Abstract
In the actinorhodin type II polyketide synthase, the first polyketide modification is a regiospecific C9-carbonyl reduction, catalyzed by the ketoreductase (actKR). Our previous studies identified the actKR 94-PGG-96 motif as a determinant of stereospecificity (Javidpour, et al., 2011). The molecular basis for reduction regiospecificity is, however, not well understood. In this study, we examined the activities of 20 actKR mutants through a combination of kinetic studies, PKS reconstitution, and structural analyses. Residues have been identified which are necessary for substrate interaction, and these observations have suggested a structural model for this reaction. Polyketides dock at the KR surface and are steered into the enzyme pocket where C7-C12 cyclization is mediated by the KR before C9-ketoreduction can occur. These molecular features can potentially serve as engineering targets for the biosynthesis of novel, reduced polyketides.
INTRODUCTION
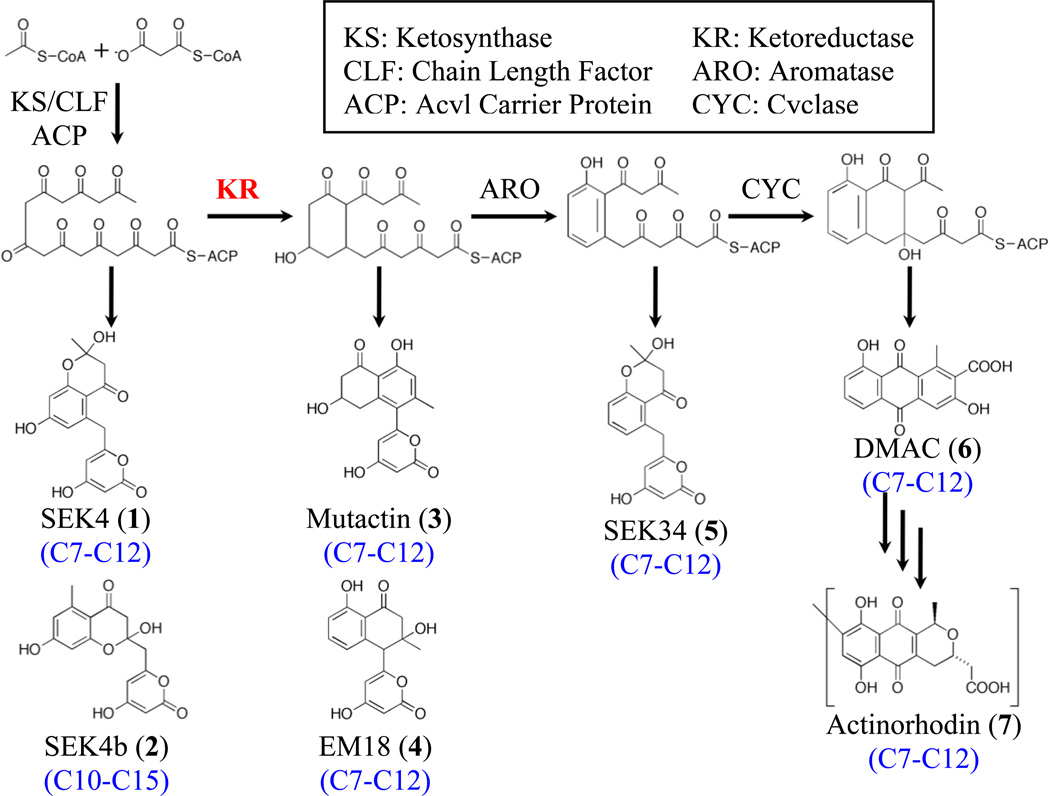
Polyketide natural products are a large, diverse class of secondary metabolites with important biological activities (Crawford and Townsend, 2010; Zhan, 2009). Many pharmaceutical compounds are polyketide-based, including antibiotics (tetracycline), anti-cancer agents (daunorubicin), and immunosuppressants (FK506) (Cane, et al., 1998). Polyketide biosynthesis is driven by the polyketide synthase (PKS), which is genetically and functionally similar to fatty acid synthase (Hopwood, 1997). PKSs are generally divided into three classes (Shen, 2003), though the focus of this study are type II PKSs found in bacteria, which biosynthesize aromatic polyketides, such as actinorhodin (Malpartida and Hopwood, 1984). In the actinorhodin (act) type II PKS (Figure 1), a linear poly-β-keto chain is biosynthesized by the minimal PKS, consisting of the dimeric ketosynthase-chain length factor (KS-CLF) and an acyl carrier protein (ACP) (Malpartida and Hopwood, 1984). Biosynthesis is initiated by an acetate starter unit derived from CLF-mediated decarboxylation of a malonyl-ACP thioester. Subsequent chain elongation occurs through decarboxylative condensation of malonyl-based units to this starter, producing the final octaketide (16-carbon) intermediate (Das and Khosla, 2009). The first modification of the nascent polyketide chain is catalyzed by the actKR, which regiospecifically reduces a single carbonyl group (typically at C9) to a hydroxyl group. This modification is thought to occur after cyclization between C7 and C12 occurs, which forms the first ring, a process that may also be directed by the ketoreductase (McDaniel, et al., 1993). Heterologous expression and in vitro reconstitution experiments have demonstrated that, in the absence of modifying enzymes, the act minimal PKS produces both the correctly C7-C12 cyclized SEK4 as well as the aberrantly C10-C15 cyclized SEK4b. When actKR is present, however, the dominant product becomes mutactin (C7-C12 cyclized), further suggesting that correct cyclization is dependent on reductase activity (Khosla, et al., 1993; McDaniel, et al., 1994; Zawada and Khosla, 1999).
Figure 1. Schematic of the Actinorhodin Type II PKS.
The minimal PKS synthesizes a 16-carbon polyketide intermediate that, in the absence of KR, spontaneously forms unreduced products SEK4 1 (C7-C12 first-ring cyclization) and SEK4b 2 (C10-C15 first-ring cyclization). KR regiospecifically reduces C9 yielding a C7-C12 cyclized polyketide that forms mutactin 3 as the major product. That all downstream products, including SEK34 5, DMAC 6, and actinorhodin 7, are C7-C12-cyclized suggests that KR also plays a role in the determination of cyclization specificity.
Type II ketoreductases can potentially be utilized in organic synthesis applications and in engineered biosynthesis (Broussy, et al., 2009; Grau, et al., 2007; Jacobsen, et al., 1998; Truppo, et al., 2007). This would require an in-depth understanding of KR-substrate interactions, including the factors that dictate regio- and stereospecificity. Our previous structural and biochemical analyses led to the identification of the actKR 94-PGG-96 motif as a determinant of stereospecificity as well as residues that interact with the inhibitor emodin (Javidpour, et al., 2011; Korman, et al., 2008). Furthermore, docking studies suggested a region on the actKR surface that could act as the site of interaction between KR and ACP (Javidpour, et al., 2011). In order to fully realize the potential of KR for engineered reduction, a detailed assessment of the molecular basis for C9-regiospecificity and the possible role of actKR in first-ring cyclization is required.
The aim of this work is to identify actKR residues that mediate interactions with the natural polyketide substrate, the NADPH cofactor and the carrier protein, as well as residues that direct C9-regiospecific ketoreduction. We present the molecular features of actKR involved in recognizing and directing the polyketide intermediate for C9-ketoreduction. This is supported by kinetic and in vitro PKS reconstitution assay data for 20 actKR mutants. High-resolution crystallographic structures for three of the mutants are also presented. We targeted specific actKR residues for mutagenesis based on previous structural and biochemical analyses of wild-type (WT) and P94L actKR, both in the presence or absence of the inhibitor emodin (Javidpour, et al., 2011; Korman, et al., 2004; Korman, et al., 2008). These mutations can be subdivided into three groups associated with surface, substrate binding pocket, and the α6-α7 loop. Significantly, we have shown that a single mutation in the actKR substrate pocket can switch the stereospecificity between S-dominant and R-dominant, which contradicts the results of type I polyketide KR mutations (Baerga-Ortiz, et al., 2006; Castonguay, et al., 2008; Holzbaur, et al., 1999; Holzbaur, et al., 2001; Keatinge-Clay, 2007; Keatinge-Clay and Stroud, 2006; O'Hare, et al., 2006; Ostergaard, et al., 2002; Siskos, et al., 2005). These results form the basis for a new model of KR-mediated C7-C12 first-ring cyclization that occurs within the enzyme binding pocket prior to ketoreduction.
RESULTS
Overall Strategy
Based on previous structural and biochemical analyses of wild-type and P94L actKR (Javidpour, et al., 2011; Korman, et al., 2004; Korman, et al., 2008), we proposed three groups of mutations: surface mutations to evaluate protein-protein interactions and long-range effects on enzyme activity, substrate binding pocket mutations to assess stereospecificity and catalytic efficiency, and finally mutations in the α6-α7 loop to determine if this flexible region plays a significant role in catalysis. Trans-1-decalone and tetralol were used as substrate analogs to kinetically assay catalytic efficiency and substrate specificity of the reduction (Tables 1, 2, and S1). Trans-1-decalone, which has previously been used as a model substrate to assay ketoreductase activity in both fatty acid synthase and PKS (Dutler, et al., 1971; Ostergaard, et al., 2002), was shown to be a more effective substrate for the actKR than 1-tetralone (Korman, et al., 2008).
Table 1.
Kinetic Parameters for the Reduction of trans-1-Decalone and interaction with NADPH and ACP by Wild-Type and Mutants of Actinorhodin Ketoreductase
| kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) |
Kd (µM) NADPH |
KD (µM) ACP |
|
|---|---|---|---|---|---|
| WT | 2.55 ± 0.12 | 0.76 ± 0.09 | 3.23 ± 0.32 | - | - |
| D109E | 1.17 ± 0.12 | 0.71 ± 0.18 | 1.64 ± 0.45 | - | - |
| D109R | 0.078 ± 0.01 | 1.25 ± 0.40 | 0.062 ± 0.021 | - | - |
| S144C | Inactive | - | - | ||
| T145A | 0.18 ± 0.008 | 0.42 ± 0.06 | 0.42 ± 0.06 | - | - |
| G146V | Inactive | - | - | ||
| V151A | Inactive | - | - | ||
| V151L | 1.24 ± 0.07 | 0.97 ± 0.12 | 1.28 ± 0.18 | - | - |
| A154G | 1.33 ± 0.16 | 0.50 ± 0.18 | 2.68 ± 1.03 | - | - |
| F189A | 0.049 ± 0.005 | 8.63 ± 2.01 | 0.0057 ± 0.0015 | - | - |
| F189W | Substrate Inhibition | - | - | ||
| M194W | 0.43 ± 0.06 | 3.22 ± 0.65 | 0.13 ± 0.03 | - | - |
| V198A | 0.24 ± 0.01 | 15.50 ± 1.49 | 0.016 ± 0.002 | - | - |
| V198Ga 1.3 ± 0.1 |
0.32 ± 0.06 | 5.36 ± 1.12 | 0.060 ± 0.017 | - | - |
| Y202Aa 2.0 ± 0.3 |
1.80 ± 0.09 | 1.07 ± 0.09 | 1.69 ± 0.16 | - | - |
| Y202F | 2.01 ± 0.17 | 0.40 ± 0.11 | 5.01 ± 1.44 | - | - |
| WTb | 2.00 ± 0.06 | 1.19 ± 0.02 | 1.68 ± 0.08 | 1.52 ± 0.09 | 0.20 ± 0.08 |
| R38A | 0.16 ± 0.03 | 1.45 ± 0.02 | 0.11 ± 0.02 | 4.30 ± 0.40 | 1.52 ± 0.12 |
| R65A | 1.97 ± 0.07 | 1.24 ± 0.02 | 1.59 ± 0.07 | 1.83 ± 0.21 | 0.53 ± 0.11 |
| R93A | 1.13 ± 0.05 | 1.21 ± 0.01 | 0.94 ± 0.05 | 6.43 ± 0.68 | 0.53 ± 0.07 |
| R177A | 1.99 ± 0.03 | 1.26 ± 0.01 | 1.58 ± 0.03 | 1.17 ± 0.10 | 0.49 ± 0.12 |
| R220A | 1.78 ± 0.06 | 1.18 ± 0.02 | 1.51 ± 0.06 | 1.99 ± 0.16 | 0.61 ± 0.10 |
Kinetic parameters determined using Hill equation: v = (Vmax × [S]n)/(K’ + [S]n). The Hill coefficient (n) is listed under the mutation identifier.
Kinetic parameters determined independently. The relative rate parameters for the arginine mutations below should be compared to this value for WT.
Table 2.
see also Table S1 and Figure S1. Oxidation of S-(+)-Tetralol and R-(−)-Tetralol by Wild-Type and Mutants of Actinorhodin Ketoreductase
| General Mutation Location |
S-(+)-Tetralol kcat/Kma |
R-(−)-Tetralol kcat/Kma |
S:R Activity Ratiob | |||
|---|---|---|---|---|---|---|
| s−1 mM−1 | % | s−1 mM−1 | % | |||
| WT | 0.035 | 100 | 0.010 | 100 | 3.5 | |
| D109E | Surface | 0.0063 | 18 | 0.0012 | 12 | 5.3 |
| D109R | Surface | Inactive | ||||
| S144C | Inactive | |||||
| T145A | .0041 | 12 | Inactive | NA | ||
| G146V | Inactive | |||||
| V151A | Inactive | |||||
| V151L | Substrate Inhibition | 0.026 | 260 | NA | ||
| A154G | 0.018 | 51 | 0.0021 | 21 | 8.6 | |
| F189A | 0.0070 | 20 | Inactive | NA | ||
| F189W | 0.029 | 83 | 0.0026 | 26 | 11.2 | |
| M194W | 0.034 | 97 | Inactive | NA | ||
| V198A | 0.023 | 66 | 0.020 | 200 | 1.2 | |
| V198G | 0.0012 | 3 | 0.00083 | 8 | 1.5 | |
| Y202A | α6-α7 Loop | 0.039 | 111 | 0.0078 | 22 | 5.0 |
| Y202F | α6-α7 Loop | 0.016 | 46 | 0.0048 | 14 | 3.3 |
Catalytic specificity (kcat/Km) expressed as percentage of wild-type value.
Ratio of S-tetralol to R-tetralol kcat/Km values.
Summary of analyses See also Table S1 and Figure S1.
Extensive studies of type I polyketide KRs have shown that their mutation often results in a mixture of enantiomeric reduced products and that multiple residues, located both within and distal to the active site, determine the stereospecificities of type I KRs (Baerga-Ortiz, et al., 2006; Castonguay, et al., 2008; Holzbaur, et al., 1999; Holzbaur, et al., 2001; Keatinge-Clay, 2007; Keatinge-Clay and Stroud, 2006; O'Hare, et al., 2006; Ostergaard, et al., 2002; Siskos, et al., 2005). In contrast, our previous studies of the type II polyketide KRs have shown that whereas wild-type actKR displays a 3:1 preference for S- over R-stereochemistry based on catalytic specificities for oxidation of tetralol enantiomers, the P94L mutation is sufficient to switch the in vitro stereospecificity of actKR to S-dominant, with no detectable activity toward R-tetralol. (Javidpour, et al., 2011). The term S-dominant herein is used to describe actKR mutants that display higher preference for S- over R-tetralol as compared to the wild-type enzyme. Our previous results also implied that long-ranged effects might be important for stereospecificity. We therefore constructed binding pocket mutants to determine how these residues affect KR stereospecificity. ActKR mutants were also assayed through in vitro reconstitution of the extended minimal PKS (act min PKS plus actKR). With the exception of evaluation of surface arginine mutants where actACP was used for accurate ACP-KR binding measurement, actACP was substituted with the orthologous frenN ACP for all reconstitution assays. The frenN ACP can be expressed at higher levels than the actinorhodin carrier protein and forms a stable protein complex (Khosla, et al., 1993). Products from mutant KR reconstitution experiments were compared to those from both the act min PKS or the extended minimal complex. Activity, relative to that of wild-type actKR, was assessed by comparing the ratio of the area of the mutactin peak to the average of both SEK4 and SEK4b peaks.
Surface Residue Mutants Display Greatly Diminished Activity and KR-ACP Interactions
Surface residues were mutated to assess whether R38A, R65A, R93A and D109E/R of actKR form a patch that interacts with ACP (Figure 2A). Previous in silico docking and mutational analyses suggested that the arginine residues, along with D109, form a charged groove that promotes complementary interactions with both helix II and the phosphopantetheine (PPant) group of the ACP (Korman, et al., 2008; Tang, et al., 2006). The D109 β-carboxyl group is within hydrogen-bonding distance of both the R65 and R93 side-chains. This interaction may maintain the structure of the phosphate-binding groove.
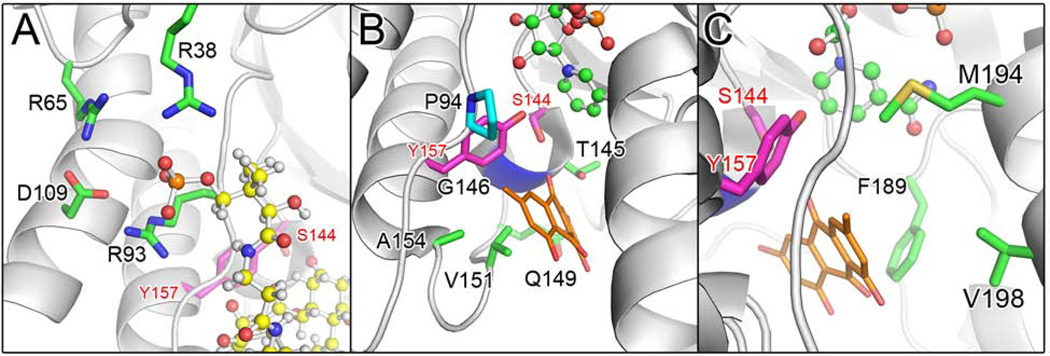
Figure 2. see also Figure S2 ActKR Residues Targeted for Site-Directed Mutagenesis.
(A) Arginines 38, 65, and 93 (green) are proposed to stabilize the PPT phosphate group during the binding of polyketide intermediate (yellow ball and sticks).
(B) Pocket residues (green) near catalytic S144 and Y157 (magenta) that interact with cocrystallized emodin (orange). NADPH (green ball and sticks) and P94 (teal) are shown for perspective and the location of G146 is highlighted in blue.
(C) Residues F189, M194, and V198 (green) relative to S144, Y157, and emodin.
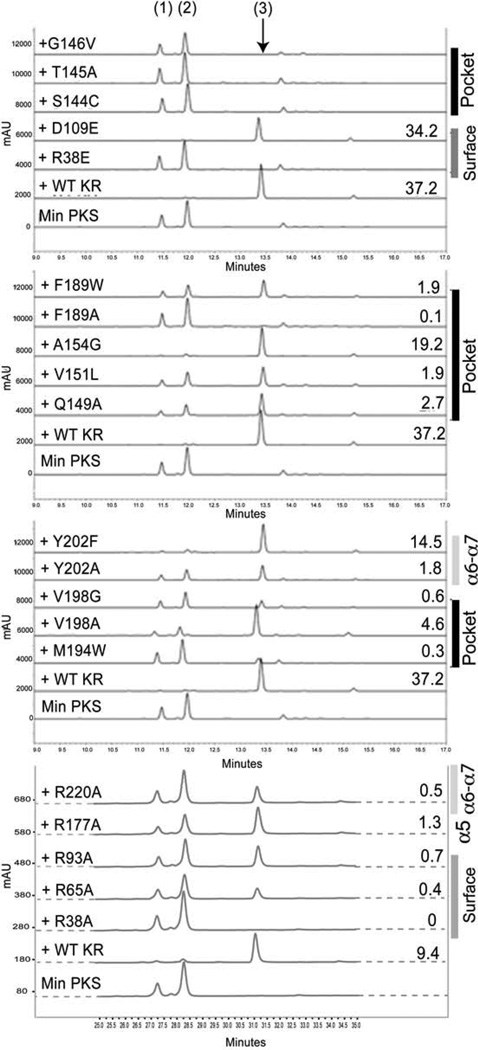
PKS reconstitution analysis showed that mutants R65A and R93A produced less mutactin than wild-type actKR, based on the respective mutactin:SEK4(b) ratios, while reconstitution with R38A actKR led to no detectable level of mutactin (Figure 3). To determine the dissociation constants (Kd) between actACP and actKR, separate fluorescence binding assays were measured (Figure S3). Here, the R38A actKR mutant displayed the weakest interaction (Kd 1.52 µM vs. 0.20 µM for wild-type). Mutants R65A and R93A both displayed a Kd value of 0.53 µM. The D109E actKR mutation had little effect on mutactin production (Figure 3), catalytic turnover in the trans-1-decalone assay, or activity when assayed with S- and R-tetralol (Table 1). The D109R mutant, in comparison, shows a 30-fold reduction in trans-1-decalone turnover and is inactive when assayed with S- and R-tetralol (Tables 2 and S1). The conservative D109E mutation is therefore, tolerated, while D109R strongly influences catalysis and may destabilize the adjacent arginine patch. NADPH binding to both wild-type actKR and the three arginine mutants (R38A, R65A, and R93A) was also determined using a fluorescence binding assay. A Kd of 1.52 µM for wild-type actKR was obtained, a value similar to that previously determined (5 µM, Korman et al., 2008). The R38A and R93A actKR mutants showed approximately 3-fold and 8-fold reductions in NADPH binding respectively, while R65A was unchanged. These results suggest that residues R38, R65, R93, and D109 are important for KR-ACP binding and activity. Mutations targeting the surface residues lead to disruption of the KR-ACP interaction and decreased activity toward the PPant-tethered polyketide substrates.
Figure 3. see also Figure S3 HPLC Chromatograms of Products from In Vitro PKS Reconstitution Assays.
The traces are divided into four panels for clarified viewing, with each panel including the traces for minimal PKS alone and with wild-type actKR as references. Products include SEK4 1, SEK4b 2, and mutactin 3. For each active KR, the number to the right of a trace denotes the respective mutactin:SEK4 + SEK4b ratio. The general mutation locations are also listed to the right of the panels.
KR Pocket Mutants: Non-Polar Side-Chains are Necessary for Substrate Interactions
The amphipathic nature of the polyketide intermediate requires both hydrophilic and hydrophobic residues for substrate-binding and the prevention of premature polyketide cyclization (Keatinge-Clay, et al., 2004). With this in mind, a number of substrate binding residues were identified, based on actKR-emodin co-crystal structures (Korman, et al., 2008). These mutated residues include S144C, T145A, G146V, V151A/L, A154G, F189A/W, M194W, and V198A/G (Figure 2B and 2C).
Mutants S144C and G146V were specifically designed to assess their role in C9-oxyanion stabilization during polyketide reduction. The S144C mutant was constructed to test substitution of the serine β-hydroxyl with a weaker hydrogen-bonding functional group. Residue G146 is located behind catalytic residues S144 and Y157. Mutation of this glycine to the larger, non-polar valine residue is expected to sterically hinder polyketide binding in the active site by disrupting the arrangement of S144 and Y157. As expected, mutants S144C and G146V did not produce any detectable levels of mutactin (Figure 3). Both mutations exhibited low activity toward S- and R-tetralol, and were inactive when assayed with trans-1-decalone (Tables 1, 2, and S1).
V151 is crucial for substrate recognition. The V151A mutant was inactive toward both trans-1-decalone and tetralol (Tables 1, 2, and S1). V151 is located ~5 Å from Y157 near the base of the monomer cleft (Figure 2B). The branched side-chain of residue 151 extends into the binding pocket where non-polar interactions orient the substrate prior to reduction. In contrast, the conservative V151L mutation displayed kcat and Km values for trans-1-decalone and R-tetralol turnover that were comparable to those of wild-type actKR, supporting the hypothesis that a large hydrophobic side-chain is required at this position to steer substrate-binding and sustain reductase activity. V151L did, however, produce 20-fold less mutactin than wild-type actKR (Figure 3). Here the leucine side-chain may interfere with polyketide-binding, further highlighting the requirement for precise steric control within the active site. This may explain the high sequence conservation of V151 amongst type II PKS KRs.
The value for kcat/Km of A154G actKR in the trans-1-decalone assay was similar to that of the wild-type enzyme, while the activity using both S- and R-tetralol substrates was also relatively high compared to the other mutants (Tables 2 and S1). F189 mutations (F189A and F189W), however, displayed the lowest activity toward trans-1-decalone (Table 1). Activity of both mutants was also significantly reduced where R-tetralol was used as the substrate, though turnover of the S-enantiomer by the F189W mutant enzyme approached wild-type levels. The F189W mutant also produced much lower levels of mutactin when compared to wild-type actKR, while mutactin production in F189A was almost undetectable (Figure 3). Assay results for the F189 mutants suggest that, similar to V151, residue F189 is crucial for enzyme-substrate interaction.
Mutation M194W abolished mutactin production, and reduced trans-1-decalone turnover to 5% of wild-type. The actKR containing this methionine-tryptophan mutation was completely selective for the S-enantiomer of tetralol. Here the bulky tryptophan may sterically clash with the NADPH nicotinamide, residue P94, or with polyketide binding.
The V198G mutation drastically reduces mutactin production and shows markedly decreased activity towards both S- and R-tetralol (Table 2). A V198A mutation in comparison only shows moderate changes in these assays, with a small increase in R-tetralol selectivity. Progressive reduction of the hydrophobic side chain at this position clearly exerts strong effects on enzyme activity and fidelity of polyketide chain processing.
Aromatic Interaction in the α6-α7 Loop Region May Be Important for Substrate Recognition
The type II PKS KRs possess a distinctive 11-residue insertion in the α6-α7 loop (actKR residues 199–209) (Korman, et al., 2008). This loop is the least conserved region in the short-chain dehydrogenase/reductase (SDR) superfamily of proteins, to which actKR belongs, and may contribute to SDR substrate specificity (Oppermann, et al., 2003). Residue Y202 is highly conserved among the type II KRs (Fig S2A). Our previous studies suggested that α6-α7 loop residues may play an important role in recognizing polyketide intermediates (Javidpour, et al., 2011). We therefore constructed mutants Y202A and Y202F in order to evaluate the respective contributions of an aromatic side-chain and a phenolic hydroxyl group toward substrate recognition and activity.
The Y202 mutants displayed activities toward trans-1-decalone and tetralol that are comparable to those of wild-type (Tables 1, 2, and S1). However, mutactin production by the Y202A mutant is reduced, suggesting that complete removal of the aromatic side chain negatively influences substrate turnover. Y202 may be involved in π-π interactions with W206 that stabilize the α6-α7 loop conformation and might therefore play a structural rather than a catalytic role. This may also explain why aromatic pairs are highly conserved amongst type II PKS KRs (Fig. S2A) at these positions.
Two further residues, R177 that packs against the α6-α7 loop from an adjacent KR subunit in the tetramer, and R220 that packs against Y202, were both mutated to alanine (Tables 1–2, S1, Fig. 3). Neither mutation had a significant effect in any of the activity assays, or NADPH or ACP binding.
Structural Analysis of V151L, F189W, and Y202F ActKR
Three of the mutants that displayed intriguing assay results were crystallized and their structures solved; V151L (for its R-tetralol specificity), F189W (for its S-tetralol specificity), and Y202F (to probe the long-range effect of the α6-α7 loop). Overall, the structures are very similar to the NADPH-bound structures of wild-type and P94L actKR (Javidpour, et al., 2011). The alignment root-mean-square deviations (RMSD) of the mutant structures relative to wild-type are: 0.41, 0.44, and 0.35 Å for V151L F189W, and Y202F, respectively, and 0.41, 0.55, and 0.40 relative to P94L. The alignments of the catalytic residues (N114, S144, Y157, and K161) and the NADPH cofactor have similar positions and conformations (Figure 4A). The three mutant structures show that while mutations significantly altered observed activities and regio-and stereospecificities in three different assays, the overall protein conformations are not changed.
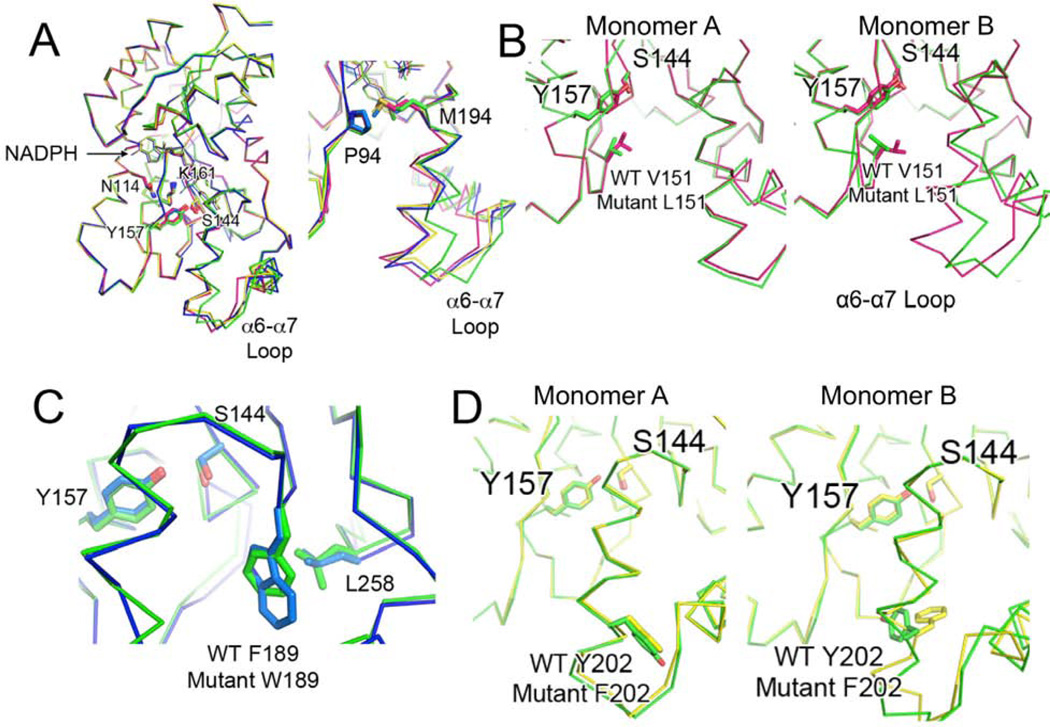
Figure 4. see also Figure S2 Mutant ActKR Structural Analysis.
(A) (Left) Alignment of monomer B of V151L (magenta), F189W (blue), and Y202F (yellow) to that of wild-type actKR (green). Although the catalytic residues and NADPH cofactors align well, the mutant monomers are in a closed conformation relative to wild-type, as seen by the shifts in the labeled α6-α7 loops and extended M194 side-chains (right).
(B) Residue 151 extends into the substrate pocket similarly in monomer A of wild-type (green) and V151L (magenta) (left), but L151 in the mutant monomer B points more toward the α6-α7 loop, creating a smaller substrate pocket (right)
(C) The aromatic plane of W189 in the F189W mutant (blue) coincides with that of wild-type F189 (green). L258 may hydrophobically interact with W189, limiting the substrate pocket size.
(D) Residue 202 overlaps when monomer A of wild-type (green) and Y202F (yellow) are aligned (left), but in monomer B (right), the wild-type Y202 points away from the substrate pocket while the mutant F202 points within the pocket.
Despite the high overall similarity, there are structural differences that distinguish the three mutants from the previously characterized crystal structures (Korman, et al., 2004) (Korman, et al., 2008). Alignment of the B subunits shows that in the V151L, F189W, and Y202F mutants, the α6-α7 loop adopts a more closed conformation than the wild-type (Figure 4 and Figure S2B). In these mutants M194 becomes extended and is potentially involved in non-polar interactions with P94. When V151L is aligned to wild-type actKR, L151 of subunit A extends into the substrate pocket in a similar conformation to V151. In the mutated subunit B, however, L151 points toward the α6-α7 loop, thus creating a closed conformation and a smaller substrate pocket (Figure 4B). Similarly, in F189W the tryptophan aromatic ring overlaps with the wild-type residue F189, while L258 interacts with W189, again resulting in a smaller substrate pocket in monomer A. In the wild-type enzyme, the Y202 side-chain potentially forms a water-mediated hydrogen bond with the H153 backbone carbonyl and consequently points away from the α6-α7 loop. In the F202 mutant, however, the phenylalanine ring rotates back into the substrate binding pocket, possibly due to loss of this hydrogen bonding interaction (Figure 4D). The V151L, F189W, and Y202F mutations therefore result in a smaller substrate pocket for monomer B, which contributes to the observed low mutant enzyme activities. The altered pocket shape also imposes more steric hindrance in V151L, resulting in the observed substrate inhibition (Tables 2 and S1).
Docking Analyses: The Molecular Basis for V151L and F189W Tetralol Assay Results
Mutant V151L displayed unusual activity toward tetralol, with almost threefold higher activity toward the R-stereoisomer than that of wild-type (Tables 2 and S1). When assayed with S-tetralol, however, we observed substrate inhibition at high (> 2 mM) S-tetralol concentrations. Both tetralol stereoisomers were docked within the V151L actKR substrate binding pocket using the program GOLD (Verdonk, et al., 2003). The docking results yielded similar positions and conformations for each stereoisomer, whereas for the wild-type actKR structure, R-tetralol does not consistently dock. In the V151L mutant, the R-tetralol aromatic ring consistently docks within the opening of the substrate binding cleft (Figure S1A), consistent with the high kcat/Km of this mutant for R-tetralol relative to the wild-type enzyme. In contrast, the simulations showed that S-tetralol docks with the aromatic ring near residues T145, F189, and L258, which avoids clashes with L151. S-tetralol may, therefore, be lodged between these three residues, causing the enzyme to be “trapped” in a substrate-bound state (ES) by the high affinity for S-tetralol and resulting in the observed substrate inhibition phenomenon. To the best of our knowledge, such a phenomenon has not been reported for type I polyketide KRs before. This result also demonstrates that the type II polyketide KR is unique in its ability to switch stereospecificity through a single mutation.
To ascertain the molecular basis for the high F189W stereospecificity, S- and R-tetralol were again docked within the enzyme binding pocket. Analysis suggests that the stereospecificity may arise from a combination of steric hindrance, (as for P94L or M194W actKR), as well as from the W189 indole group, which may adopt a different rotamer conformation upon substrate-binding. This allows π-π interactions with the S-tetralol benzyl moiety, but not with the R-stereoisomer (Figure S1B). The adoption of a different rotameric state is supported by the crystallographic data, wherein there is well-defined 2Fo-Fc electron map density for W189 in subunit A of the mutant, but weaker density for the residue in subunit B. This suggests the presence of multiple rotamers in subunit B. The steric effect exerted by W189 thus steers the mutant toward S-dominant stereospecificity.
DISCUSSION
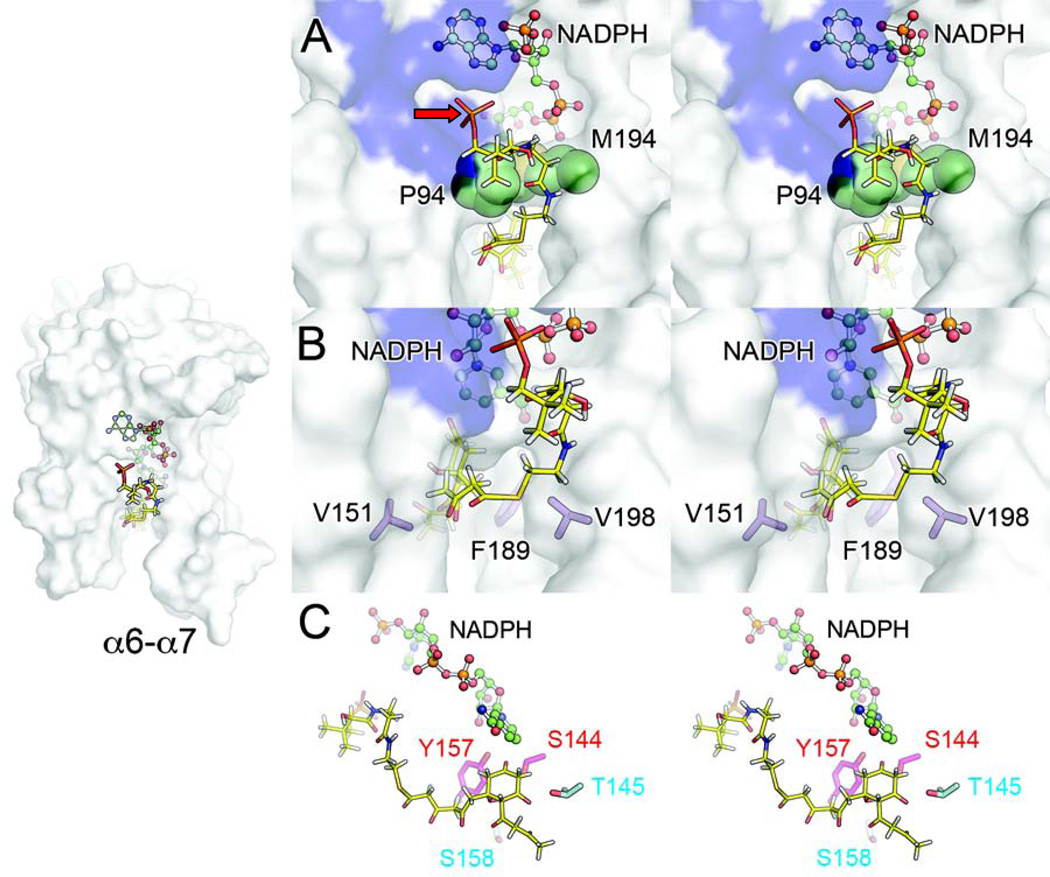
Based on the results of kinetic and reconstitution assays, structural analyses, docking simulations, and incorporation of previous studies (Javidpour, et al., 2011; Korman, et al., 2004; Korman, et al., 2008), we propose the following model for polyketide docking, entrance into the substrate pocket, C7-C12 first-ring cyclization, and stereospecific ketoreduction by a type II polyketide KR that accounts for the observed properties of wild-type and mutant actKR (Figure 5).
Figure 5. Stereo Images of Proposed Interactions between Polyketide and Wild-Type ActKR.
The polyketide chain is represented in yellow and actKR in transparent white surface.
(A) Ligand docking results suggest that after the PPT phosphate binds within the arginine patch groove (blue surface), the polyketide chain enters the substrate pocket underneath the gate residues P94 and M194 (green spheres) in order to access the active site.
(B) The polyketide intermediate is steered toward the active site by the bulky residues V151, F189, and V198 (purple sticks), which presumably constrain chain movement.
(C) Ligand docking also suggests that T145 (teal) may stabilize the C11-oxyanion after C12 deprotonation through hydrogen-bonding, thus lowering the C12-proton pKa and allowing C7-C12 cyclization to occur. S158 (teal) may stabilize the resulting C7-oxyanion through water-mediated hydrogen-bonding. Both T145 and S158 are highly conserved among type II KRs.
PPant-Tethered Polyketide Docking
Docking results described in this paper, combined with studies of the homologous S. coelicolor SCO1815 KR (Javidpour, et al., 2011; Tang, et al., 2006), identify an arginine patch as the putative docking site of the PPant phosphate (Figure 2A). In the tetrameric actKR structure, the arginine patch is accessible on each monomer and is not hindered by any neighboring molecule. The assay results for the R38A, R65A, and R93A mutants, which displayed decreased reconstitution activity and interaction with ACP, lend further support to the importance of the arginine patch for enzyme activity.
Access to the ActKR Substrate Pocket
The docked PPant carrying the polyketide intermediate must now access the actKR active site. Two openings are seen in the surface representation of wild-type actKR subunit A, separated by P94 and the M194, which hereafter are referred to as “gate” residues (Figure 5A). The importance of M194 is supported by assays of the M194W mutant, which produced less mutactin than expected based on the trans-1-decalone assay. The mutant tryptophan may sterically hinder ]polyketide docking, or block access to the active site. Entrance to the enzyme pocket from the opening immediately proximal to the ]arginine patch is further blocked by the NADPH pyrophosphate moiety. The polyketide must, therefore, enter the pocket through the cleft below the P94 and M194 gate residues.
Steering of Polyketide toward the Active Site and Monomer-Closing
The polyketide substrate must be directed by the “steering” residues, which include the hydrophobic V151, F189, and V198. These residues must be non-polar and not glycine as suggested by the assay results for V198G, which produced less mutactin than expected. The V198 side-chain protrudes into the enzyme cleft, effectively steering the PPant arm and polyketide intermediate toward the active site residues (Figure 5B). V198 may also participate in non-polar interactions with the end of the PPant arm distal from ACP. Furthermore, the V151 and F189 side-chains define the narrow cleft and limit substrate movement. This is supported by assays of the V151 mutants: V151A is inactive toward trans-1-decalone, whereas V151L is active and leads to mutactin production, suggesting the need for a bulky residue at this position. Similarly, the need for a bulky residue at position 189 is supported by assays of F189A and F189W. The latter mutant produced more mutactin when compared to F189A, despite the presence of the bulky tryptophan.
KR-Mediated C7-C12 First-Ring Cyclization and C9-Ketoreduction
Having beem steered towards the active site, the linear polyketide is then positioned such that the C11-carbonyl can interact through hydrogen-bonding with T145, which is highly conserved among type II KRs. This interaction could sufficiently lower the pKa of the C12 α-protons (Gerlt, et al., 1991) such that a proton can be abstracted by water in the substrate binding pocket. This leads to C7-C12 cyclization, with the C7-oxyanion stabilized by water-mediated hydrogen-bonding to the highly conserved S158. This mechanism is supported by assays of T145A, which displayed activity toward trans-1-decalone (Table 1), but intriguingly did not produce any detectable level of mutactin. Other mutants less active toward trans-1-decalone than T145A were able to produce some mutactin (Figure 3). We therefore propose that T145 is involved in C7-C12 polyketide cyclization through hydrogen-bonding with the C11-carbonyl, which lowers the pKa of the C12-protons via enolate stabilization.
The model presented in this paper helps to explain how the actKR affects cyclization specificity such that all downstream products of the PKS are cyclized between C7 and C12. In the absence of KR, approximately equal amounts of SEK4 (C7-C12 cyclized) and SEK4b (C10-C15 cyclized) are produced by the minimal PKS (Figure 1). The KR may, therefore, serve as a scaffold that steers the polyketide substrate toward T145 for C7-C12 cyclization, which then forces the C9-carbonyl within the active site for regiospecific ketoreduction (Figure 5C). If the polyketide intermediate were not cyclized, the C9-carbonyl would be able to adopt a number of orientations. In addition, C7-C12 cyclization would restrict the mode of polyketide binding within the active site, which explains why actKR may produce only one enantiomer of mutactin in vivo (Javidpour, et al., 2011). The absence of hydrogen bond donation from T145A would prevent directed-cyclization from occurring, which in turn would compromise ketoreduction. As a result, the ACP-bound polyketide may pull out of the KR pocket and spontaneously cyclize in solution to yield SEK4 and SEK4b.
We propose that during ketoreduction, actKR achieves a closed-state when the α6-α7 loop moves toward the opposite side of the enzyme cleft, with π-π interactions between the highly conserved residues 202 and 206 maintaining the loop conformation as it closes in on the polyketide substrate (Figure S2). This motion effectively traps the polyketide within the enzyme, yet leaves openings for water to enter the binding site. The monomer opens once catalysis is complete, allowing the ACP-bound reduced polyketide to leave the enzyme. In this regard, the binding site of actKR is distinct from that of actKS-CLF (Keatinge-Clay, et al., 2004), where an amphipathic tunnel is needed for polyketide elongation toward the KS-CLF gate residues (Tang, et al., 2003). The situation is arguably more complex for the actKR, however, as a tunnel would prevent KR from mediating cyclization. The actKR pocket must, therefore, be large enough to accommodate a monocyclized polyketide, yet still maintain tight interactions with the substrate so that release does not occur before reduction. The bulky residues, V151, F189, and V198, are necessary to both steer and cradle the polyketide in position until catalysis is complete.
SIGNIFICANCE
Type II polyketides include many antibiotics and chemotherapeutic agents that are of interest to the pharmaceutical industry (Saleem, et al., 2010). The type II polyketide KR is an important modification enzyme that introduces chemical diversity in polyketides by reducing intermediates with high regio- and stereospecificity and possibly mediating a specific C7-C12 first-ring cyclization. Previous biochemical and structural analyses have identified the “PGG” motif as important for in vitro stereospecificity (Javidpour, et al., 2011; Korman, et al., 2008). In this work, we depart from the status quo to elucidate the roles of surface, binding pocket, and loop-region residues of actKR through a suite of informative tools: site-directed mutagenesis, kinetic assays, and PKS reconstitution assays. Highly consistent results from three different assays of 20 mutants offer a rich array of information about the type II KR, such as the identification of bulky hydrophobic pocket residues that are necessary to serve as “gate” and “steering” residues for substrate binding. We have also shown that a single mutation (V151L) converts stereospecificity to R-dominant, as opposed to other single mutations that result in S-dominant stereospecificity. Furthermore, the T145 mutant offered the first experimental support to the suggestion that this type II KR fosters first-ring cyclization between C7 and C12. The results support a proposed model for polyketide docking, entrance into the KR enzyme pocket, cyclization, and ketoreduction that accounts for the high specificity of actKR. The molecular features identified in this work will serve as protein engineering targets for rational control of KR specificity to produce new polyketides with pharmaceutical potential.
EXPERIMENTAL PROCEDURES
ActKR, ActKS-CLF, Holo-FrenN ACP and MAT Protein Expression and Purification
Recombinant wild-type or mutant actKR, actKS-CLF, holo-ACP and MAT proteins were express and purified as described previously (Korman, et al., 2004; Korman, et al., 2008; Matharu, et al., 1998); (Li, et al., 2003); (Kumar, et al., 2003). Additional details can be found in the Supplemental Information.
In Vitro trans-1-Decalone, Tetralol, and In Vitro PKS Reconstitution Assays for ActKR Activity
Steady-state kinetic parameters for wild-type and mutant actKR were determined by monitoring reduction of trans-1-decalone in the presence of NADPH, by monitoring oxidation of S-(+)- or R-(−)-tetralol in the presence of NADP+, and PKS reconstitution assay. Steady-state kinetic data are listed in Tables 1, 2 and S1. Additional details can be found in the Supplementary Information.
In Vitro Tetralol Assay for ActKR Stereospecificity
Steady-state kinetic parameters for wild-type and mutant actKR were determined by monitoring oxidation of S-(+)- or R-(−)-tetralol in the presence of NADP+. Summarized kinetic data are listed in Table 2 and detailed steady-state data in Table S1. Additional details can be found in the Supplemental Information.
ACP Fluorescence Binding Assay
To gauge interaction between ACP and either wild-type, or mutant KR, actACP was titrated into a solution of actKR containing 25 µM NADPH and 1 mM DTT. Fluorescence emission spectra were collected over the range of 320–380 nm at a 295-nm excitation wavelength (tryptophan fluorescence). For NADPH binding, increasing concentrations of NADPH (0–38µM) were titrated in actACP (1µM) and the NADPH fluorescence was excited at 340nm and monitored between 400–500nm. Additional details can be found in the Supplemental Information.
Protein Crystallization, Data Collection, and Structure Solution
Experimental procedures and statistics for structure determination of the V151L, F189W, and Y202F actKR mutants can be found in the Supplemental Information.
Supplementary Material
Highlights.
ActKR surface arginines are important for ACP-binding and activity toward polyketides
In contrast to the S-specific P94L actKR, mutant V151L displays R-stereospecificity
ActKR is proposed to mediate C7-C12 polyketide cyclization prior to C9-ketoreduction
ACKNOWLEDGEMENTS
We thank Justin Nowell, and Eunahn Suh for assisting with KR expression, purification, and spectrophotometric assays. This work is supported by the Pew Foundation and National Institute of General Medical Sciences (NIGMS R01GM076330, ARRA-GM076330-S4). Portions of this research were carried out at the Advanced Light Source, which is supported by the U.S. Department of Energy Office of Science by Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Structure coordinates have been deposited with PDB codes 4DBZ, 4DC0, and 4DC1 for V151L, F189W, and Y202F actKR, respectively.
REFERENCES
- Baerga-Ortiz A, Popovic B, Siskos AP, O'Hare HM, Spiteller D, Williams MG, Campillo N, Spencer JB, Leadlay PF. Directed mutagenesis alters the stereochemistry of catalysis by isolated ketoreductase domains from the erythromycin polyketide synthase. Chem Biol. 2006;13:277–285. doi: 10.1016/j.chembiol.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Broussy S, Cheloha RW, Berkowitz DB. Enantioselective, ketoreductase-based entry into pharmaceutical building blocks: ethanol as tunable nicotinamide reductant. Org Lett. 2009;11:305–308. doi: 10.1021/ol802464g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- Castonguay R, Valenzano CR, Chen AY, Keatinge-Clay A, Khosla C, Cane DE. Stereospecificity of ketoreductase domains 1 and 2 of the tylactone modular polyketide synthase. J Am Chem Soc. 2008;130:11598–11599. doi: 10.1021/ja804453p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Townsend CA. New insights into the formation of fungal aromatic polyketides. Nat Rev Microbiol. 2010;8:879–889. doi: 10.1038/nrmicro2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Khosla C. Biosynthesis of aromatic polyketides in bacteria. Acc Chem Res. 2009;42:631–639. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutler H, Coon MJ, Kull A, Vogel H, Waldvogel G, Prelog V. Fatty acid synthetase from pig liver. 1. Isolation of the enzyme complex and characterization of the component with oxidoreductase activity for alicyclic ketones. Eur J Biochem. 1971;22:203–212. doi: 10.1111/j.1432-1033.1971.tb01533.x. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Kozarich JW, Kenyon GL, Gassman PG. Electrophilic Catalysis Can Explain the Unexpected Acidity of Carbon Acids in Enzyme-Catalyzed Reactions. Journal of the American Chemical Society. 1991;113:9667–9669. [Google Scholar]
- Grau BT, Devine PN, DiMichele LN, Kosjek B. Chemo- and enantioselective routes to chiral fluorinated hydroxyketones using ketoreductases. Org Lett. 2007;9:4951–4954. doi: 10.1021/ol701810v. [DOI] [PubMed] [Google Scholar]
- Holzbaur IE, Harris RC, Bycroft M, Cortes J, Bisang C, Staunton J, Rudd BA, Leadlay PF. Molecular basis of Celmer's rules: the role of two ketoreductase domains in the control of chirality by the erythromycin modular polyketide synthase. Chem Biol. 1999;6:189–195. doi: 10.1016/S1074-5521(99)80035-0. [DOI] [PubMed] [Google Scholar]
- Holzbaur IE, Ranganathan A, Thomas IP, Kearney DJ, Reather JA, Rudd BA, Staunton J, Leadlay PF. Molecular basis of Celmer's rules: role of the ketosynthase domain in epimerisation and demonstration that ketoreductase domains can have altered product specificity with unnatural substrates. Chem Biol. 2001;8:329–340. doi: 10.1016/s1074-5521(01)00014-x. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. Genetic Contributions to Understanding Polyketide Synthases. Chem Rev. 1997;97:2465–2498. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- Jacobsen JR, Keatinge-Clay AT, Cane DE, Khosla C. Precursor-directed biosynthesis of 12-ethyl erythromycin. Bioorg Med Chem. 1998;6:1171–1177. doi: 10.1016/s0968-0896(98)00081-9. [DOI] [PubMed] [Google Scholar]
- Javidpour P, Korman TP, Shakya G, Tsai SC. Structural and Biochemical Analyses of Regio- and Stereo-Specificities Observed in a Type II Polyketide Ketoreductase. Biochemistry. 2011 doi: 10.1021/bi200335f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javidpour P, Korman TP, Shakya G, Tsai SC. Structural and biochemical analyses of regio- and stereospecificities observed in a type II polyketide ketoreductase. Biochemistry. 2011;50:4638–4649. doi: 10.1021/bi200335f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT, Maltby DA, Medzihradszky KF, Khosla C, Stroud RM. An antibiotic factory caught in action. Nat Struct Mol Biol. 2004;11:888–893. doi: 10.1038/nsmb808. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT, Stroud RM. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Khosla C, McDaniel R, Ebert-Khosla S, Torres R, Sherman DH, Bibb MJ, Hopwood DA. Genetic construction and functional analysis of hybrid polyketide synthases containing heterologous acyl carrier proteins. J Bacteriol. 1993;175:2197–2204. doi: 10.1128/jb.175.8.2197-2204.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korman TP, Hill JA, Vu TN, Tsai SC. Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity. Biochemistry. 2004;43:14529–14538. doi: 10.1021/bi048133a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korman TP, Tan YH, Wong J, Luo R, Tsai SC. Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase. Biochemistry. 2008;47:1837–1847. doi: 10.1021/bi7016427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Koppisch AT, Cane DE, Khosla C. Enhancing the modularity of the modular polyketide synthases: transacylation in modular polyketide synthases catalyzed by malonyl-CoA:ACP transacylase. J Am Chem Soc. 2003;125:14307–14312. doi: 10.1021/ja037429l. [DOI] [PubMed] [Google Scholar]
- Li Q, Khosla C, Puglisi JD, Liu CW. Solution structure and backbone dynamics of the holo form of the frenolicin acyl carrier protein. Biochemistry. 2003;42:4648–4657. doi: 10.1021/bi0274120. [DOI] [PubMed] [Google Scholar]
- Malpartida F, Hopwood DA. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature. 1984;309:462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- Matharu AL, Cox RJ, Crosby J, Byrom KJ, Simpson TJ. MCAT is not required for in vitro polyketide synthesis in a minimal actinorhodin polyketide synthase from Streptomyces coelicolor. Chem Biol. 1998;5:699–711. doi: 10.1016/s1074-5521(98)90663-9. [DOI] [PubMed] [Google Scholar]
- McDaniel R, Ebert-Khosla S, Fu H, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: influence of a downstream enzyme on the catalytic specificity of a minimal aromatic polyketide synthase. Proc Natl Acad Sci U S A. 1994;91:11542–11546. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- O'Hare HM, Baerga-Ortiz A, Popovic B, Spencer JB, Leadlay PF. High-throughput mutagenesis to evaluate models of stereochemical control in ketoreductase domains from the erythromycin polyketide synthase. Chem Biol. 2006;13:287–296. doi: 10.1016/j.chembiol.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Oppermann U, Filling C, Hult M, Shafqat N, Wu X, Lindh M, Shafqat J, Nordling E, Kallberg Y, Persson B, et al. Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem Biol Interact. 2003:143–144. doi: 10.1016/s0009-2797(02)00164-3. 247–253. [DOI] [PubMed] [Google Scholar]
- Ostergaard LH, Kellenberger L, Cortes J, Roddis MP, Deacon M, Staunton J, Leadlay PF. Stereochemistry of catalysis by the ketoreductase activity in the first extension module of the erythromycin polyketide synthase. Biochemistry. 2002;41:2719–2726. doi: 10.1021/bi0117605. [DOI] [PubMed] [Google Scholar]
- Saleem M, Nazir M, Ali MS, Hussain H, Lee YS, Riaz N, Jabbar A. Antimicrobial natural products: an update on future antibiotic drug candidates. Nat Prod Rep. 2010;27:238–254. doi: 10.1039/b916096e. [DOI] [PubMed] [Google Scholar]
- Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- Siskos AP, Baerga-Ortiz A, Bali S, Stein V, Mamdani H, Spiteller D, Popovic B, Spencer JB, Staunton J, Weissman KJ, et al. Molecular basis of Celmer's rules: stereochemistry of catalysis by isolated ketoreductase domains from modular polyketide synthases. Chem Biol. 2005;12:1145–1153. doi: 10.1016/j.chembiol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Tang Y, Lee HY, Kim CY, Mathews I, Khosla C. Structural and functional studies on SCO1815: a beta-ketoacyl-acyl carrier protein reductase from Streptomyces coelicolor A3(2) Biochemistry. 2006;45:14085–14093. doi: 10.1021/bi061187v. [DOI] [PubMed] [Google Scholar]
- Tang Y, Tsai SC, Khosla C. Polyketide chain length control by chain length factor. J Am Chem Soc. 2003;125:12708–12709. doi: 10.1021/ja0378759. [DOI] [PubMed] [Google Scholar]
- Truppo MD, Pollard D, Devine P. Enzyme-catalyzed enantioselective diaryl ketone reductions. Org Lett. 2007;9:335–338. doi: 10.1021/ol0627909. [DOI] [PubMed] [Google Scholar]
- Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- Zawada RJ, Khosla C. Heterologous expression, purification, reconstitution and kinetic analysis of an extended type II polyketide synthase. Chem Biol. 1999;6:607–615. doi: 10.1016/s1074-5521(99)80112-4. [DOI] [PubMed] [Google Scholar]
- Zhan J. Biosynthesis of bacterial aromatic polyketides. Curr Top Med Chem. 2009;9:1958–1610. doi: 10.2174/156802609789941906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.