mediated RhoA transcription in an SCF E3 ligase-independent manner, it is very likely that Skp2 may interact with other coregulators and recruit them to the Myc transcription complex for RhoA activation. Of note, p300 is known to serve as a coactivator for Myc on numerous Myc target genes, and the interaction between p300 and Skp2 has recently been unraveled35. To gain further insight into how Skp2 regulates RhoA transcription, we determined whether Skp2 interacts with p300 and recruits p300 to the Myc transcription complex. We found that Skp2 interacted with Myc and p300, and the Myc immunocomplex contained both Skp2 and p300 (Supplementary information, Fig. S6a-c). Notably, the endogenous Myc complex also consisted of Skp2, p300 and Max, and the presence of these proteins in Myc immunocomplex was reduced upon Myc knockdown (Fig. 5a and Supplementary information, Fig. S6d), suggesting Myc forms a complex with Skp2, p300 and Max. Of importance, Skp2 knockdown significantly reduced Myc and p300 interaction, but not Myc and Max interaction (Fig. 5b).
Figure 5. Skp2 cooperates with Myc to regulate RhoA transcription by recruiting p300.
(a) Endogenous Myc interacts with endogenous p300, Miz1, Skp2, and Max in vivo. 293T total cell lysates were immunoprecipitated with Myc antibody, followed by Western blot analysis. (b) Co-immunoprecipitation assay in 293T cells with GFP or Skp2 knockdown. The relative intensity of p300 was quantified using ImageQuant software and normalized with the levels of immunoprecipitated Myc in GFP or Skp2-silenced cells. (c) Co-immunoprecipitation assay in 293T cells transfected with the indicated plasmids. (d) RhoA reporter assay in 293T cells transfected with the indicated plasmids. The number indicates the amount (μg) of the plasmids used in this assay. The results were shown as mean ± s.d. of one representative experiment (from 3 independent experiments) performed in triplicate. (e) Western blot analysis of p300 and RhoA expression in MDAMB-231 cells with GFP or p300 knockdown. Two p300 lentiviral shRNAs were used in this assay. (f) ChIP assay in PC-3 cells with GFP or Skp2 knockdown followed by real-time PCR analysis using various antibodies for immunoprecipitation as indicated. The quantified results were presented as mean ± s.d. (n=3).
As expected, we found that Skp2-LRR interacted with Myc and p300 and cooperated with them to activate RhoA transcription as Wt Skp2 did (Fig. 5c, 5d, and Supplementary information, Fig. S6d). p300 could induce and synergized with Myc and Skp2 to activate RhoA transcription, while its knockdown profoundly reduced RhoA protein expression (Fig. 5d, e). Importantly, the ChIP assay revealed that endogenous p300 was also present in the RhoA promoter, and the recruitment of p300 to the RhoA promoter was impaired in Skp2-knockdown cells (Fig. 5f and Supplementary Fig. 6e). These results support the notion that Skp2 recruits p300 to the Myc complex to activate RhoA transcription independently of its SCF E3 ligase activity.
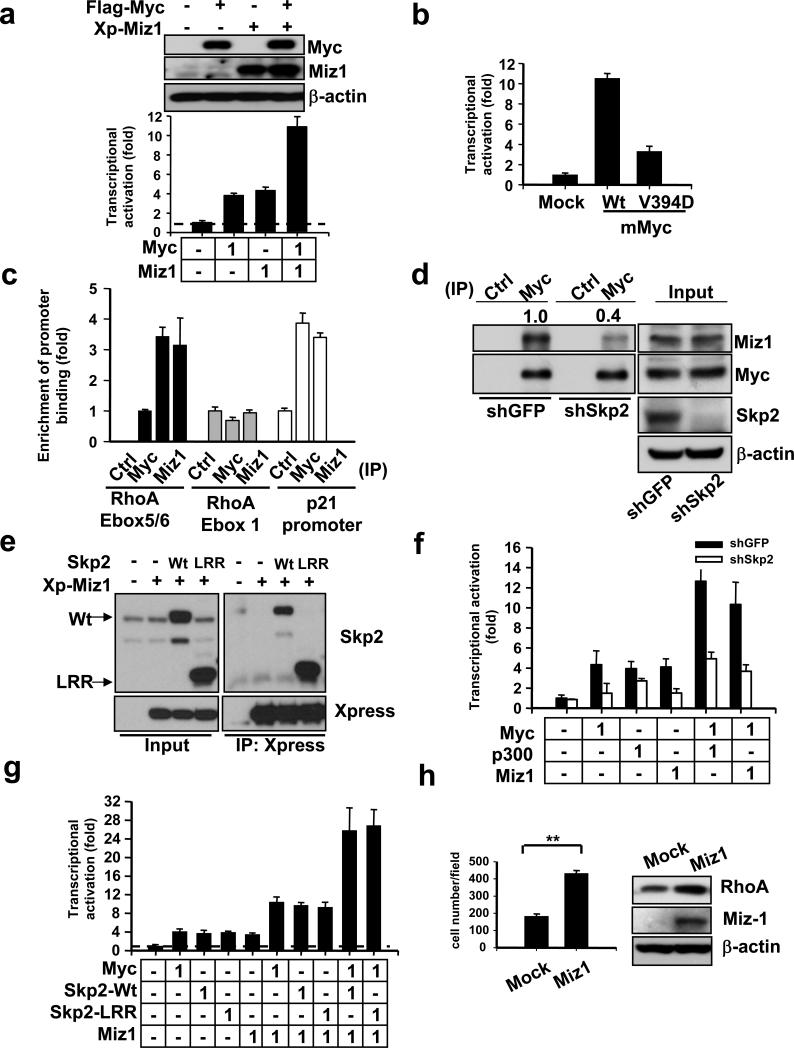
Miz1 was originally identified as a Myc interaction protein that causes cell growth arrest by inducing expression of the cell cycle inhibitors p15 and p2136-38. In line with these reports36-38, we found that Miz1 knockdown promoted cancer cell growth (Supplementary information, Fig. S2b). We examined the effect of Miz1 on Myc-mediated RhoA transcription. Surprisingly, we found that that Miz1 induced and synergized with Myc to activate RhoA transcription (Fig. 6a). To further support the role of Miz1 in Myc-mediated RhoA transcription, we used the Myc V394D mutant defective in Miz1 binding for the RhoA transcription assay39. MycV394D compromised its ability to induce RhoA transcription (Fig. 6a and Supplementary information, Fig. S6f, g), suggesting Miz1 plays an important role in Myc-mediated RhoA transcription.
Figure 6. Skp2 cooperates with Myc to regulate RhoA transcription by recruiting Miz1.
(a) RhoA reporter assay and Western blot analysis in 293T cells transfected with the indicated plasmids. The number indicates the amount (μg) of the plasmids used in this assay (lower panel). Western blot analysis showing the expression of constructs as indicated (upper panel). (b) The effect of MycV394D mutant on RhoA transcription. RhoA reporter assay in 293T cells transfected with the mouse Wt Myc and MycV394D mutant. (c) ChIP assay in PC-3 cells followed by real-time PCR analysis using various antibodies for immunoprecipitation as indicated. The quantified results were presented as mean ± s.d. (n=3). (d) Endogenous immunoprecipitation assay in 293T cells with GFP or Skp2 knockdown. The relative intensity of Miz1 was quantified using ImageQuant software and normalized with the levels of immunoprecipitated Myc in GFP or Skp2-silenced cells. (e) Co-immunoprecipitation assay in 293T cells transfected with the indicated plasmids. (f) RhoA reporter assay in 293T cells with GFP or Skp2 knockdown transfected with plasmids as indicated. (g) RhoA reporter assay in 293T cells transfected with the indicated plasmids. (h) Matrigel cell invasion assay and Western blot analysis in MDA-MB-231 cells infected with MSCV or MSCV-Miz1. The quantified results were presented as the mean ± s.d. (n=4). Results in panel a, b, f, g were shown as mean ± s.d. of one representative experiment (from 3 independent experiments) performed in triplicate.
The ChIP assay revealed that endogenous Miz1 was also present in the same RhoA promoter as were Myc, Max, Skp2, and p300 (Fig. 1d, 3c, 6c, and Supplementary information, Fig. S6e). As a control, we showed Miz1, like Myc, was present in the p21 core promoter region39, but not in the distal E-box1 region of RhoA promoter (Fig. 6c). Notably, endogenous Miz1 was also present in the endogenous Myc/Skp2/p300 immunocomplex (Fig. 5a). The recruitment of Miz1 to the Myc complex and the RhoA promoter relied partly on Skp2, since Skp2 knockdown reduced the ability of Miz1 to interact with Myc and its binding to the RhoA promoter (Fig. 5f, 6d). This result was explained, in part, by the fact that Skp2 was able to interact with Miz1 (Fig. 6e). As a consequence, RhoA transcriptional activation induced by Myc, Miz1, and p300 was all impaired in Skp2-knockdown cells (Fig. 6f). Similarly, we found that the synergetic effect of Myc with p300 or Miz1 on RhoA transcription was diminished upon Skp2 knockdown (Fig. 6f).
Skp2-LRR mutant interacted with Miz1 and synergized with Miz1 and Myc to induce RhoA transcription as Wt Skp2 did (Fig. 6e, g). Likewise, Miz1 overexpression induced RhoA protein expression, while its knockdown reduces it (Fig. 6h and Supplementary information, Fig. S6h). Despite Miz1 overexpression causes cell cycle arrest36,37,40, it promoted cell invasion (Fig. 6g). Our results therefore support the notion that Skp2 induces RhoA transcription, cell migration, and cell invasion by recruiting p300 and Miz1 to the Myc complex independently of SCF-Skp2 E3 ligase activity.
The Myc/Skp2/Miz1 complex is critical for cancer metastasis
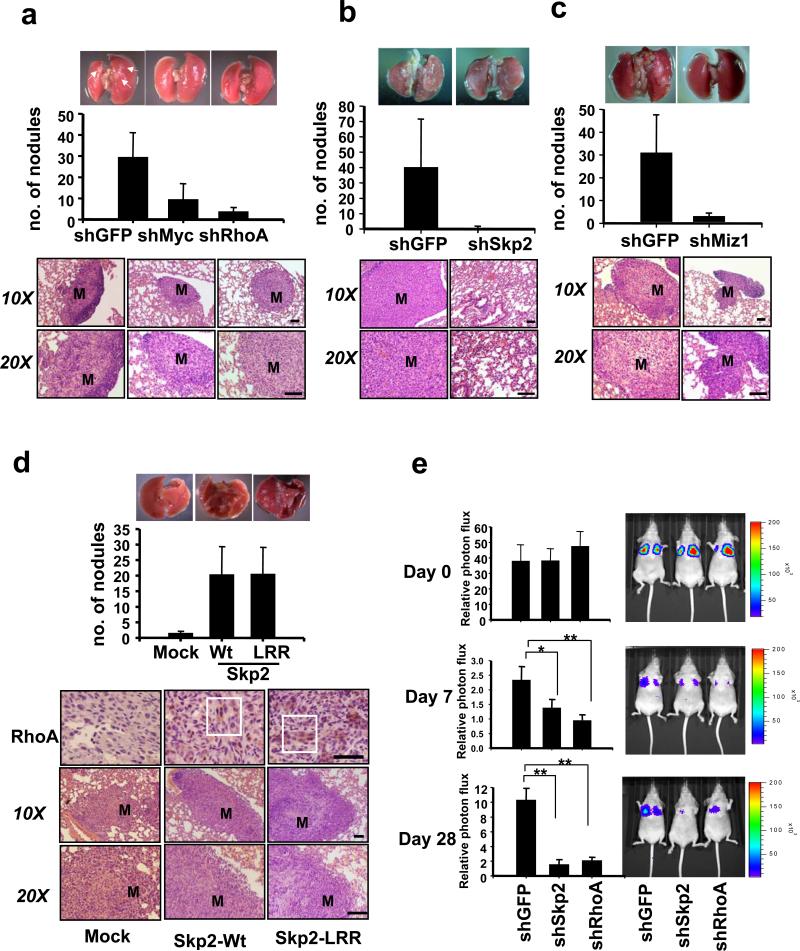
We used a well-established metastasis animal model to determine the role of the Myc/Skp2/Miz1 complex and RhoA in breast cancer metastasis41-43. Deficiency of Myc or Skp2 profoundly restricted breast cancer metastasis to the lung, while overexpression of Myc or Skp2 remarkably promoted it (Fig. 7a, b, d, and Supplementary information, Fig. S7a). Although Miz1 is thought to be a tumour suppressor whose knockdown enhanced cell proliferation (Supplementary information, Fig. S2b), Miz1 knockdown significantly suppresses cancer metastasis to the lung (Fig. 7c). Of importance, RhoA deficiency phenocopied metastasis defects in the knockdown of the Myc/Skp2/Miz1 complex (Fig.7a). Consistently, deficiency of Myc, Skp2, or Miz1 reduced RhoA protein expression in metastatic breast tumours within the lung, while overexpression of Myc or Skp2 enhanced it (Fig. 7d and Supplementary information, Fig. S7). Our results underscore the critical role of the Myc/Skp2/Miz1 complex in RhoA expression, cell invasion, and cancer metastasis.
Figure 7. The Myc/Skp2/Miz1 complex regulates cancer metastasis in mouse model.
(a-c) Lung metastasis assay and histological analysis from nude mice injected with MnDA-MB-231 cells with GFP, Myc, RhoA, Skp2 or Miz1 knockdown. (n = 6 for each group) (d) Lung metastasis assay and histological analysis from nude mice injected with MDA-MB-231 cells infected with pBabe, pBabe-Skp2-Wt or pBabe-Skp2-LRR. IHC of RhoA protein expression in metastatic breast tumour in the lung obtained from nude mice injected with MDA-MB-231 cells with pBabe, pBabe-Skp2-Wt or pBabe-Skp2-LRR. (n = 5 for each group) “M” represents the metastatic nodule. The white box indicates that RhoA protein expression was enhanced. (e) MDA-MB-231-Luc cells with GFP, Skp2, or RhoA knockdown were injected to the nude mice, and the kinetics of breast cancer metastasis to the lung were measured by bioluminescence and quantified. Representative bioluminescent images were shown in day 0, 7, and 28. The results are presented as mean values ± s.d. (n = 7 for each group).* indicates p<0.05; ** indicates p<0.01. Scale bar = 100 μm.
We next determined whether Skp2 also exhibits SCF-Skp2 E3 ligase-independent function in cancer metastasis. Strikingly, Skp2-LRR promoted breast cancer metastasis to the lung as Wt Skp2 did, correlated with its ability to induce RhoA expression in metastatic breast tumours in the lung (Fig. 7d). However, Skp2-LRR compromised its effects on promoting cell cycle progression, cell proliferation, and primary tumour formation compared with Wt Skp2 (Supplementary information, Fig. S8), supporting the current view of the SCF-Skp2 E3 ligase-dependent role in cell cycle progression and tumourigenesis30,31,34. Accordingly, our results strongly support a novel SCF-Skp2 E3 ligase-independent role in cancer metastasis.
To gain further insight into how Skp2 regulates cancer metastasis, we monitored the kinetics of breast cancer metastasis to the lung by using the bioluminescence imaging. We found that the circulation of breast cancer cells with control or Skp2 silencing to the lung were comparable 2 hours (day 0) after the tail vein injection (Fig. 7e), suggesting that Skp2 do not impact on the arrival of breast cancer cell to the lung. However, the bioluminescence signal in the lung was reduced in Skp2 knockdown cells within the first week, and the profound reduction in bioluminescence signal was also observed at later time points (Fig. 7e and Supplementary information, Fig. S9), indicating that Skp2 positively regulates the lung colonization of breast cancer cells and subsequent metastasis. Supporting the role of Skp2 in RhoA expression and cancer metastasis, we found that RhoA knockdown phenocopied the effects of Skp2 knockdown on these processes (Fig.7e and Supplementary information, Fig. S9). These results suggest that Skp2 and RhoA affect the colonization of breast cancer cells to the lung, in turn regulating cancer metastasis.
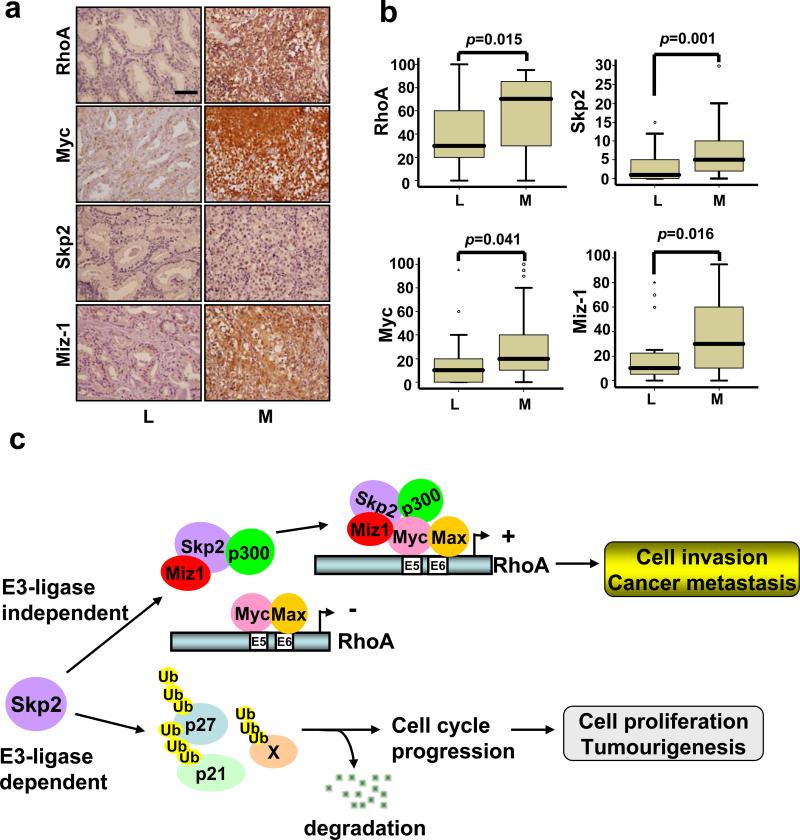
The Myc/Skp2/Miz1 complex is overexpressed and correlated with RhoA expression in metastatic human cancers
Since the Myc/Skp2/Miz1 overexpression is correlated with RhoA expression in vitro and these components are critical for cancer metastasis, we sought to determine whether there is a positive correlation between RhoA expression and the Myc/Skp2/Miz1 complex in human cancer samples. Analyzing prostate cancer samples (64 cases), we found that the expression of RhoA (p=0.015), Myc (p=0.041), Skp2 (p=0.001), and Miz1 (p=0.016), was significantly correlated with prostate cancer metastasis compared with primary prostate cancer (Fig. 8a, b and Supplementary information, Fig. S10a). RhoA expression was also significantly correlated with Myc (p=0.024), Skp2 (p=0.007), and Miz1 (p<0.001) in prostate cancers (Supplementary information, Fig. S10b). Interestingly, we found that there was also a positive correlation between the components of the Myc/Skp2/Miz1 complex (e.g. Myc and Skp2, p=0.001; Myc and Miz1, p<0.001; Skp2 and Miz1, p<0.001, Supplementary information, Fig. S10b). Our study therefore underscores the clinical relevance of the Myc/Skp2/Miz1 complex and RhoA expression in cancer metastasis.
Figure 8. The Myc/Skp2/Miz1 complex is overexpressed and correlated with RhoA expression in metastatic prostate cancer samples.
(a) The expression of RhoA, Myc, Skp2, and Miz1 in representative cases of primary prostate adenocarcinomas that remained localized (L) or developed metastasis (M). The labeling indices of RhoA, Myc, Skp2, and Miz1 were appeared lower in the localized tumour but indicated significant overexpression in the case with metastasis. Original magnification, × 400; Scale bar = 100 μm. (b) The expression of RhoA, Myc, Skp2, and Miz1 in primary prostate cancer versus metastatic prostate cancer. (c) The working model for SCF-Skp2 E3 ligase-dependent and -independent functions.
Discussion
Our study provides the first cue as to how the RhoA transcription is induced. We found that the RhoA transcription is orchestrated by the Myc/Skp2/Miz1/p300 transcription complex. Knockdown or deficiency of these components impairs the RhoA expression, resulting in defects in cell migration and invasion. This finding is further underscored by the important clinical observations that RhoA expression was positively correlated with the Myc/Skp2/Miz1 complex in metastatic prostate cancer, which may provide the relevant molecular mechanism by which RhoA overexpression occurs during cancer progression and metastasis.
Myc ubiquitination by HectH9 and Skp2 is shown to be critical for Myc target genes involved in cell cycle progression and proliferation18-20. However, our study suggests that the Myc ubiquitination by these two E3 ligases is not required for RhoA gene expression. Although Myc is thought to displays an oncogenic activity partly through antagonizing Miz1 transcriptional activity towards p21 and p15 gene expression, it has been unclear whether Miz1 has any impact on Myc transcriptional activation. The current dogma favors the notion of a mutual antagonism between the oncogenic Myc and tumour suppressor Miz1 in gene regulation15-17,44. Surprisingly, our results show that Miz1 synergizes with Myc to induce RhoA transcription, providing a new light into the novel regulatory mechanism between Myc and Miz1. Although Miz1 overexpression causes cell cycle arrest and inhibits cell proliferation36,37,40, we found that Miz1 cooperates with Myc to induce RhoA expression, cancer cell invasion, and is required for cancer metastasis, consistent with our finding that Myc/Miz1/RhoA overexpression is positively correlated with each other in metastatic cancer samples. Our study therefore reveals a previously unrecognized function of Miz1 in RhoA transcription, cell invasion, and cancer metastasis.
Skp2 exerts its oncogenic activity towards promoting cell proliferation and tumourigenesis dependently of SCF-Skp2 E3 ligase activity (Supplementary information, Fig. S8)30,31,34. Our study, surprisingly, reveals a SCF-Skp2 E3 ligase-independent function for Skp2 in RhoA transcription, cell invasion, and cancer metastasis. We propose a novel SCF E3 ligase-independent function for Skp2, specifically, that Skp2 recruits Miz1 and p300 coregulators to the Myc complex, in turn facilitating RhoA transcription, cell invasion, and cancer metastasis (Fig. 8c). In contrast, SCF-Skp2 E3 ligase is required for Skp2's function in cell cycle progression, proliferation, and primary tumour formation by promoting the ubiquitination and degradation of its substrates such as p27.
In summary, our study reveals that the Myc/Skp2/Miz1 transcription complex is critical for RhoA transcription, cell migration, invasion, and cancer metastasis. Our findings that overexpression of the Myc/Skp2/Miz1 complex promotes cancer cell invasion and metastasis, whereas their deficiency restricts cancer metastasis suggest that targeting this transcription complex may provide appealing therapeutic strategies for treatment of cancer patients with metastasis.
Methods
Mice, cell culture, and reagents
Mouse embryonic fibroblasts (MEFs) from wild-type and Skp2−/− mice were prepared as previously described. 293T, PC-3 (from ATCC), MDA-MB-231, and Rat1 cells (from Dr. M. J.M. Sedivy) were cultured in DMEM containing 10% fetal bovine serum (FBS). To clone Xp-Skp2, Skp2 was amplified by PCR using pcDNA3-Skp2 as a template and inserted into a pcDNA4/HisMax-TOPO vector (Invitrogen). To clone the full-length RhoA promoter, human RhoA promoter cDNA (nucleotides −1850 to +1; numbered relative to the transcription initiation site) was amplified from the genomic DNA of 293T cells and inserted into the pGL3 luciferase vector. The RhoA mutant promoters were generated by using the site-directed mutagenesis kit (Stratagene) according the manufacturer's standard procedure with the full-length RhoA promoter as a template. pBabe-Skp2 was generated by subcloning Skp2 into the EcoRI site of the pBabe vector.MSCV-PIG-RhoA and MSCV-Myc were generated by subcloning RhoA and c-Myc into the MSCV-PIG vector. (His)6-ubiquitin and HA-p27 constructs were from Drs. D. Bohmann and M. Pagano, respectively. Myc-Twist and pcDNA-p300 were obtained from Drs. R. Weinberg and W. Sellers, respectively. HectH9 and the Skp2-LRR mutant were obtained from Dr. M. Eliers and W. Tansey, respectively. pBabe-MycER and mouse pcDNA3.1-c-MycV394D was obtained from Drs. El-Deiry Wafik and Libo Yao, respectively. To clone Xp-Miz1, the human Skp2 cDNA was amplified by RT-PCR from 293T cells and inserted into the pcDNA4/HisMax-TOPO vector. MG-132 was obtained from Calbiochem.
RhoA reporter assay
293T cells were transfected by using Lipofectamine 2000 (Invitrogen) for 48 h, and the luciferase activity was measured by using the dual luciferase system (Promega). Each sample was analyzed in triplicates and experiments were performed at least three times. The relative luciferase activity was calculated as a ratio between Firefly luciferase activity and Renilla luciferase activity.
Immunoprecipitation and immunoblotting
Immunoprecipitation (IP) and immunoblotting (IB) were performed essentially as described previously with some modifications. For protein-protein interaction, cells were lysed by E1A lysis buffer (250 mM NaCl, 50 mM HEPES, [pH 7.5], 0.1% NP40, 5 mM EDTA), with a protease inhibitor cocktail (Roche). The following antibodies were used for IP, IB, or IF: anti-Skp2 (IP: 1:250; IB: 1:1000, Zymed), anti-Skp2 (IB: 1:1000, Santa Cruz), anti-Skp1 (IB: 1:2000, BD Transduction Lab), anti-Cul-1 (IB: 1:2000, BD Transduction Lab), anti-Xpress (IP/IF: 1:500; IB: 1:5000, Invitrogen), anti-HA (IB/IF: 1:1000, Covance, Upstate), anti-β-actin (IB: 1:1000, Sigma), anti-p27 (IP: 1:100; IB: 1:1000, BD Transduction Lab), anti-Myc (IP: 1: 100, IB: 1:1000, Santa Cruz), anti-p300 (IP: 1:100; IB:1:1000, Santa Cruz), anti-Miz1 (IP: 1:100; IB:1:1000, Santa Cruz), anti-HectH9 (IB: 1:1000, Abcam) and anti-RhoA (IB:1:2000, Santa Cruz).
RhoA activity assay
RhoA activity assay was performed by using Rho activation assay kit (Millipore) according to the manufacturer's instructions. Equal amount of protein was used to pull down active Rho using GST-Rhotekin Rho binding domain, which binds selectively to GTP-Rho, not GDP-Rho. After precipitation, samples were analyzed using Western blotting with a specific anti-RhoA antibody.
Viral infection
Retroviral plasmid were transfected into Phoenix packing cells for 2 days, and the virus-containing medium was harvested and used to infect target cells. For lentiviral short hairpin RNA (shRNA) infection, 293T cells were cotransfected Skp2, Myc, RhoA or GFP control shRNA with packing plasmids (deltaVPR8.9) and envelope plasmid (VSV-G) using Lipofectamine 2000 reagent according to the manufacturer's instructions. RhoA-lentiviral shRNA-1 (5’-GAAAGCAGGTAGAGTTGGCTT-3’), RhoA-lentiviral shRNA-2 (5'-GTACATGGAGTGTTCAGCAAA-3'), Skp2-lentivivral shRNA-1 (5'-GATAGTGTCATGCTAAAGAAT-3'), Skp2-lentivivral shRNA-2 (5'-GCCTAAGCTAAATCGAGAGAA-3'), Myc-lentiviral shRNA-1 (5’-CCTGAGACAGATCAGCAACAA-3’), Myc-lentiviral shRNA-2 (5'-CAGTTGAAACACAAACTTGAA-3'), p300-lentivivral shRNA-1 (5'-CAATTCCGAGACATCTTGAGA-3’), p300-lentivivral shRNA-2 (5'-CCAGCCTCAAACTACAATAAA-3’), Miz1-lentivivral shRNA-1 (5’-GTGTTCACTTTAAGGCTCATA-3’), Miz1-lentivivral shRNA-2 (5’- CGAGTACTTCAAGATGCTCTT-3’), HectH9-lentivivral shRNA-1 (5’- GCGTCTGTGTTGTGGAGGTTT-3’), HectH9-lentivivral shRNA-2 (5’-CCACACTTTCACAGATACTAT-3’), HectH9-lentivivral shRNA-3 (5’-GCTCCCACTATAACCTCACTT-3’), HectH9-lentivivral shRNA-4 (5’-CGACGAGAACTAGCACAGAAT-3’) and GFP shRNA (5'-GCAAGCTGACCCTGAAGTTC-3') were transfected with packing plasmids into 293T cells for 2 days, and virus-particles containing Skp2, Myc, RhoA, or GFP shRNA were used to infect mammalian cells. All the infected cells were cultured in medium containing appropriate antibiotics.
ChIP assay
The ChIP assay was performed according to the manufacturer's instructions (Upstate). A DNA/protein complex was sheared by sonication. One percent of sheared DNA/protein complex was kept as an input DNA sample. Anti-Skp2 (clone 8D9, Zymed), anti-Myc (clone N262, Santa Cruz), anti-Max (clone C17, Santa Cruz), anti-p300 (clone N15, Santa Cruz), anti-Miz1 (clone H190, Santa Cruz), and anti-RNA polymerase II (clone 6H5-3, UPSTATE) or normal mouse/rabbit IgG (Upstate, Sigma) were used for immunoprecipitation. Enrichment of promoter binding levels were analyzed by real-time PCR, normalized by input, and were expressed as the fold increase over the control. Primers used for real-time PCR were: RhoA E-box5/6, 5’- CTTCGCGTGCGTGAAGAGTTG-3’ and 5’-CATCCACTATTGCTCAGGAGC-3’; RhoA E-box1, 5’-GAGAAGCCTTCCCTGACCACC-3’ and 5’- GCTCACTAATCAGGGCATTCAT-3’; p21 core promoter, 5’- ACCGGCTGGCCTGCTGGAACT-3’ and 5’-TCTGCCGCCGCTCTCTCACCT-3’. The quantified results were presented as the mean ± s.d.
In vivo ubiquitination assay
In vivo ubiquitination assay was performed as described45. In brief, 293T cells were transfected with the indicated plasmids for 24 h, treated with 20 μM MG-132 for 6 h, and lysed by the denatured buffer (6M guanidine-HCl, 0.1M Na2HPO4/NaH2PO4, 10 mM imidazole). The cell extracts were then incubated with nickel beads for 3 h, washed, and subjected to Western blot analysis.
Cell migration and invasion assays
The cell migration assay was done in a 24-well transwell plate with 8-μm polyethylene terephthalate membrane filters (Falcon cell culture insert, Becton-Dickinson)] separating the lower and upper culture chambers. In brief, MEFs, MDA-MB-231 cells, and PC-3 cells were plated in the upper chamber at 1 × 104, 5 × 104, and 1 × 105 cells per well, respectively, in serum-free DMEM. The bottom chamber contained DMEM with 10% FBS. MEFs and PC-3 cells were allowed to migrate for 18 hours, while MDA-MB-231 cells were allowed to migrate for 6 hours. After the incubation period, the filter was removed, and non-migrant cells on the upper side of the filter were detached using a cotton swab. Filters were fixed with 4% formaldehyde for 15 min and cells located in the lower filter were stained with 0.1% crystal violet for 20 min and counted from three random fields. The quantified results were presented as the mean ± s.d. The cell invasion assay was essentially similar to the cell migration assay, the an exception that the membrane filter was precoated with diluted Matrigel (Matrigel:serum-free DMEM,1:6) prior to the assay.
Immunohistochemistry and statistical analysis
Immunohistochemical analysis was performed on representative tissue blocks of primary prostatic adenocarcinomas from 64 patients. Among them, 37 patients presented with localized disease after follow-up for at least 5 years and 27 patients developed metastatic disease either at diagnosis of adenocarcinoma or within 2 years. The sections were heated for epitope retrieval, quenched in 3% H2O2 to abolish endogenous peroxidase, and then incubated with 10% normal horse serum to block non-specific immunoreactivity. The primary antibodies were detected by using the DAKO EnVision kit (DAKO) for RhoA (26C4, 1:200, Santa Cruz Biotechnology), Myc (9E10, 1:200, Zymed Laboratories), Skp2 (2C8D9, 1:100, Zymed Laboratories), and Miz-1 (1:400, Atlas Antibodies). To evaluate the expression of all markers, the percentage of tumour cells with moderate or strong nuclear and/or cytoplasmic immunoreactivity was recorded to obtain the labeling index for each case. The correlations among the four markers were calculated by using Pearson's correlation test, and the differences in the labeling indices of the markers tested between cases with and without metastatic disease were compared by Student's t-test.
Metastasis assay
To study RhoA, Myc, Skp2 or Miz1 deficiency in a breast cancer metastasis model, MDA-MB-231 cells silenced with GFP, RhoA, Myc, Skp2, or Miz1 lentiviral shRNAs were injected into the tail veins of athymic female nude mice. Twelve weeks later, mice (n=5-7 for each group) were sacrificed and lung tissues were analyzed for the occurrence of metastasis. To study the role of overexpression of Myc, Wt Skp2, and Skp2-LRR in metastasis, MDA-MB-231 cells were infected with retroviruses expressing empty vector, Myc, Skp2, or Skp2-LRR and selected by 2 μg/ml puromycin for 4 days. These viable cells were injected into the tail vein of athymic male nude mice. Eight weeks later, mice (n=6~7 for each group) were sacrificed and lung tissues were obtained for analysis of cancer metastasis. To study the kinetics of breast cancer metastasis to the lung, MDAMB-231-Luc cells stably overexpresssed with luciferase gene silenced with GFP, Skp2, or RhoA shRNA were injected into the lateral tail vein (2 × 106 cells) of athymic female nude mice (7 mice for each group). For bioluminescence imaging, mice were anesthetized and given 150 mg/g of D-luciferin in PBS by i.p. injection. Five minutes after injection, bioluminescence was detected by an IVIS imagining system analyzed using the Living Image software (Xenogen).
BrdU-incorporation assay
BrdU-incorporation assays were performed by using the In Situ Cell Proliferation Kit, FLUOS (Roche). In brief, COS-1 cells transfected with various plasmids were serum-starved (0.1 % FBS) for 24 h, refreshed with 10% FBS for 16 h, incubated with 20 μM BrdU for 1 h, and harvested for quantification of BrdU-incorporation. The percentage of cell numbers in S phase was scored, the representative images were shown as indicated (left panel), and the results are presented as mean values ± s.d. (right panel).
Cell growth, apoptosis, and in vivo tumourigenesis assay
For cell growth assay, 5 × 103 cells were seeded in 12 wells in triplicate, harvested, and stained with trypan blue on different days. Viable cells were counted directly under the microscope. For apoptosis assay, cells were cultured for 2 days, harvested, labeled with Annexin-V-FITC, and followed by flow cytometry analysis. For in vivo tumourigenesis assay, cells were subcutaneously injected into athymic female nude mice, and tumour size was measured weekly with a calliper, and tumour volume was determined with the standard formula: L × W2 × 0.52, where L is the longest diameter and W the shortest diameter.
Primer sequences for real-time RT–PCR
Human CAD: 5′-CAGGTTTGCCAGCTGAGGA-3′ (forward) and 5′-TGCCTGTCTCGGTACTGGTG-3′ (reverse); Human ODC: 5′-TGTAGGAAGCGGCTGTAC-3′ (forward) and 5′-GCTATGATTCTCACTCCAGAG-3′ (reverse), Human RhoA: 5′-GAGCACACAAGGCGGGAG-3′ (forward) and 5′-CTTGCAGAGCAGCTCTCGTAG-3′ (reverse), Human GAPDH: 5′-GATTCCACCCATGGCAAATTC-3′ (forward) and 5′-CTTCTCCATGGTGGTGAAGAC-3′ (reverse), Mouse RhoA: 5′-GAGTTGGCTTTATGGGACAC-3′ (forward) and 5′-GAAATGCTTGACTTCTGGAGTC-3′ (reverse), Mouse GAPDH: 5′-GACAAAATGGTGAAGGTCGGTG-3′ (forward) and 5′-CTGGAACATGTAGACCATG-3′ (reverse).
Statistical analysis
Unless otherwise noted, data were presented as means ± s.d. of three or more independent results, and statistical significance was assessed by two-tailed paired Student's-t-test. (P < 0.05 was considered significant).
Supplementary Material
Acknowledgements
We thank Drs. R. Weinberg, W. Sellers, D. Bohmann, W. Tansey, J. M. Sedivy, W. Wei, M. Pagano, M. Eilers, W. S. El-Deiry, L. Yao, and D. Sarbassov for reagents. We also thank Drs. D. Sarbassov, M. H. Lee, and M. G. Lee for comments and suggestions. Special thanks extended to S. Patterson for editing and critical reading of the manuscript. This work was supported by Research Trust Scholar funds from M.D. Anderson Cancer Center, the National Cancer Institute's Prostate Cancer Specialized Program of Research Excellence (SPORE) at M. D. Anderson Cancer Center, and DOD Prostate Cancer New Investigator Award (to H.K.L.).
Footnotes
Author Contributions
C.H.C. and H.K.L designed the experiments and wrote the manuscript; C.H.C., H.K.L., S.W.L., J.W., W.L.Y., C.Y.W., and J.W. performed the experiments; H.Y. K., C.F.L., and H.Y.H performed the immunochemistry and analyzed the data; M.C.H and P.P.P. provided the cell lines and reagents. K. I. N. provided the Skp2−/− mice.
References
- 1.Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 4.Lahoz A, Hall A. DLC1: a significant GAP in the cancer genome. Genes Dev. 2008;22:1724–1730. doi: 10.1101/gad.1691408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moscow JA, et al. Examination of human tumors for rhoA mutations. Oncogene. 1994;9:189–194. [PubMed] [Google Scholar]
- 6.Rihet S, et al. Mutation status of genes encoding RhoA, Rac1, and Cdc42 GTPases in a panel of invasive human colorectal and breast tumors. J Cancer Res Clin Oncol. 2001;127:733–738. doi: 10.1007/s004320100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellizzi A, et al. RhoA protein expression in primary breast cancers and matched lymphocytes is associated with progression of the disease. Int J Mol Med. 2008;22:25–31. [PubMed] [Google Scholar]
- 8.Faried A, Faried LS, Usman N, Kato H, Kuwano H. Clinical and prognostic significance of RhoA and RhoC gene expression in esophageal squamous cell carcinoma. Ann Surg Oncol. 2007;14:3593–3601. doi: 10.1245/s10434-007-9562-x. [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi A, et al. Up-regulation of small GTPases, RhoA and RhoC, is associated with tumor progression in ovarian carcinoma. Lab Invest. 2003;83:861–870. doi: 10.1097/01.lab.0000073128.16098.31. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X, et al. Overexpression of RhoA induces preneoplastic transformation of primary mammary epithelial cells. Cancer research. 2009;69:483–491. doi: 10.1158/0008-5472.CAN-08-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pille JY, et al. Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol Ther. 2005;11:267–274. doi: 10.1016/j.ymthe.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 12.Valastyan S, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Kamai T, et al. RhoA is associated with invasion and lymph node metastasis in upper urinary tract cancer. BJU Int. 2003;91:234–238. doi: 10.1046/j.1464-410x.2003.03063.x. [DOI] [PubMed] [Google Scholar]
- 14.Faried A, et al. Correlation between RhoA overexpression and tumour progression in esophageal squamous cell carcinoma. Eur J Surg Oncol. 2005;31:410–414. doi: 10.1016/j.ejso.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Kleine-Kohlbrecher D, Adhikary S, Eilers M. Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- 16.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 17.Eilers M, Eisenman RN. Myc's broad reach. Genes & development. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adhikary S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 20.von der Lehr N, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 21.Ansieau S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Smit MA, Peeper DS. Deregulating EMT and senescence: double impact by a single twist. Cancer cell. 2008;14:5–7. doi: 10.1016/j.ccr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 24.Prochownik EV. c-Myc: linking transformation and genomic instability. Curr Mol Med. 2008;8:446–458. doi: 10.2174/156652408785747988. [DOI] [PubMed] [Google Scholar]
- 25.Ricci MS, et al. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Molecular and cellular biology. 2004;24:8541–8555. doi: 10.1128/MCB.24.19.8541-8555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes & development. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser P, Flick K, Wittenberg C, Reed SI. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 29.Bres V, et al. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat Cell Biol. 2003;5:754–761. doi: 10.1038/ncb1023. [DOI] [PubMed] [Google Scholar]
- 30.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–333. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, et al. The ETS protein MEF is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCFSkp2. Molecular and cellular biology. 2006;26:3114–3123. doi: 10.1128/MCB.26.8.3114-3123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanbrough M, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 34.Lin HK, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nature cell biology. 2009;11:420–432. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitagawa M, Lee SH, McCormick F. Skp2 suppresses p53-dependent apoptosis by inhibiting p300. Mol Cell. 2008;29:217–231. doi: 10.1016/j.molcel.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 36.Seoane J, et al. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nature cell biology. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 37.Staller P, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nature cell biology. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 38.Wanzel M, et al. A ribosomal protein L23-nucleophosmin circuit coordinates Mizl function with cell growth. Nature cell biology. 2008;10:1051–1061. doi: 10.1038/ncb1764. [DOI] [PubMed] [Google Scholar]
- 39.Wu S, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 40.Wanzel M, et al. A ribosomal protein L23-nucleophosmin circuit coordinates Miz1 function with cell growth. Nature cell biology. 2008 doi: 10.1038/ncb1764. [DOI] [PubMed] [Google Scholar]
- 41.Gupta GP, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y, et al. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer cell. 2009;15:416–428. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta GP, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19506–19511. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wanzel M, Herold S, Eilers M. Transcriptional repression by Myc. Trends Cell Biol. 2003;13:146–150. doi: 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 45.Yang WL, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science (New York, N.Y. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.