Abstract
Cancers of the oesophagus, gastro-oesophageal junction and stomach (upper gastrointestinal tract cancers; UGICs) pose a major health risk around the world. Collectively, the 5-year survival rate has remained <15% and therapeutic improvements have been very slow and small. Therefore, novel molecules for early diagnosis, prognosis, prediction, and therapy are urgently needed. The role that microRNA (miRNA) molecules seem to play in UGICs are worth pursuing. miRNAs are small noncoding RNA molecules that regulate ~60% of coding genes in humans and, therefore, are pivotal in mediating and regulating many physiologic processes. miRNAs are deregulated in many disease states, particularly in cancer, making them important targets. Here, we review the building body of evidence regarding the alterations of miRNAs in UGICs. By suppressing translation and/or promoting degradation of mRNAs, miRNAs can contribute to carcinogenesis and progression of UGICs. In-depth studies of miRNAs in UGICs might yield novel insights and potential novel therapeutic strategies.
Introduction
Cancers of the upper gastrointestinal tract (UGICs) include those originating in the oesophagus, gastro-oesophageal junction and stomach. The incidence of adenocarcinoma involving the lower third of the oesophagus, gastro-oesophageal junction and proximal stomach has risen considerably in the past 30 years.1 It is estimated that 38,780 new cases and 25,610 deaths are likely to occur in 2012 in the USA alone.2 Gastric cancer is the fourth most common cancer and the second leading cause of cancer-related death in the world.3 Helicobacter pylori infection, gastrin levels, germline mutations, dietary factors and other chronic gastric conditions are all factors involved in the development of gastric cancer. Our understanding of the molecular basis of carcinogenesis and progression of UGICs has lagged behind compared with many other tumour types. This knowledge deficit is creating a barrier in the development of effective therapeutics, and progress against UGICs has been unsatisfactory and slow. Consequently, outcomes for patients with UGICs have remained dismal, with 5-year survival rates <15%. Improved understanding of the role of microRNAs (miRNAs) in UGICs could lead to novel prevention strategies, early detection and improved therapeutics.
MicroRNAs
miRNAs are short (20–24 nucleotides) stable RNA molecules that are not translated into proteins but regulate 60% of coding genes by binding to mRNA molecules (to prevent their translation and/or promote degradation). More than 1,000 miRNAs have been identified, and they are involved in nearly all physiologic processes (as well as having a role in diseases like cancers). Novel functions and mechanisms by which miRNAs regulate genes are constantly being discovered.4–6
miRNA genes are located in intergenic regions but can also be in exonic or intronic regions of other genes.7 In addition, miRNA genes can be found in the introns of protein-coding or non-protein-coding genes.8 Each miRNA can target a large number of genes (mRNAs). In addition, each mRNA can be targeted by several miRNAs.9 The many ways in which miRNAs engage mRNA have been described and new mechanisms are regularly being discovered.8 Most commonly, miRNAs can either degrade an mRNA (when perfect complementarity is established) or inhibit its translation (when imperfect complementarity is established). The outcome is a reduction in the amount of a particular protein inside the cell.4,10 In addition, some miRNAs can directly bind to the (open reading frame of the) DNA or modify the methylation status of a gene.11 A mature miRNA can target mRNA binding proteins (functioning like a decoy).12
The genes that encode miRNAs are frequently located inside or close to fragile sites of chromosomes and are subject to considerable deregulation in cancer.10 Alterations in the expression of miRNAs in cancer are related to deletions, mutations, polymorphisms, promoter hypermethylation and/or histone acetylation of miRNA genes as well as alterations in the miRNA processing machinery. Amplifications, translocations and/or transcript activations can also lead to changes in miRNAs.
Many miRNAs have been identified to act as oncogenes, tumor suppressors and important modulators in cellular pathways. The oncogenic or tumour suppressor function of miRNAs depends on the outcome of the target mRNA. Increased activity of oncomiRNAs leads to inhibition of tumour suppressor genes, facilitating cell proliferation and tumour progression. Decreased activity of tumour-suppressor miRNAs (tsmiRs) therefore leads to increased oncogene translation, contributing to tumour formation.13 Certain miRNAs are involved in the regulation of metastasis (metastamiRs); these miRNAs can positively or negatively regulate cancer cell migration, invasion and metastasis.14,15
Some miRNAs are upregulated or downregulated in specific types of cancer (for miR-122 in hepatocellular carcinoma (refs: 1. Kota, J, Chivukula, RR, O’Donnell, KA, Wentzel, EA, Montgomery, CL, Hwang, HW, Chang, TC, Vivekanandan, P, Torbenson, M, Clark, KR, Mendell, JR, Mendell, JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009;137(6): 1005–1017.http://www.ncbi.nlm.nih.gov/pubmed/19524505, Haussecker, D, Kay, MA. miR-122 continues to blaze the trail for microRNA therapeutics. Mol Ther 2010;18(2): 240–242.http://www.ncbi.nlm.nih.gov/pubmed/20125164,). Further research may yield tissue-specific miRs. This level of specificity could enable tumour-type-specific targeting. Some miRNAs are actively secreted in blood and other body fluids and could potentially be used for the early diagnosis of cancer and monitoring of therapy; circulating miRNAs have been described in UGICs.16 Some miRNAs participate in genetic exchange among cells.17 miRNA profiling can make a distinction between normal and malignant tissue7 but, more importantly, since tumour-type-specific miRNA profiles (miRNomes) have been discovered, such observations lend themselves to establishing miRNomics as a diagnostic tool for uncovering the origin of tumours.8 For example, miR-122 appears to be highly enriched in the liver, suggesting that the discovery of more organ-specific miRNAs could improve our ability to target these processes.18 For greater understanding of the mechanisms of deregulated miRNA, we recommend a previous report.19
miRNAs and oesophageal cancer
Microarray or real-time PCR and in situ hybridization have been used to determine and validate expression of miRNA in cell lines and tumour specimens. Many reports have described deregulated miRNA, their targets as well as their functions in the major oesophageal cancer histologic subtypes: oesophageal adenocarcinoma (Table 1) and oesophageal squamous cell carcinoma (Table 2).20
Table 1.
miRNAs and their targets in oesophageal adenocarcinoma
| miRNA | Targets | References |
|---|---|---|
| OncomiRs | ||
| miR-21** | PDCD4 | 21,23,24 |
| miR-196a* | ANXA1/SPRR2C/S100 | 18,19 |
| miR-192* | NR | 26 |
| miR-106b-25 | P21/Bim | 17 |
| miR-99b/199a | NR | 79 |
| miR-194 | NR | 23 |
| tsmiRs | ||
| miR-203** | NR | 24–27,29 |
| miR-205* | NR | 24,25,27,29 |
| Let-7a/b/c | NR | 28 |
| miR-200a/b/c | ZEB1, ZEB2 | 20 |
| miR-31/miR375 | NR | 22 |
| miR-345/494/193a | NR | 28 |
Reported consistently by more than four individual groups;
Reported by two groups. Abbreviation: NR, not reported
Table 2.
Deregulated miRNAs, potential targets and functions in oesophageal squamous carcinoma
| miRNA | Targets | Functions | References |
|---|---|---|---|
| OncomiRs | |||
| miR-21** | PTEN, PDC4 | ↑Proliferation/invasion | 29–31 |
| miR-25 | E-cadherin | ↑metastasis | 32 |
| miR-1322 | ECRG2(SPINK7) | ↑proliferation | 17 |
| miR-223 | FBXW7 | ↑Poor prognosis | 37 |
| miR-31 | EMP1/KSR2/RGS4 | ↑Progression | 38 |
| miR-92a | E-cadherin | ↑Metasta/poor prog. | 33 |
| miR-296 | CyclinD1, BCL-2 | ↑Progression | 39 |
| miR-10b | KLF-4 | ↑Migration/invasion | 34 |
| miR-373 | LATS2 | ↑Proliferation | 35 |
| miR-196a | ANXA1 | ↑Proliferation | 19 |
| miR-129 | N/A | 40 | |
| miR-17-92 cluster | TNF-α | ↑Proliferation | 41 |
| miR-200c/miR-21 | PP2R1B | Cisplatin resistance | 42 |
| miR-200b/c/miR-429 | AP-2α | Cisplatin resistance | 43 |
| TSmiRs | |||
| miR-375** | PDK1, IGF1R | ↓growth/metas. | 23,36, 38 |
| miR-133a | CD47 | ↓Lymph node metas. | 39,40 |
| Let-7 | HMGA2 | ↓proliferation | 6,43 |
| miR-29c | Cyclin E | Cell cycle arrest | 47 |
| miR-34a | 44–46 | ||
| miR-223 | ARTN | ↓Migration/invasion | 49 |
| miR-205 | ZEB2/EMT | ↓Migration/invasion | 41,42 |
| miR-141 | YAP1 | ↓Chemo-resistance | 97 |
| miR-203 | NP63 | ↓proliferation | 57 |
| miR-148a | n/a | ↑Chemo-sensitivity | 58 |
| miR-210 | FGFRL1 | Cell cycle arrest | 48 |
| miR-205* | E-cadherin | ↓Inhibit EMT | 49 |
| miR-10a | Homebox gene | ↓Migration/invasion | 50 |
| miR-143/145 cluster | FSCN1 | ↓Lymph node metas | 59 |
| miR-34a/b/c/129-2 | 52 |
Reported consistently by more than four individual groups;
Reported by two groups
Oesophageal adenocarcinoma
OncomiRs
miRNA profiling and functional studies have been used to understand the mechanisms of progression of Barrett oesophagus to oesophageal adenocarcinoma. Several reports discuss the alterations in miRNAs in Barrett oesophagus and oesophageal adenocarcinoma and have identified targets and characterized their functional roles in the carcinogenesis of oesophageal adenocarcinoma. For example, miR-106b-25 polycistron is progressively upregulated in high-risk Barrett oesophagus to oesophageal adenocarcinoma; this upregulation is associated with genomic amplification and overexpression of MCM7 locus at chromosome 7q22.1.21 The upregulated polycistron targets and inhibits P21 and Bim proteins, which affect cell cycle and apoptosis, therefore contributing to oesophageal adenocarcinoma carcinogenesis.21 Two independent reports have described upregulation of miR-196a in a progressive manner from Barrett oesophagus to Barrett oesophagus with dysplasia and finally to oesophageal adenocarcinoma; miR-196a targets annexin A1 (ANXA1), small proline-rich protein 2C (SPRR2C) and S100A9.22,23 Expression of programmed cell death receptor 4 (PDCD4), a tumour suppressor gene, decreased progressively and significantly with progression of Barrett oesophagus to oesophageal adenocarcinoma, but miR-21 expression was upregulated in high-grade dysplasia with Barrett oesophagus and oesophageal adenocarcinoma, consistent with PDCD4 deregulation; functional studies have confirmed that miR-21 is a negative regulator of PDCD4 in vivo and play an oncogenic role in EAC.25 Therefore, miR-21, miR-196a, and miR-192, which are upregulated in oesophageal adenocarcinoma but not in normal samples, and have been termed as oncomiRs.27,28
tsmiRs
Two miRNA alterations in Barrett oesophagus and oesophageal adenocarcinoma has been identified that might mediate progression to oesophageal adenocarcinoma: miR-31 showed frequent downregulation only in HGD and EAC cases suggesting association with transition from BM to HGD, while miR-375, showed marked downregulation exclusively in EACs and in none of the BM or HGD lesions, suggesting its association with progression to invasive carcinoma. it is, therefore, proposed that miR-31 and miR-375 might be specifically associated with early and late-stage malignant progression, respectively.26 However, documenting the functional significance of such discoveries is difficult in this setting because of the lack of mature genetically engineered mouse models. Some of the work is, therefore, preliminary. For example, several studies have found alterations in miRNAs in Barrett oesophagus and oesophageal adenocarcinoma, but the target mRNAs have not been identified. miR-200 family members were downregulated in oesophageal adencarcinoma compared with Barrett oesophagus, and a significant inverse correlation was noted between miR-200 family expression and zinc finger E-box binding homeobox 1 (ZEB1) and ZEB2 expression in Barrett oesophagus and oesophageal adenocarcinoma.24 miR-203, miR-205, Let-7a/b/c, miR-345, miR-494, miR-193a and miR-375, are downregulated in oesophageal adenocarcinoma and could be considered tsmiRNAs.28–32 In-depth studies of these miRNAs will be important.
Osophageal squamous cell carcinoma
OncomiRs
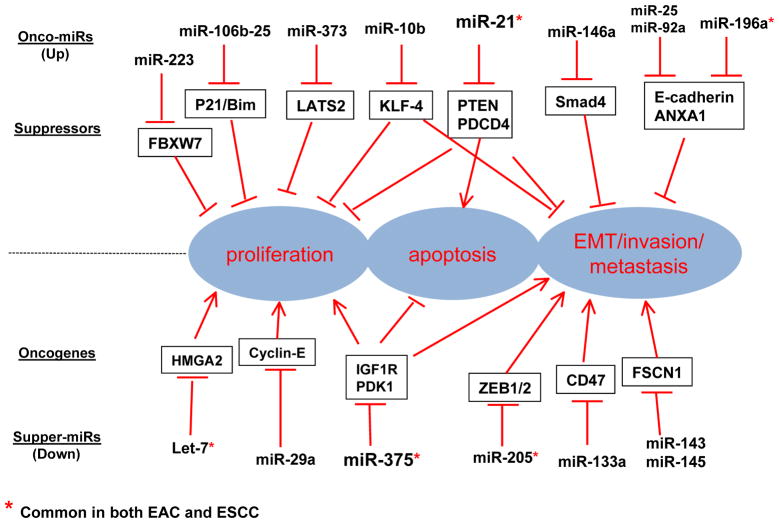
A large number of miRNAs and their targets have been identified that are upregulated in oesophageal squamous cell carcinoma. These oncomiRs target important tumour suppressors. miR-21 has been reported by several independent groups to be an important oncomiR in oesophageal squamous cell carcinoma.33–35 miR-21 targets phosphatase and tensin homolog (PTEN) and PDCD4 and seems to promote progression of oesophageal squamous cell carcinoma; miR-21 should be explored as a therapeutic target for preclinical or clinical trials for subset of patients with high expression levels of this miRNA. Two reports note an upregulation of miR-25 and miR-92a in oesophageal squamous cell carcinoma, both of which target CDH1. This gene encodes E-cadherin, which is an important protein for maintaining normal epithelial phenotype—its downregulation can lead to epithelial–mesenchymal transition (EMT) and metastasis.36,37 Both miRNAs promote migration and invasion through reduced expression of E-cadherin, and are associated with poor prognosis.36,37 Upregulation of miR-10b has been reported in 95% of oesophageal squamous cell carcinomas, along with reduced expression of KLF4 (Krüppel-like factor 4), an important tumour suppressor in the gastrointestinal tract.38 LATS2 (large tumour suppressor homolog 2) is a member of the LATS tumour suppressor family and is involved in Hippo signalling, which controls organ size and has a critical role in liver and gastrointestinal tract carcinogenesis. Frequent loss of heterozygosity of LATS has been reported in human oesophageal cancers, and miR-373 inhibits LATS2 in vitro and in vivo.39 Therefore, miR-373 might have an oncogenic role worthy of further pursuit to gain therapeutic advantage.39 Many more potential oncogenic miRNAs have been reported in oesophageal squamous cell carcinoma (Table 2, Figure 1).20,22,40–46
Figure 1.
miRNAs, Targets and functions in Esophageal Cancers
* Common in both EAC and ESCC
tsmiRs
Similarly, many downregulated miRNAs and their targets have been identified in oesophageal squamous cell carcinoma. Among these, miR-375 is the most promising tsmiR that is consistently downregulated either in tumour tissues or plasma from patients with oesophageal squamous cell carcinoma compared with healthy controls.27,47–49 Hypermethylation of miR-375 promoter is the reported mechanism of its downregulation.49 IGF1R and PDK1 are important components of IGF1 and the phosphatidylinositol 3-kinase (PI3K) pathways, which are frequently overexpressed in many malignancies and have a crucial role in promoting cell proliferation, survival and metastasis.47 miR-375 could interact with the 3′-untranslated region of IGF1R and PDK1 and downregulate their expression. Therefore, IGF1R and PDK1 are two major targets of miR-375.47,49 Another report suggests that miR-133a, a tumour suppressor, inhibits tumorigenesis and growth in vivo and inhibits CD47; overexpression of CD47 correlates with poor prognosis.50 miR-133a in concert with miR-145 and miR-133b target FSCN1 (actin-binding oncogenic protein, Fascin homolog1) and thereby inhibit cell proliferation and invasion in oesophageal sqaumous cell carcinoma cells.51 An in vitro study by Matsushima et al. demonstrated that the downregulation of miR-205 results in reduced cellular invasion and migration by targeting ZEB2; it also reduces EMT by decreasing E-cadherin expression.52,53 Let-7a and miR-34a, reportedly involved in cancer stem cell regulation,6 are downregulated in oesophageal squamous cell carcinoma.54–56 Other tsmiRs in oesophageal squamous cell carcinoma include: miR-29c, which targets cyclin E without affecting cyclin dependent kinase (CDK) CDK2 and CDK6;57 miR-210, which targets FGFRL1,58 and miR-223, which targets ARTN59 (Table 252,53,55,60).
miRNAs and gastric cancer
OncomiRs
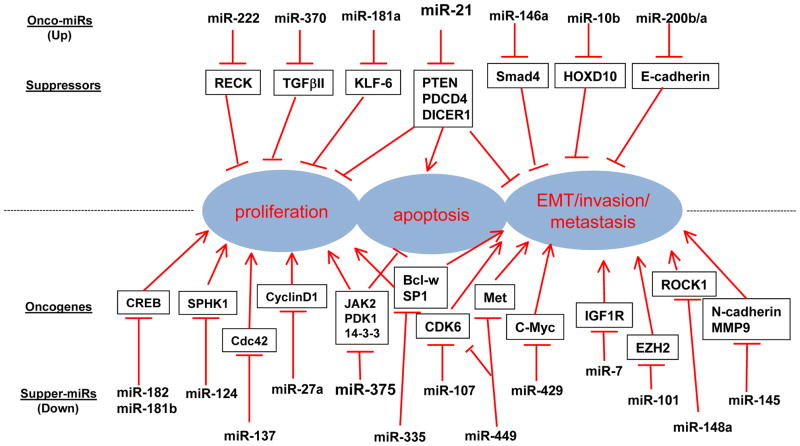
Many overexpressed oncomiRs affect apoptosis and proliferation in gastric cancer. Overexpression of miR-181a in a gastric cancer cell line led to increased cell proliferation and inhibition of apoptosis by repression of tumour suppressor KLF6.61 Several investigators have reported upregulation of miR-21 in gastric cancer (its targets, as mentioned above, are PDCD4 and PTEN).62–64 miR-196a/196b are markedly overexpressed in tumor tissues and serum of gastric cancer patients.65,66 Increased miR-146a in gastric cancer directly targets SMAD4; and ectopic expression of miR-146a could improve proliferationis and inhibit apoptosis of gastric cancer cells.67 Overexpression of miR-191 has been reported in the gastric cancer cell line MGC803 and in gastric cancer tissue; this miRNA regulates NDST1 (N-deacetylase/N-sulfotransferase 1).68 Overexpression of miR-370 in gastric cancer tissue has been observed, leading to downregulation of TGFBR2 (TGF-β type II receptor).69 miR-126 overexpression leads to inhibition of SOX2, which seems to contribute gastric cancer carcinogenesis.70 Thus it seems that many oncomiRs are worthy of pursuit and might become therapeutic targets in the future. More oncomiRNAs in gastric cancer are shown in Table 3 and Figure 2.59,71–76
Table 3.
Deregulated miRNAs, targets and potential functions in gastric cancer
| miRNA | Targets | Functions | References |
|---|---|---|---|
| OncomiRs | |||
| miR-21** | PTEN, PDCD4, DICE | ↑ Proliferation/invasion | 51–53,80 |
| miR-181a | R1 | ↑ prolif.↓apop. | 50 |
| KLF-6 | |||
| miR-196a* | n/a | ↑migraton/invasion/EMT | 54 |
| miR-93 | n/a | Poor prognosis | 70 |
| miR-196b | ETS2 | ↑EMT | 55,81 |
| miR-222 | RECK | ↑ Proliferation | 71 |
| miR-10b | HOXD10 | ↑ invasion/metastasis | 82 |
| miR-663 | P21 | 72 | |
| miR-146a | Smad4 | 56 | |
| miR-200b/a | E-cadherin | ↑EMT | 73 |
| miR-27 | n/a | ↑EMT, metastasis | 83 |
| miR-191 | NDST1 | 57 | |
| miR-370 | TGF-βII | 58 | |
| miR-223 | EPB41L3 | ↑invasion, metastasis | 56 |
| miR-126 | SOX2 | 59 | |
| miR-17-5P/20a | n/a | ↑ in plasma, marker | 74 |
| miR-107/196a/21/9 | DICER1 | ↑lymph node metastasis | 75 |
| TSmiRs | |||
| miR-7* | IGF1R | ↓metastasis | 84 |
| Let-7a* | RAB40C | ↓CSC | 70–72 |
| Let-7f | MYH9 | ↓invasion, metastasis | 89 |
| miR-181b | CREB | Prognostic marker | 60 |
| miR-101(microdeletion) | EZH2 | ↓EMT | 62 |
| miR-124 | SPHK1 | ↓proliferation | 63 |
| miR-145 | N-cadherin, MMP9 | ↓invasion, metastasis | 85 |
| miR-182 | CREB | ↓proliferation | 61 |
| miR-148a | ROCK1 | ↓invasion, metastasis | 88 |
| miR-29a | P42.3 | 91 | |
| miR-335 | Bcl-w/SP1 | ↓invasion, metastasis | 87 |
| miR-429 | C-MYC | ↓ metastasis | 65 |
| miR-296 | Scribble | 66 | |
| miR-146a | EGFR, IRAK1 | Prognostic marker | 92 |
| miR-27a | CyclinD1 | ↓proliferation | 74 |
| miR-10b | MAPRE1 | 93 | |
| miR-375 | Jak2, PDK1/14-3-3δ | 66,67 | |
| miR-622 | ING1 | ↓invasion/migration | 94 |
| miR-107 | CDK6 | ↓proliferation/invasion | 95 |
| miR-497 | BCL2 | ↑drug sensitivity | 96 |
| miR-137 down by methy | Cdc42 | ↓proliferation | 73 |
| miR-449 | MET, SIRT1, CDK | 74 | |
| miR-486 | 6 | 69 | |
| miR-495/551a | OLFM4 | ↓invasion/migration | 97 |
| miR-409-3P* | PRL-3 | ↓prolif./metastasis | 64,86 |
| miR-200b/c/429 let-7g/miR-34 |
PHF10, RDX BCL2/XIAP |
Drug resistance Chemo-sensitivity/↓CSC |
98 99 |
Reported consistently by more than four individual groups;
Reported by two groups
Figure 2.
miRNAs, Targets and functions in Gastric Cancers
tsmiRs
miR-181b and miR-182 were significantly downregulated in human gastric adenocarcinoma tissue samples compared to the adjacent normal gastric tissues. Functinally, overexpression of miR-181b suppressed the proliferation and colony formation rate of gastric cancer cells and decreased the expression of cAMP responsive element binding protein 1 (CREB1) by binding its 3′ untranslated region. Thereby, miR-181b and miR-182 may function as a tumor suppressor in gastric adenocarcinoma cells through negative regulation of CREB1.77,78 miR-101 was downregulated in gastric cancer owing to microdeletions at miR-101 genomic loci that subsequently led to EZH2 overexpression and CDH1 dysfunction, especially in intestinal-type gastric cancer.79 miR-124 inhibits gastric cancer cell proliferation by inducing cell cycle inhibitors P21 and P27 proteinsby targeting oncogenic SPHK1.80 The miR-409-3p cluster also inhibits proliferation by targeting the transcriptional regulator PHF10.81 MYC, an important modulator for cell growth and apoptosis, is downregulated by miR-429.82 miR-296 is progressively lost during tumour progression and leading to aberrant overexpression of SCRIB83 miR-375 is downregulated in gastric cancer and its targets are oncogenes JAK2, PDK1 and YWHAZ (14-3-3 zeta).84,85 miR-486, reported as a tsmiR in gastric cancer by several investigators, negatively regulates the antiapoptotic glycoprotein OLFM4.86 Let-7a, a tsmiRNA, has reduced expression in gastric cancer tissues and cell lines;87–89 its target is RAB40C. miR-137 is a negative regulator of CDC42 and is downregulated in gastric cancer as a result of hypermethylation.90 Quantitative PCR analyses have confirmed the loss of miR-449 in gastric cancer tissue compared with normal tissue, and miR-449 facilitates translation of p53, p21 as well as the apoptosis markers cleaved CASP3 and PARP by decreasing its oncogenic targets, MET, CCNE2, SIRT1 and CDK6.91 More details are shown in Table 3 and Figure 2.92–100
miRNAs and metastasis of UGICs
MetastamiRs in oesophageal cancer
Invasion and metastasis are critical for cancer progression.101 In oesophageal squamous cell carcinoma, several upregulated miRNAs (including miR-2536, miR-92a37 and miR-205.52,53) target CDH1 to promote EMT. In addition to regulating EMT, many metastamiRs regulate targets that promote invasion and metastasis of tumour cells (Figure 1). miR-10b, which is overexpressed in 95% of cancers, promotes migration and invasion through the tumour suppressor KLF4 in human esophageal cancer cell lines,38 while miR-10a controls cell migration and invasion by targeting homebox genes.53 miR21 is also involved in invasion and metastasis of oesophageal squamous cell carcinoma by targeting PDCD4 in vitro; furthermore, patients with oesophageal squamous cell carcinoma with lymph-node metastasis or venous invasion showed higher expression of miR-21 in comparison with patients without metastasis or venous invasion. Anti-miRNA-21 transfected cells showed marked reductions in cellular proliferation and invasion.35 miR-133a targets CD47 to inhibit tumorigenesis and metastasis in vivo.50 miR-375 inhibits invasion and metastasis by targeting IGFLR1 and PDK1.47,49 In addition, reduced expression of the miR-143/145 cluster (which regulates FSCN1) in oesophageal squamous cell carcinoma is associated with lymph-node metastases.60 miR-223 inhibits tumour migration and invasion by targeting ubiquitin ligase FBXW7 and ARTN in this cancer.40,59
A significant inverse correlation between miR-200 family expression and ZEB1 and ZEB2 expression in oesophageal adenocarcinoma has been reported.24 The expression of three miRNAs has been associated with lymph-node metastasis (miR-99b and miR-199a-3p, and miR-199a-5p).102
MetastamiRs in gastric cancer
miR-21 promotes invasion and lymph-node metastasis in gastric cancer.62,76 miR-196a promotes EMT.65 Overexpression of mir-196b has been reported to induce migration and invasion by inducing EMT, increasing expression of VIM (vimentin) and MMP2, and reducing CDH1 expression.103 miR-10b is markedly increased in lymph-node metastasis positive gastric cancer tissues compared with lymphoma node metastasis-free tumor tissues, and were correlated to dowregulation of HOXD10 expression. Functionally, miR-10 overexpression promotes invasion by stimulating RhoC and AKT by targeting HOXD10.104 Upregulation of miR-27 increased the expression of genes associated with EMT, including ZEB1, ZEB2, SLUG and VIM, while decreasing expression of CDH1.105
miR-7 has shown anti-metastatic properties by targeting IGF1R and SNAI1 and increasing expression of CDH1; hence this pathway could be a potential therapeutic target.106 miR-145 suppresses tumour metastasis by directly targeting CDH2 (N-cadherin) and MMP9 but not MMP2.107 miR-409-3p is frequently downregulated in gastric cancer and it reduces tumour cell migration and invasion in vitro and metastases in vivo.108 miR-409 targets the pro-metastatic gene RDX to suppress metastasis. miR-335 is downregulated in gastric cancer and it targets BCLW and SP1 to suppress tumour cell invasion and metastasis.109 ROCK1 promotes invasion and metastasis in many tumour types and miR-148a directly binds to ROCK1 3′ untranslated region and inhibits ROCK1 expression, therefore suppressing metastasis.110 miR-429 targets MYC to inhibit invasion.82 Overexpression of let-7f can inhibit invasion and migration by targeting MYH9 (a tumour metastasis associated gene).111
Therapeutic implications of miRNAs in UGICs
miRNAS as therapeutic targets
Synthetic miRNA mimics that have been developed for other cancer types include small interfering RNA (siRNA)-like oligoribonucleotide duplexes112 and chemically modified oligoribonucleotides.113 miRNAs can be inhibited in vitro and in vivo by various modified antisense oligonucleotides called antagomiRs; these molecules might, therefore, have a role in cancer treatment. 114 Intensive efforts have been made to develop miRNA-based therapeutics in preclinical models of breast cancer, pancreatic cancer and prostate cancer to modify oncogene or tumour suppressor functions.6,115,116 miRNA-based therapeutics are lagging behind for UGICs, but this is a promising area of research.
Modifying chemoresistance
In oesophageal squamous cell carcinoma, cisplatin induces expression of AP-2a, which confers chemosensitivity by promoting apoptosis; AP-2a is a target of the miR-200b/200c/miR-429 cluster, which negatively regulates AP-2a to induce cisplatin resistance.46 A similar study suggests that overexpression of miR-200c causes cisplatin resistance in oesophageal squamous cell carcinoma cells by upregulating the AKT pathway.45 miR-141 sensitizes oesophageal squamous cell carcinoma cells to cisplatin by targeting the 3′-untranslated region of YAP1, which is known to have a crucial role in apoptosis induced by DNA-damaging agents. 117 miR-148a upregulation in both oesophageal squamous cell carcinoma and oesophageal adenocarcinoma cells increases sensitivity to cisplatin and 5-fluorouracil.118,119 More detailed studies are needed to elucidate these mechanisms.
The miR-200b/200c/miR-429 cluster sensitized gastric cancer cells (by increasing apoptosis) to vincristine and cisplatin by targeting BCL-2 and XIAP.99 Overexpression of miR-497 sensitized SGC7901/VCR and A549/CDDP gastric cancer cells to vincristine (VCR), 5-fluorouracil (5-FU), cisplatin (CDDP) by targeting BCL-2 in vitro.97 In vivo experiments and subsequent clinical development is needed.
miRNAs as biomarkers in UGICs
Diagnostic markers
miRNAs are stably expressed in serum, plasma, urine, saliva and other bodily fluids,16 and this property can be exploited. However, little work has been done in this regard in UGICs. In oesophageal squamous cell carcinoma, miR-1322 levels are higher in serum and tumour tissues than in the controls, so this miRNA could serve as a biomarker.20 OncomiRNAs (miR-21/miR-184/miR-221) and one tsmiR (miR-375) have been studied in the plasma of 50 patients with oesophageal squamous cell carcinoma and 20 healthy volunteers; the plasma level of miR-21 tended to be higher in ESCC patients (P=0.0649), while that of miR-375 was significantly lower (P<0.0001) and the miR-21/miR-375 ratio was significantly higher (P<0.0001) in ESCC patients than in controls.48
In gastric cancer, three serum miRNAs (miRs-221/744/376c) could distinguish patients with gastric cancer from healthy controls with 82.4% sensitivity and 58.8% specificity.120 Moreover, miR-221 and miR-376c demonstrated significantly positive correlation with poor differentiation of GC.120 In a validation experiment, plasma levels of miR-451 and miR-486 were higher in patients with gastric cancer compared with healthy controls, with high area under the curve (AUC) values (0.96 and 0.92).121 A genome-wide microRNA profile identified high serum levels of miR-378 in patients with gastric cancer, and validation yielded a high receiver operating characteristic AUC (0.86).122 A quantitative real-time PCR analysis identified five serum miRNAs (miR-1, miR-20a, miR-27a, miR-34 and miR-423-5p) as biomarkers for gastric cancer, and levels correlated with tumour stage.123
Prognostic markers
Although prognostic biomarkers are of less value than predictive or early detection markers, here we describe the literature briefly. miR-21 seems to be a reliable poor prognosticator for oesophageal adenocarcinoma, oesophageal squamous cell carcinoma and gastric cancer.29,124,125 High levels of miR-92a, miR-99b and miR-199a in both oesophageal adenocarcinoma and oesophageal squamous cell carcinoma are also poor prognosticators.102 Reduced expression of miR-375, miR-203 and miR-205 is associated with late stages of oesophageal adenocarcinoma and oesophageal squamous cell carcinoma.26,30 High serum levels of miR-31 in patients with oesophageal squamous cell carcinoma portend poor prognosis.41 The serum ratio of miR-21:miR-375 correlates with disease recurrence.48
In gastric cancer, overexpression of miR-93 is associated with short survival.71 Overexpression of miR-10b, miR-107/196a and miR-223 correlates with metastasis status.76 High expression of circulating miR-17-5p/20a was an independent poor prognostic factor.75 Low-level expression of Let-7a/let-7g/let-7f are associated with poor prognosis.87,100 miR-181b and miR-182 are also novel poor prognosticators.77 Low expression levels of miR-125a-3p were found to be associated with enhanced malignant potential such as tumour size, lymph node and liver metastasis and poor prognosis, and this study suggests miR-125a-3p is a potent prognostic marker in gastric cancer.126 miR-409-3p was found to be downregulated frequently in patients with gastric cancer, and its expression was associated with distant metastasis.108 Collectively, many miRNAs demonstrate markedly different expression levels between patients with cancer and control groups, and might serve as prognostic markers for monitoring disease status. However, large case–control studies are needed to validate these individual miRNA markers as useful clinical tools.
miRNAs and cancer stem cells
Emerging evidence indicates that deregulation of miRNAs has an important role in regulating cancer stem cells of organs such as the breast,127,128 brain (glioma)129 and prostate.6 Research to establish the role miRNAs have in regulating cancer stem cells in UGICs is needed.
Conclusions
miRNAs are ubiquitous and plentiful, and are crucial post-transcriptional regulators of human gene expression. Research over the last ten year has identified numerous miRNAs that have diverse roles at multiple steps of tumour progression and metastasis. However, these discoveries have not been translated in to the clinics to help patients with UGICs. Considerably more research is needed, including the demonstration of functional consequences in genetically engineered mouse models. The clinical potential is enormous as miRNAs might provide tools for diagnosis, early detection, prediction and monitoring of therapy, and as therapeutic targets. Overall, this area of research has huge potential and should be actively pursued. Furthermore, discoveries made through the ENCODE (encyclopedia of DNA elements) project seem extremely relevant to miRNA research.130–133 The interim results of the ENCODE project suggest that only up to 3% of the genome codes for proteins and 76% of the genome codes for RNA elements (including miRNAs), which probably regulate the protein coding genes. Many other discoveries, which are outside the scope of this Review, demonstrate that we must focus on the non-protein-coding genome with the same vigour as we have focussed on the protein-coding genome.
Key points.
Worldwide, cancers of the oesophagus, gastro-oesophageal junction and stomach (UGICs) are common and outcomes of patients with UGICs have remained dismal
miRNAs are non-coding, single-stranded RNAs of ~22 nucleotides and consist of a novel class of gene regulators that negatively regulate their targets.
Many miRNA have been identified to act as oncogenes, tumor suppressors and important modulators in the process of invasion and metastasis of UGICs.
Increased activity of oncomiRNAs leads to inhibition of tumour suppressor genes, facilitating cell proliferation and tumour progression; while decreased activity of tumour-suppressor miRNAs (tsmiRs) leads to increased oncogene translation, contributing to tumour progression.
Certain miRNAs are involved in the regulation of metastasis (metastamiRs) as well as in modulation of chemoresistance in UGICs.
Circulating miRNAs provide potential biomarkers for ealier diagnosis and prognosis in patients of UGICs.
Further research could lead to exploitation of miRNAs as therapeutic or diagnostic targets for UCICs.
Review criteria.
A literature search was carried out in PubMed for papers published from 2002 to 2012. The search terms used were “miRNA”, “miRNA and Cancer”, “miRNA and esophageal cancer” miRNA and EAC” and “miRNA and ESCC” and “miRNA and GC”. miRNA and cancer progression, apoptosis, metastasis, chemoresistance, target and biomarker were also used as search terms. The search was restricted to English language papers.
Acknowledgments
This work was supported in part by the Dallas, Park, Smith, and Cantu family funds; the Kevin Fund; the Sultan Fund; the River Creek Foundation; and the Aaron and Martha Schecter Private Foundation. This work was also supported by the Multidisciplinary Research Program at The University of Texas MD Anderson Cancer Center and by the National Institutes of Health through MD Anderson Cancer Center Support Grant CA016672. Also supported by CA138671 (JAA)
Biographies
Shumei Song, M.D., Ph.D; Assistant Professor; Department of Gastrointestinal Medical Oncology, U. T. M.D. Anderson Cancer Center.
After obtaining her Ph.D at Biochemistry and Molecular Biology in Beijing University in 1999, she continued her five year’s postdoctal fellow training in cancer biology in U.T.M D Anderson Cancer Center. As a result of her academic growth and achievements, she was promoted to Assistant Professor in 2008. During this period, she have actively and productively pursued research of the molecular mechanisms of GI track tumors and have been very astute in identifying areas of GI cancer research which are novel and important especially in the area of esophageal adenocarcinoma.
Jaffer A. Ajani, M.D. is a Professor in the Departments of Gastrointestinal Oncology and Molecular Epidemiology at the University of Texas M. D. Anderson Cancer Center, Houston, TX, USA
Graduated from Government Medical College in Nagpur, India and completed training in Orthopedics (India), Family Medicine (USA), Internal Medicine (USA), and Medical Oncology (USA). He has spent more than 20 years on conducting trials of combined modality therapies for localized gastric and esophageal cancers. This has led to the development of first strategies of preoperative therapy for resectable upper gastrointestinal cancers. He has also conducted numerous phase II chemotherapy trials and 4 randomized trials over the years. In the past 5 years, he has established an infrastructure for translational work in gastric and esophageal cancer.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. Journal of the National Cancer Institute. 2008;100:1184–1187. doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Wu WK, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29:5761–5771. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 4.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu WKK, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29:5761–5771. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 6.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature medicine. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 11.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eiring AM, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Camarillo C, et al. MetastamiRs: Non-Coding MicroRNAs Driving Cancer Invasion and Metastasis. Int J Mol Sci. 2012;13:1347–1379. doi: 10.3390/ijms13021347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichikawa D, Komatsu S, Konishi H, Otsuji E. Circulating microRNA in digestive tract cancers. Gastroenterology. 2012;142:1074–1078. e1071. doi: 10.1053/j.gastro.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 18.Nana-Sinkam P, Croce CM. MicroRNAs in diagnosis and prognosis in cancer: what does the future hold? Pharmacogenomics. 2010;11:667–669. doi: 10.2217/pgs.10.57. [DOI] [PubMed] [Google Scholar]
- 19.Song JH, Meltzer SJ. MicroRNAs in Pathogenesis, Diagnosis, and Treatment of Gastroesophageal Cancers. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.05.003. S0016–5085(12)00683-X [pii] [DOI] [PubMed] [Google Scholar]
- 20.Zhang T, et al. MicroRNA-1322 regulates ECRG2 allele specifically and acts as a potential biomarker in patients with esophageal squamous cell carcinoma. Molecular carcinogenesis. 2012 doi: 10.1002/mc.21880. [DOI] [PubMed] [Google Scholar]
- 21.Kan T, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luthra R, et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 23.Maru DM, et al. MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–1948. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith CM, et al. miR-200 family expression is downregulated upon neoplastic progression of Barrett’s esophagus. World J Gastroenterol. 2011;17:1036–1044. doi: 10.3748/wjg.v17.i8.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fassan M, et al. PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch. 2011;458:413–419. doi: 10.1007/s00428-011-1046-5. [DOI] [PubMed] [Google Scholar]
- 26.Leidner RS, et al. The microRNAs, MiR-31 and MiR-375, as candidate markers in Barrett’s esophageal carcinogenesis. Genes, chromosomes & cancer. 2012;51:473–479. doi: 10.1002/gcc.21934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathe EA, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fassan M, et al. MicroRNA expression profiling in human Barrett’s carcinogenesis. Int J Cancer. 2011;129:1661–1670. doi: 10.1002/ijc.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feber A, et al. MicroRNA expression profiles of esophageal cancer. The Journal of thoracic and cardiovascular surgery. 2008;135:255–260. doi: 10.1016/j.jtcvs.2007.08.055. discussion 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luzna P, et al. Changes of microRNAs-192, 196a and 203 correlate with Barrett’s esophagus diagnosis and its progression compared to normal healthy individuals. Diagn Pathol. 2011;6:114. doi: 10.1186/1746-1596-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wijnhoven BP, et al. MicroRNA profiling of Barrett’s oesophagus and oesophageal adenocarcinoma. The British journal of surgery. 2010;97:853–861. doi: 10.1002/bjs.7000. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, et al. MicroRNA expression signatures in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:5744–5752. doi: 10.1158/1078-0432.CCR-09-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akagi I, et al. Relationship between altered expression levels of MIR21, MIR143, MIR145, and MIR205 and clinicopathologic features of esophageal squamous cell carcinoma. Dis Esophagus. 2011;24:523–530. doi: 10.1111/j.1442-2050.2011.01177.x. [DOI] [PubMed] [Google Scholar]
- 34.Ma WJ, et al. Role of microRNA-21 and effect on PTEN in Kazakh’s esophageal squamous cell carcinoma. Mol Biol Rep. 2011;38:3253–3260. doi: 10.1007/s11033-010-0480-9. [DOI] [PubMed] [Google Scholar]
- 35.Hiyoshi Y, et al. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15:1915–1922. doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- 36.Xu X, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochemical and biophysical research communications. 2012;421:640–645. doi: 10.1016/j.bbrc.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 37.Chen ZL, et al. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. The Journal of biological chemistry. 2011;286:10725–10734. doi: 10.1074/jbc.M110.165654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Y, et al. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. The Journal of biological chemistry. 2010;285:7986–7994. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee KH, et al. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315:2529–2538. doi: 10.1016/j.yexcr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Kurashige J, et al. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. British journal of cancer. 2012;106:182–188. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang T, et al. The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond) 2011;121:437–447. doi: 10.1042/CS20110207. [DOI] [PubMed] [Google Scholar]
- 42.Hong L, et al. The prognostic and chemotherapeutic value of miR-296 in esophageal squamous cell carcinoma. Annals of surgery. 2010;251:1056–1063. doi: 10.1097/SLA.0b013e3181dd4ea9. [DOI] [PubMed] [Google Scholar]
- 43.Ogawa R, et al. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Medical molecular morphology. 2009;42:102–109. doi: 10.1007/s00795-009-0443-1. [DOI] [PubMed] [Google Scholar]
- 44.Liu M, et al. TNF-alpha is a novel target of miR-19a. International journal of oncology. 2011;38:1013–1022. doi: 10.3892/ijo.2011.924. [DOI] [PubMed] [Google Scholar]
- 45.Hamano R, et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res. 2011;17:3029–3038. doi: 10.1158/1078-0432.CCR-10-2532. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, et al. A miR-200b/200c/429-binding site polymorphism in the 3′ untranslated region of the AP-2alpha gene is associated with cisplatin resistance. PLoS One. 2011;6:e29043. doi: 10.1371/journal.pone.0029043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong KL, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 48.Komatsu S, et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. British journal of cancer. 2011;105:104–111. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X, Lin R, Li J. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Digestive diseases and sciences. 2011;56:2849–2856. doi: 10.1007/s10620-011-1711-1. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki S, et al. CD47 expression regulated by the miR-133a tumor suppressor is a novel prognostic marker in esophageal squamous cell carcinoma. Oncology reports. 2012;28:465–472. doi: 10.3892/or.2012.1831. [DOI] [PubMed] [Google Scholar]
- 51.Kano M, et al. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. International journal of cancer. 2010;127:2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 52.Matsushima K, et al. MiRNA-205 modulates cellular invasion and migration via regulating zinc finger E-box binding homeobox 2 expression in esophageal squamous cell carcinoma cells. J Transl Med. 2011;9:30. doi: 10.1186/1479-5876-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsushima K, Isomoto H, Kohno S, Nakao K. MicroRNAs and esophageal squamous cell carcinoma. Digestion. 2010;82:138–144. doi: 10.1159/000310918. [DOI] [PubMed] [Google Scholar]
- 54.Li J, et al. Transcriptional activation of microRNA-34a by NF-kappa B in human esophageal cancer cells. BMC Mol Biol. 2012;13:4. doi: 10.1186/1471-2199-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, et al. CpG island methylation status of miRNAs in esophageal squamous cell carcinoma. International journal of cancer. 2012;130:1607–1613. doi: 10.1002/ijc.26171. [DOI] [PubMed] [Google Scholar]
- 56.Liu Q, et al. Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma. Mol Biol Rep. 2012;39:1239–1246. doi: 10.1007/s11033-011-0854-7. [DOI] [PubMed] [Google Scholar]
- 57.Ding DP, et al. miR-29c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis. 2011;32:1025–1032. doi: 10.1093/carcin/bgr078. [DOI] [PubMed] [Google Scholar]
- 58.Tsuchiya S, et al. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) The Journal of biological chemistry. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li S, et al. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci. 2011;18:24. doi: 10.1186/1423-0127-18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu R, et al. The cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog 1. PLoS One. 2012;7:e33987. doi: 10.1371/journal.pone.0033987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang X, et al. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumour Biol. 2012 doi: 10.1007/s13277-012-0414-3. [DOI] [PubMed] [Google Scholar]
- 62.Zhang BG, et al. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncology reports. 2012;27:1019–1026. doi: 10.3892/or.2012.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao Z, Yoon JH, Nam SW, Lee JY, Park WS. PDCD4 expression inversely correlated with miR-21 levels in gastric cancers. Journal of cancer research and clinical oncology. 2012;138:611–619. doi: 10.1007/s00432-011-1140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamanaka S, et al. MicroRNA-21 inhibits Serpini1, a gene with novel tumour suppressive effects in gastric cancer. Dig Liver Dis. 2012;44:589–596. doi: 10.1016/j.dld.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsai KW, et al. Aberrant expression of miR-196a in gastric cancers and correlation with recurrence. Genes, chromosomes & cancer. 2012;51:394–401. doi: 10.1002/gcc.21924. [DOI] [PubMed] [Google Scholar]
- 66.Tsai KW, et al. Epigenetic regulation of miR-196b expression in gastric cancer. Genes, chromosomes & cancer. 2010;49:969–980. doi: 10.1002/gcc.20804. [DOI] [PubMed] [Google Scholar]
- 67.Xiao B, et al. Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis. Oncology reports. 2012;27:559–566. doi: 10.3892/or.2011.1514. [DOI] [PubMed] [Google Scholar]
- 68.Shi X, Su S, Long J, Mei B, Chen Y. MicroRNA-191 targets N-deacetylase/N-sulfotransferase 1 and promotes cell growth in human gastric carcinoma cell line MGC803. Acta Biochim Biophys Sin (Shanghai) 2011;43:849–856. doi: 10.1093/abbs/gmr084. [DOI] [PubMed] [Google Scholar]
- 69.Lo SS, et al. Overexpression of miR-370 and downregulation of its novel target TGFbeta-RII contribute to the progression of gastric carcinoma. Oncogene. 2012;31:226–237. doi: 10.1038/onc.2011.226. [DOI] [PubMed] [Google Scholar]
- 70.Otsubo T, et al. MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis. PLoS One. 2011;6:e16617. doi: 10.1371/journal.pone.0016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen L, Jiang M, Yuan W, Tang H. Prognostic value of miR-93 overexpression in resectable gastric adenocarcinomas. Acta gastro-enterologica Belgica. 2012;75:22–27. [PubMed] [Google Scholar]
- 72.Li N, et al. Increased miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and targeting RECK. FEBS letters. 2012;586:722–728. doi: 10.1016/j.febslet.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 73.Yi C, et al. MiR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene. 2012 doi: 10.1038/onc.2011.629. onc2011629 [pii] [DOI] [PubMed] [Google Scholar]
- 74.Ahn SM, et al. Smad3 regulates E-cadherin via miRNA-200 pathway. Oncogene. 2011 doi: 10.1038/onc.2011.484. onc2011484 [pii] [DOI] [PubMed] [Google Scholar]
- 75.Wang M, et al. Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol Med Report. 2012;5:1514–1520. doi: 10.3892/mmr.2012.828. [DOI] [PubMed] [Google Scholar]
- 76.Inoue T, Iinuma H, Ogawa E, Inaba T, Fukushima R. Clinicopathological and prognostic significance of microRNA-107 and its relationship to DICER1 mRNA expression in gastric cancer. Oncology reports. 2012;27:1759–1764. doi: 10.3892/or.2012.1709. [DOI] [PubMed] [Google Scholar]
- 77.Chen L, et al. MicroRNA-181b targets cAMP responsive element binding protein 1 in gastric adenocarcinomas. IUBMB Life. 2012;64:628–635. doi: 10.1002/iub.1030. [DOI] [PubMed] [Google Scholar]
- 78.Kong WQ, et al. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J. 2012;279:1252–1260. doi: 10.1111/j.1742-4658.2012.08519.x. [DOI] [PubMed] [Google Scholar]
- 79.Carvalho J, et al. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. The Journal of pathology. 2012 doi: 10.1002/path.4032. [DOI] [PubMed] [Google Scholar]
- 80.Xia J, et al. miR-124 inhibits cell proliferation in gastric cancer through down-regulation of SPHK1. The Journal of pathology. 2012 doi: 10.1002/path.4030. [DOI] [PubMed] [Google Scholar]
- 81.Li C, et al. MicroRNA-409-3p regulates cell proliferation and apoptosis by targeting PHF10 in gastric cancer. Cancer letters. 2012;320:189–197. doi: 10.1016/j.canlet.2012.02.030. [DOI] [PubMed] [Google Scholar]
- 82.Sun T, Wang C, Xing J, Wu D. miR-429 modulates the expression of c-myc in human gastric carcinoma cells. Eur J Cancer. 2011;47:2552–2559. doi: 10.1016/j.ejca.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 83.Vaira V, et al. miR-296 regulation of a cell polarity-cell plasticity module controls tumor progression. Oncogene. 2012;31:27–38. doi: 10.1038/onc.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding L, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 85.Tsukamoto Y, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer research. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 86.Oh HK, et al. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res. 2011;17:2657–2667. doi: 10.1158/1078-0432.CCR-10-3152. [DOI] [PubMed] [Google Scholar]
- 87.Yang Q, et al. Low-level expression of let-7a in gastric cancer and its involvement in tumorigenesis by targeting RAB40C. Carcinogenesis. 2011;32:713–722. doi: 10.1093/carcin/bgr035. [DOI] [PubMed] [Google Scholar]
- 88.Golestaneh AF, et al. miRNAs expressed differently in cancer stem cells and cancer cells of human gastric cancer cell line MKN-45. Cell Biochem Funct. 2012 doi: 10.1002/cbf.2815. [DOI] [PubMed] [Google Scholar]
- 89.Zhu Y, Zhong Z, Liu Z. Lentiviral vector-mediated upregulation of let-7a inhibits gastric carcinoma cell growth in vitro and in vivo. Scandinavian journal of gastroenterology. 2011;46:53–59. doi: 10.3109/00365521.2010.510566. [DOI] [PubMed] [Google Scholar]
- 90.Chen Q, et al. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Digestive diseases and sciences. 2011;56:2009–2016. doi: 10.1007/s10620-010-1536-3. [DOI] [PubMed] [Google Scholar]
- 91.Bou Kheir T, et al. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cui Y, et al. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. PLoS One. 2011;6:e25872. doi: 10.1371/journal.pone.0025872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kogo R, Mimori K, Tanaka F, Komune S, Mori M. Clinical significance of miR-146a in gastric cancer cases. Clin Cancer Res. 2011;17:4277–4284. doi: 10.1158/1078-0432.CCR-10-2866. [DOI] [PubMed] [Google Scholar]
- 94.Kim K, et al. Epigenetic regulation of microRNA-10b and targeting of oncogenic MAPRE1 in gastric cancer. Epigenetics. 2011;6:740–751. doi: 10.4161/epi.6.6.15874. [DOI] [PubMed] [Google Scholar]
- 95.Guo XB, et al. Down-regulation of miR-622 in gastric cancer promotes cellular invasion and tumor metastasis by targeting ING1 gene. World J Gastroenterol. 2011;17:1895–1902. doi: 10.3748/wjg.v17.i14.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feng L, Xie Y, Zhang H, Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol. 2012;29:856–863. doi: 10.1007/s12032-011-9823-1. [DOI] [PubMed] [Google Scholar]
- 97.Zhu W, et al. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2012;29:384–391. doi: 10.1007/s12032-010-9797-4. [DOI] [PubMed] [Google Scholar]
- 98.Li Z, et al. miR-495 and miR-551a inhibit the migration and invasion of human gastric cancer cells by directly interacting with PRL-3. Cancer letters. 2012;323:41–47. doi: 10.1016/j.canlet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 99.Zhu W, et al. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer chemotherapy and pharmacology. 2012;69:723–731. doi: 10.1007/s00280-011-1752-3. [DOI] [PubMed] [Google Scholar]
- 100.Kim CH, et al. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC medical genomics. 2011;4:79. doi: 10.1186/1755-8794-4-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Geiger TR, Peeper DS. Metastasis mechanisms. Biochimica et biophysica acta. 2009;1796:293–308. doi: 10.1016/j.bbcan.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 102.Feber A, et al. MicroRNA prognostic signature for nodal metastases and survival in esophageal adenocarcinoma. The Annals of thoracic surgery. 2011;91:1523–1530. doi: 10.1016/j.athoracsur.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liao YL, et al. Transcriptional regulation of miR-196b by ETS2 in gastric cancer cells. Carcinogenesis. 2012;33:760–769. doi: 10.1093/carcin/bgs023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Z, Zhu J, Cao H, Ren H, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. International journal of oncology. 2012;40:1553–1560. doi: 10.3892/ijo.2012.1342. [DOI] [PubMed] [Google Scholar]
- 105.Zhang Z, Liu S, Shi R, Zhao G. miR-27 promotes human gastric cancer cell metastasis by inducing epithelial-to-mesenchymal transition. Cancer Genet. 2011;204:486–491. doi: 10.1016/j.cancergen.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 106.Zhao X, et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. 2012 doi: 10.1038/onc.2012.156onc2012156. [pii] [DOI] [PubMed] [Google Scholar]
- 107.Gao P, et al. The molecular mechanism of microRNA-145 to suppress invasion-metastasis cascade in gastric cancer. Oncogene. 2012 doi: 10.1038/onc.2012.61onc201261. [pii] [DOI] [PubMed] [Google Scholar]
- 108.Zheng B, et al. MicroRNA-409 suppresses tumour cell invasion and metastasis by directly targeting radixin in gastric cancers. Oncogene. 2011 doi: 10.1038/onc.2011.581onc2011581. [pii] [DOI] [PubMed] [Google Scholar]
- 109.Xu Y, et al. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng B, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–7583. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 111.Liang S, et al. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS One. 2011;6:e18409. doi: 10.1371/journal.pone.0018409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 113.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Molecular and cellular biology. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 115.Pramanik D, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Molecular cancer therapeutics. 2011;10:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nature biotechnology. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seki N. A commentary on MicroRNA-141 confers resistance to cisplatin-induced apoptosis by targeting YAP1 in human esophageal squamous cell carcinoma. J Hum Genet. 2011;56:339–340. doi: 10.1038/jhg.2011.26. [DOI] [PubMed] [Google Scholar]
- 118.Hummel R, et al. Chemotherapy-induced modification of microRNA expression in esophageal cancer. Oncology reports. 2011;26:1011–1017. doi: 10.3892/or.2011.1381. [DOI] [PubMed] [Google Scholar]
- 119.Hummel R, et al. Mir-148a improves response to chemotherapy in sensitive and resistant oesophageal adenocarcinoma and squamous cell carcinoma cells. J Gastrointest Surg. 2011;15:429–438. doi: 10.1007/s11605-011-1418-9. [DOI] [PubMed] [Google Scholar]
- 120.Song MY, et al. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One. 2012;7:e33608. doi: 10.1371/journal.pone.0033608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Konishi H, et al. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. British journal of cancer. 2012;106:740–747. doi: 10.1038/bjc.2011.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu H, et al. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer letters. 2012;316:196–203. doi: 10.1016/j.canlet.2011.10.034. [DOI] [PubMed] [Google Scholar]
- 123.Liu R, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 124.Mori Y, et al. MicroRNA-21 induces cell proliferation and invasion in esophageal squamous cell carcinoma. Mol Med Report. 2009;2:235–239. doi: 10.3892/mmr_00000089. [DOI] [PubMed] [Google Scholar]
- 125.Zheng Y, et al. MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients. Cancer Biomark. 2011;10:71–77. doi: 10.3233/CBM-2011-0231. [DOI] [PubMed] [Google Scholar]
- 126.Hashiguchi Y, et al. Down-regulation of miR-125a-3p in human gastric cancer and its clinicopathological significance. International journal of oncology. 2012;40:1477–1482. doi: 10.3892/ijo.2012.1363. [DOI] [PubMed] [Google Scholar]
- 127.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 128.Yu F, et al. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- 129.Godlewski J, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer research. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 130.Ecker JR, et al. Genomics: ENCODE explained. Nature. 2012;489:52–55. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- 131.Birney E. The making of ENCODE: Lessons for big-data projects. Nature. 2012;489:49–51. doi: 10.1038/489049a. [DOI] [PubMed] [Google Scholar]
- 132.Schadt E, Chang R. Genetics. A GPS for navigating DNA. Science. 2012;337:1179–1180. doi: 10.1126/science.1227739. [DOI] [PubMed] [Google Scholar]
- 133.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]