SUMMARY
Replication Protein A (RPA) is an essential eukaryotic single-stranded DNA binding protein with a central role in DNA metabolism. RPA directly participates in DNA double-strand break repair by stimulating 5’-3’ end resection by the Sgs1/BLM helicase and Dna2 endonuclease in vitro. Here we investigated the role of RPA in end resection in vivo, using a heat-inducible degron system that allows rapid conditional depletion of RPA in Saccharomyces cerevisiae. We found that RPA depletion eliminated both the Sgs1-Dna2 and Exo1-dependent extensive resection pathways, and synergized with mre11Δ to prevent end resection. The short single-stranded DNA tails formed in the absence of RPA were unstable due to 3’ strand loss and the formation of fold-back hairpin structures that required resection initiation and Pol32-dependent DNA synthesis. Thus, RPA is required to generate ssDNA, and also to protect ssDNA from degradation and inappropriate annealing that could lead to genome rearrangements.
INTRODUCTION
Accidental double-strand breaks (DSBs) are highly cytotoxic lesions that must be accurately repaired to maintain genome integrity. There are two main mechanisms to repair DSBs: nonhomologous end joining (NHEJ) and homologous recombination (HR) (Krogh and Symington, 2004). HR relies on the presence of an intact homologous duplex, usually the sister chromatid, to serve as a template for repair of the broken chromosome and is generally considered to be an error-free mechanism. In contrast, the direct ligation of DNA ends by NHEJ can lead to gain or loss of nucleotides at the junction. The association of several human cancer predisposition syndromes with defects in the recognition and repair of DSBs underscores the importance of DSB repair mechanisms (Chu and Hickson, 2009; Roy et al., 2012; Stracker and Petrini, 2011).
HR initiates by nucleolytic degradation of the 5’ strands of DSBs to yield 3’ single-stranded DNA (ssDNA) tails that function to prime DNA synthesis after Rad51-catalyzed strand invasion of an intact homologous duplex (Krogh and Symington, 2004). RPA is initially bound to the ssDNA generated by resection and is frequently used as a cytological marker for DNA end processing in vivo (Barlow et al., 2008; Gasior et al., 1998; Raderschall et al., 1999). Furthermore, RPA-bound ssDNA recruits the Mec1-Ddc2 complex (ATR-ATRIP in human) for activation of the DNA damage checkpoint in response to DSBs (Zou and Elledge, 2003). Genetic studies in S. cerevisiae have shown that the Mre11-Rad50-Xrs2 (MRX) complex and Sae2 can initiate end resection by removal of oligonucleotides from the 5’ ends to form short 3’ ssDNA tails, or act indirectly through recruitment of the Exo1 5’-3’ exonuclease, Sgs1 helicase and Dna2 endonuclease (Mimitou and Symington, 2008, 2010; Shim et al., 2010; Zhu et al., 2008). Extensive resection to generate long 3’ ssDNA tails requires Exo1 or Sgs1 in collaboration with Dna2 (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008).
Reconstitution of the Sgs1/BLM-Dna2 and WRN-Dna2 resection mechanisms in vitro revealed an essential role for replication protein A (RPA) to promote duplex DNA unwinding by Sgs1/BLM or WRN, and to enforce the 5’-3’ resection polarity of Dna2 (Cejka et al., 2010; Nimonkar et al., 2011; Niu et al., 2010; Yan et al., 2011). Dna2 can degrade both 5’ and 3’ flaps in vitro, but in the presence of RPA the 5’ endonuclease activity of Dna2 is stimulated and the 3’ endonuclease activity is attenuated (Bae and Seo, 2000; Cejka et al., 2010; Masuda-Sasa et al., 2008; Nimonkar et al., 2011; Niu et al., 2010; Yan et al., 2011). In vivo, DNA end resection is predominantly 5’-3’ although some 3’ strand loss has been reported (White and Haber, 1990; Zierhut and Diffley, 2008). The role of RPA in Exo1-mediated resection remains controversial. In one study with S. cerevisiae proteins, RPA was shown to be dispensable for Exo1 resection (Nicolette et al., 2010), but a study using the human proteins reported a stimulatory effect of RPA on Exo1 nuclease activity (Nimonkar et al., 2011).
In addition to the role in end resection, RPA is required for Rad51-mediated strand exchange in vitro. It has been suggested that RPA removes secondary structures from ssDNA to allow assembly of a contiguous Rad51-ssDNA nucleoprotein filament, and it stimulates strand exchange by sequestering protein-free ssDNA that can inhibit the Rad51 pairing reaction (Eggler et al., 2002; Sugiyama et al., 1997; Van Komen et al., 2002). However, RPA impedes Rad51 binding if added to ssDNA prior to Rad51 and the Rad52 mediator protein is required to overcome the RPA inhibitory effect (New et al., 1998; Shinohara and Ogawa, 1998; Sung, 1997). Rad52 binds directly to RPA and Rad51, and is thought to facilitate nucleation of Rad51 on RPA-coated ssDNA (Seong et al., 2008; Song and Sung, 2000; Sugiyama and Kowalczykowski, 2002). In addition, Rad52 stimulates ssDNA annealing and is able to overcome the inhibitory effect of RPA on this process (Mortensen et al., 1996; Sugiyama et al., 1998).
Because the genes encoding each subunit of the heterotrimeric RPA complex (RFA1, RFA2 and RFA3) are essential in S. cerevisiae (Brill and Stillman, 1991), investigating the biological relevance of RPA functions in HR has relied on the use of hypomorphic alleles. The rfa1-44 allele was identified in a screen for mutants with reduced frequencies of DSB-induced recombination, and shown to confer sensitivity to ultraviolet and ionizing radiation (Firmenich et al., 1995). Genetic screens for conditional mutations of RFA1 recovered several alleles (rfa1-M2, rfa1-t11 and rfa1-t48) that impart sensitivity to DNA damaging agents, and reduce mitotic and meiotic recombination (Longhese et al., 1994; Soustelle et al., 2002; Umezu et al., 1998). The rfa1-D228Y mutation was shown to suppress the requirement for Rad52 in DSB-induced single-strand annealing between direct repeats, suggesting that Rad52-catalyzed annealing of ssDNA is no longer needed when RPA is defective (Smith and Rothstein, 1999). Of the hypomorphic rfa1 mutants tested (rfa1-t11, rfa1-t48 and rfa1-D228Y), none exhibit defects in end resection suggesting they are compromised at a later step of HR (Lee et al., 1998; Smith and Rothstein, 1999; Soustelle et al., 2002). Therefore, to investigate the role of RPA in end resection we chose to conditionally ablate the Rfa1 subunit of the complex in vivo.
RESULTS
RPA is required for extensive resection
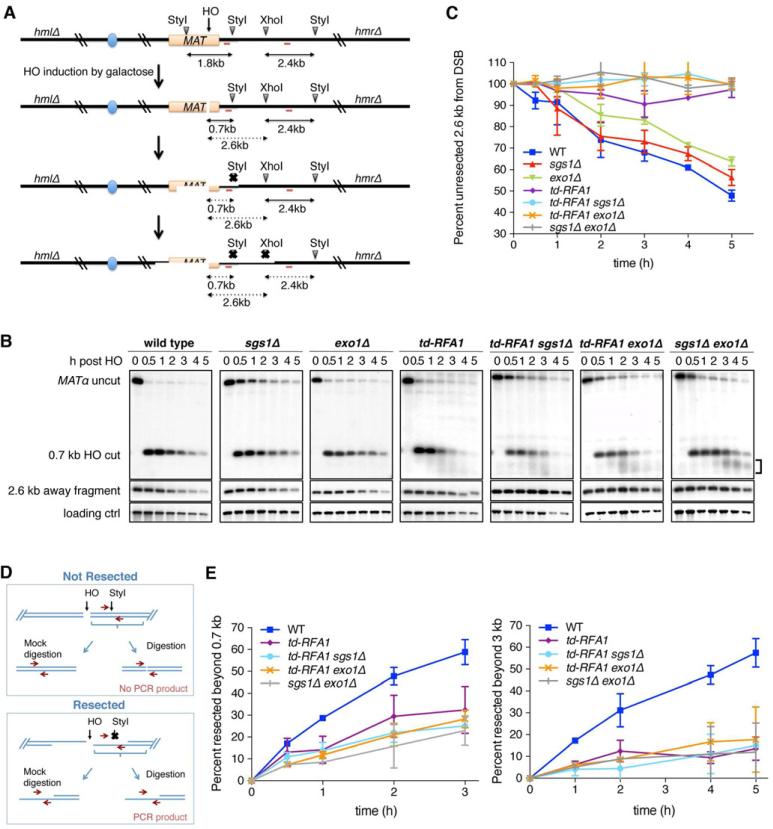
To circumvent the lethality of RPA knockout strains, we employed a heat-inducible degron system that renders the Rfa1 protein temperature degradable (td) (Figure S1) (Lisby et al., 2004; Sanchez-Diaz et al., 2004; Zou and Elledge, 2003). The promoter region of the endogenous RFA1 locus was replaced by a CUP1 driven degron cassette and the C-terminus of RFA1 was tagged with YFP (Lisby et al., 2004). UBR1 was expressed under the GAL1-10 promoter to facilitate proteolysis of the degron-fused Rfa1. The efficiency of RPA depletion at 37°C was verified by cell growth and western blot a nalysis (Figure S1). We analyzed resection of an HO endonuclease-induced DSB at the MAT locus in strains with an integrated PGAL1-HO fusion and deletions of the HML and HMR loci to prevent repair of the DSB by gene conversion (Zierhut and Diffley, 2008). Resection beyond the StyI and XhoI sites located 0.7 and 2.6 kb centromere distal to the DSB, respectively, was monitored after HO induction (Figure 1A). As resection proceeds, restriction enzyme sites in proximity to the DSB become single stranded and are resistant to digestion, which results in the disappearance of the corresponding restriction fragments over time (Zhu et al., 2008). Thus, the rate of band disappearance corresponds to the rate of resection. Resection assays were performed at 37°C, the restrictive temperature for the td-RFA1 strains. The td-Rfa1-YFP fusion protein functions as wild type at the permissive temperature, indicating that the N-terminal degron and C-terminal YFP fusions do not interfere with RPA function (Figure S1).
Figure 1. RPA is essential for both extensive resection pathways.
A. Schematic representation of the band disappearance assay for DSB end resection. Red bars show the location of the two probes used for hybridization. B. Southern blot analysis of the StyI-XhoI digested genomic DNA from indicated strains taken at different time points after HO induction, showing resection 0.7 kb and 2.6 kb from the DSB. The bracket indicates the smeared products observed in the td-RFA1 derivatives and the sgs1Ä exo1Ä mutant. C. Quantification of the 2.4 kb fragment located 2.6 kb from the DSB by the ratio at each time point to 0 h. The mean values from 3 independent trials are plotted and error bars show SD. D. Schematic representation of the quantitative PCR method to monitor HO-induced DSB end resection. E. Plot showing the ratio of resected DNA amongst HO cut DNA at each time point by qPCR analysis. The mean values from 3 independent trials are plotted and error bars show SD. See also Figures S1 and 2
To determine the role of RPA in the two extensive-resection pathways, RPA depletion was conducted in the sgs1Δ or exo1Δ background, where the alternative pathway is non functional. Similar to the sgs1Δ exo1Δ double mutant, both td-RFA1 sgs1Δ and td-RFA1 exo1Δ strains exhibited resection defects: smeared products were apparent below the HO cut band (Figure 1B), and the restriction fragment 2.6 kb distal to the HO cut site remained stable for up to 5 h after HO induction (Figure 1B, C). Consistent with Dna2 acting together with Sgs1 during extensive resection, depletion of RPA in the dna2Δ background caused a similar resection phenotype to that seen in the td-RFA1 sgs1Δ mutant (Figure S1). The dna2Δ strains used carry the pif1-m2 allele to suppress the lethality caused by loss of Dna2 (Budd et al., 2006). To better quantify resected DNA we employed a qPCR-based method (Zierhut and Diffley, 2008) (Figure 1D). Primers were designed flanking restriction enzyme sites 0.7 and 3.0 kb from the HO cut site and the PCR products from digested and mock treated genomic DNA were compared. Again, the td-RFA1 sgs1Δ and td-RFA1 exo1Δ cells showed reduced resection at both sites, similar to the sgs1Δ exo1Δ mutant (Figure 1E). These data suggest that RPA is essential for resection by both Sgs1-Dna2 and Exo1-dependent pathways. Consistent with this view, extensive resection in the td-RFA1 single mutant was similar to the sgs1Δ exo1Δ mutant (Figure 1B, C, E).
Cell cycle progression and checkpoint activation in the absence of RPA
Because end resection is low in G1 cells and is activated in S/G2, the reduced resection observed in the td-RFA1 strains could be due to cell cycle delays as a result of the replication defect (Aylon et al., 2004; Ira et al., 2004). To address this, resection of wild type and td-RFA1 strains was monitored at 37°C in G2/M arrested cell s. Although wild-type cells displayed less efficient resection under this condition compared to cycling cells, the smearing below the HO cut band and persistence of the restriction fragment 2.6 kb distal to the HO cut site were still seen in the td-RFA1 cells (Figure S2A). Furthermore, the FACS profiles of cycling cells used for resection assays showed most cells accumulated with a 2C DNA content following RPA depletion (Figure S2B). Thus, the resection defect in the absence of RPA is not due to cell cycle defects.
A previous study showed that RPA bound to ssDNA is required for recruitment of Mec1-Ddc2/ATR-ATRIP to damage sites, and to activate the Mec1/ATR kinase (Zou and Elledge, 2003). In budding yeast, Rad53/Chk2 is the effector kinase for both Mec1 and Tel1 in response to DSBs (Harrison and Haber, 2006). The G2/M arrest of RPA depleted cells following HO induction indicates that the DNA damage checkpoint is intact and Rad53 phosphorylation increased in the td-RFA1 strain following DSB formation (Figure S2C). Rad53 phosphorylation was apparent prior to HO induction suggesting activation of the replication checkpoint before RPA is completely depleted from cells. Phosphorylation of Rad53 was abolished in the td-RFA1 mec1Δ mutant indicating that low levels of RPA and short ssDNA tails are sufficient to activate Mec1-Ddc2.
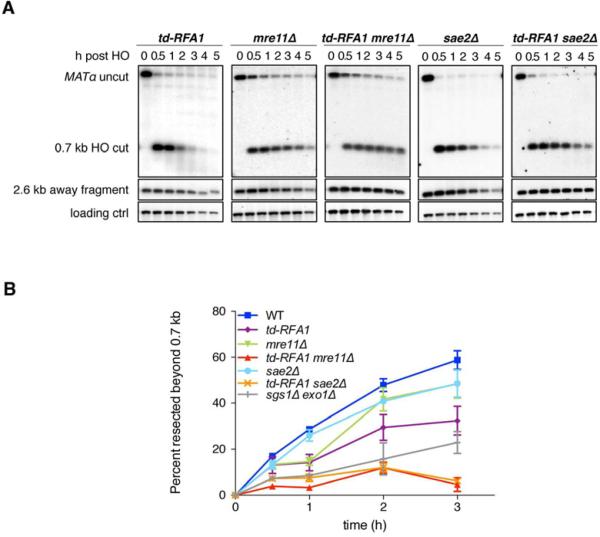
Mre11 and Sae2 are required for resection initiation in the absence of RPA
Smearing below the HO cut band signifies resection initiation in the absence of extensive resection (Figure 1). To investigate whether end processing initiates by MRX-Sae2 when RPA is depleted, resection was monitored by band disappearance and qPCR in td-RFA1 mre11Δ and td-RFA1 sae2Δ cells (Figure 2A, B). Compared to the single mutants, the td-RFA1 mre11Δ and td-RFA1 sae2Δ double mutants displayed a further reduction in end resection, to a lower level than observed for the sgs1Δ exo1Δ strain (Figure 2B). These data are consistent with MRX-Sae2 and RPA functioning at different steps of resection. At early time points (30 and 60 min) RPA was still detected by Western blot analysis (Figure S1); thus, Exo1 might be responsible for the residual resection in the absence of MRX and Sae2 (Mimitou and Symington, 2010; Shim et al., 2010).
Figure 2. MRX-Sae2 and RPA act sequentially in end resection.
A synergistic defect of td-RFA1 and mre11Δ or sae2Δ in resection was seen by both the band disappearance assay (A) and qPCR (B). Plot showing the ratio of resected DNA (0.7 kb fragment) amongst the HO cut DNA at each time point by qPCR analysis. The mean values from 3 independent trials are plotted and error bars show SD.
RPA protects the 3’ ssDNA tails from degradation
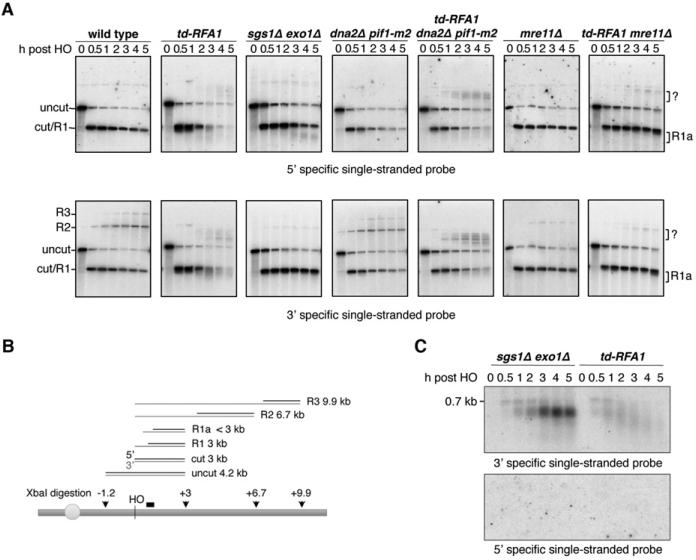
The smeared products observed in the td-RFA1 derivatives showed a broader distribution than the products seen in the sgs1Δ exo1Δ mutant suggesting resection initiation is altered. To determine whether the 5’-3’ polarity of resection is retained in the absence of RPA we performed denaturing alkaline gel electrophoresis of XbaI-digested genomic DNA, followed by Southern blot hybridization with RNA probes specific for the 5’ or 3’ strands 0.2-0.7 kb to the right of HO cut site (Figure 3A, B). End resection renders the DNA single-stranded and resistant to digestion by XbaI resulting in a ladder of higher molecular weight fragments, one end of which corresponds to the HO site and the other where the DNA is still double-stranded and cut by XbaI. The 3’ specific probe hybridizes to fragments of discrete mobility because the 3’ ends are largely intact; the 5’ specific probe hybridizes to the smeared partially processed ends that are stabilized in the absence of extensive resection (both probes also recognize the uncut and unresected cut bands). Surprisingly, smeared products below the HO cut band were detected with both probes in the td-RFA1 cells, revealing abnormal 3’ strand degradation.
Figure 3. RPA is required to shield the 3’ ssDNA tails from nucleolytic degradation.
A. Both 3’ and 5’ end degradation were observed (R1a) when Rfa1 was depleted. Alkaline electrophoresis of XbaI-digested genomic DNA from different time points after HO induction. ssDNA intermediates were detected by 3’ or 5’ strand specific RNA probes spanning the region from 0.2 kb to 0.7 kb distal of the HO cut site. B. Schematic showing alkaline gel analysis of genomic DNA digested by XbaI. Possible DNA intermediates formed after HO cutting and resection are shown. The black bar shows the location of the probe used. The thin gray lines represent fragments detected by 3’ strand specific probe. C. Only 3’ tails were detected in sgs1 exo1 and td-RFA1 cells. StyI digested genomic DNA was run on a neutral gel, transferred without denaturing and hybridized with the indicated strand specific probes. See also Figure S3
Because RPA enforces the 5’ strand specificity of resection in vitro through its interaction with Dna2 endonuclease it seemed likely that the 3’ strand degradation was due to unrestricted Dna2 activity (Bae and Seo, 2000; Cejka et al., 2010; Masuda-Sasa et al., 2008; Niu et al., 2010). The 3’ strand degradation was still evident in the td-RFA1 dna2Δ mutant, indicating that it is independent of Dna2 (Figures 3A and S3). However, the 3’ ends were stabilized in the absence of Mre11 (Figure 3A). As noted above, Mre11 is required for resection initiation in the td-RFA1 strains, suggesting that 5’ degradation precedes 3’ end removal. To test directly for the formation of 3’ or 5’ overhangs, StyI digested genomic DNA from sgs1Δ exo1Δ and td-RFA1 cells was separated by neutral gel electrophoresis, transferred without denaturation and then hybridized with 3’ or 5’ strand-specific probes (Sun et al., 1991). Although 3’ ends were detected in both strains, no signal was obtained from the 5’ specific probe (Figure 3C). Therefore, the polarity of end resection is retained in the absence of RPA, and RPA shields the 3’ ssDNA overhangs generated by MRX-Sae2 from nucleolytic attack.
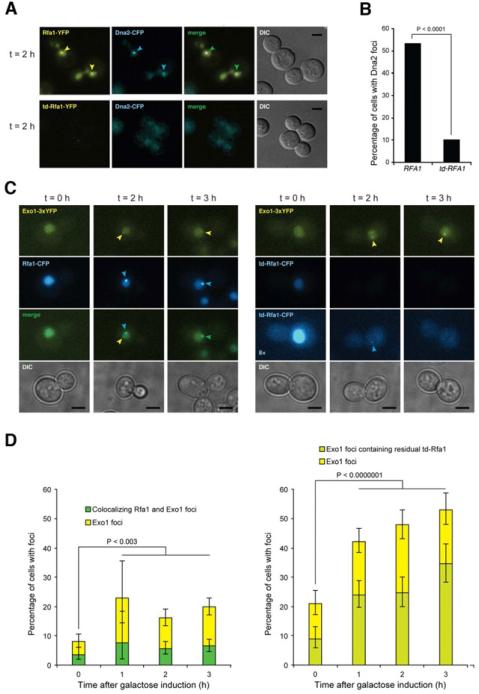
RPA is required for Dna2 and Rad51 localization to DSBs
Since previous studies have shown RPA and Dna2 physically interact (Bae et al., 2003), we considered the possibility that Dna2 is poorly recruited to DSBs in the td-RFA1 strain. To test this hypothesis, we used epifluorescence microscopy to monitor Dna2-CFP localization in td-RFA1-YFP cells and RFA1-YFP cells after HO induction. Because RPA and Dna2 also participate in DNA replication, cells were synchronized in G2/M with nocodazole for 4 h prior to HO induction to eliminate interference from replication foci. While ~50% of the cells displayed a Dna2-CFP focus co-localizing with a Rfa1-YFP focus 2 h after HO induction, only 10% of the cells had a detectable CFP focus when Rfa1 was depleted (Figure 4A, B). The residual Dna2 foci detected in the td-RFA1 strain showed very low levels of Rfa1 when the contrast of the images was increased by 8-fold. These data suggest that recruitment of Dna2 to DSB sites is largely dependent on RPA.
Figure 4. RPA is required for Dna2 but not for Exo1 localization to DSBs.
A. Epifluorescence microscopy showing DSB induced foci formation of YFP-tagged Rfa1 or td-Rfa1 and Dna2-CFP in G2/M arrested cells. Arrowheads indicate foci. Scale bar is equal to 3 m. B. Quantitation of Dna2-CFP foci. The percentage of cells with Dna2 foci was quantitated for the experiment shown in panel A, of which more than 90% co-localized with RPA. C. Exo1 focus formation is independent of Rfa1. Exo1-YFP and Rfa1-CFP or td-Rfa1-CFP were monitored before (t = 0) and 2 or 3 h after HO expression in G2-M-arrested cells. The left panel shows Rfa1-CFP cells and the right panel shows td-Rfa1-CFP cells. For the CFP channel, an 8-fold contrast enhanced version of the images from td-Rfa1-CFP cells are shown to illustrate that some cells contain weak residual Rfa1 foci that co-localize with Exo1 (t = 2), while other cells exhibit an Exo1 focus even in the complete absence of detectable td-Rfa1-CFP. D. Quantitation of Exo1 and Rfa1 foci. For the experiment in panel C, 200-600 cells were examined for Exo1 and Rfa1 foci at each time point. Error bars indicate 95% confidence intervals. Significance was determined by Fisher's exact test. See also Figure S4
A previous study showed Rad52 fails to localize to DSBs when Rfa1 is depleted. Since Rad52 is required for Rad51 recruitment to DSBs (Lisby et al., 2004), it seemed likely that Rad51 localization would also be dependent on RPA. On the other hand, if the role of Rad52 is to overcome the inhibition to Rad51 binding by RPA then in the absence of RPA a mediator might not be required. We found a significant decrease in the number of cells with DSB-induced YFP-Rad51 foci following Rfa1 depletion, indicating an essential role for RPA in the recruitment of Rad51 (Figure S4).
Although no physical interaction between RPA and Exo1 has been reported, to gain some insight into the role of RPA in Exo1-catalyzed end resection we monitored the localization of Exo1-YFP to DSBs in RFA1-CFP and td-RFA1-CFP strains. Prior to DSB formation about 8% of cells displayed spontaneous Exo1 foci of which ~40% co-localized with RPA (Figure 4C, D). Exo1 foci increased after HO induction, but with only 30% positive for RPA. Following Rfa1 depletion the number of cells with Exo1 foci increased before and after HO induction. Notably, many Exo1 foci showed no detectable Rfa1 staining, even when the contrast of the CFP channel was increased. Thus, RPA is not required for Exo1 recruitment to DSBs.
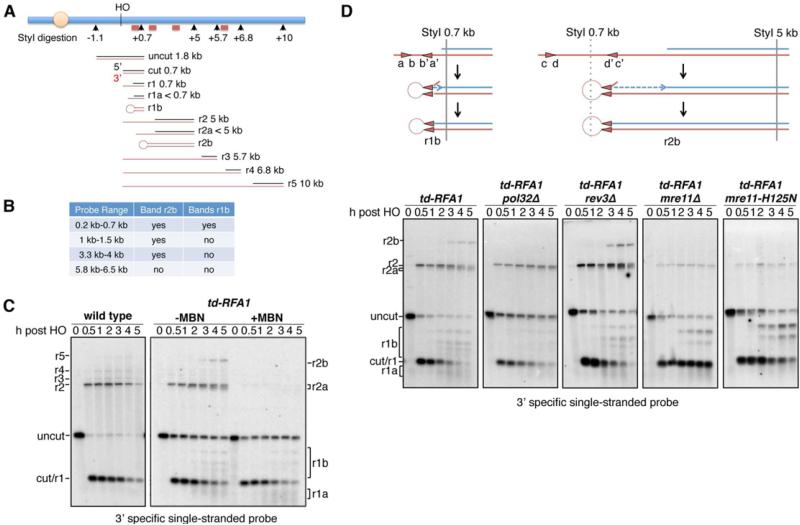
Formation of hairpin structures in the absence of RPA
In addition to smearing below the HO cut band, we detected some unexpected DNA species sized from about 5 to 6 kb in the td-RFA1 derivatives that hybridized to both 3’ and 5’ strand specific probes (Figure 3A). These cannot be due to 3’ tailed resection intermediates because the 5’ strand-specific probe would not be able to anneal to products with resection beyond the XbaI site located 3 kb from the HO cut site. To further characterize these intermediates, DNA was digested with StyI and hybridized with different probes that recognize sequences further from the DSB. Resection intermediates of 5 kb (r2), 5.7 kb (r3), 6.8 kb (r4) and 10 kb (r5) were detected in the wild-type strain by the 3’ strand specific probe close to the HO site (Figure 5). In the td-RFA1 strain, the r2 intermediate, indicative of a ssDNA tail of >0.7 kb and <5 kb, was detected, and a novel band migrating at around 9 kb (r2b). In addition, several DNA intermediates were detected between the 0.7 kb cut and 1.8 kb uncut fragments (r1b). Using three additional dsDNA probes at different positions distal to the HO cut site (Figure 5A, B), we were able to narrow down the range of the r2b band to be within 5.7 kb from the HO cut site, approximately half of its apparent size by alkaline gel electrophoresis. These findings suggest a stem-looped structure is formed during resection in the absence of RPA, which is denatured under alkaline electrophoresis conditions and appears as almost twice the size of the r2 fragment (Figure 5A). Similarly, the r1b species only hybridize with the probe located 0.2 to 0.7 kb from the HO cut site indicating they are due to fold-back structures formed within the 0.7 kb cut fragment. Consistent with this hypothesis, treatment of StyI digested DNA with mung bean nuclease (MBN), which cleaves ssDNA, was able to eliminate most of the aberrantly-sized species and cause further smearing below the HO cut fragment (Figure 5C).
Figure 5. RPA prevents the formation of fold-back structures.
A. Schematic showing alkaline gel analysis of genomic DNA digested by StyI. Possible DNA intermediates formed after HO cutting and resection are shown. The red bars show the locations of the probes used. The red lines represent fragments detected by the 3’ strand specific probe close to the HO cut site. B. Four different dsDNA probes were used to determine the range of fragments r1b and r2b. C. Alkaline electrophoresis of the StyI-digested genomic DNA from different time points after HO induction. ssDNA intermediates were detected by the 3’ strand specific RNA probe. r1b and r2b bands can be eliminated by mung bean nuclease treatment of StyI-digested DNA. D. Schematic showing the formation of hairpins. Alkaline electrophoresis of the StyI-digested genomic DNA from different mutants showing their contribution to hairpin formation. See also Figures S3 and S5
Previous studies have shown that hairpin capped DNA ends are stable in the absence of the MRX complex and Sae2 (Lobachev et al., 2002; Rattray et al., 2005). The td-RFA1 mre11Δ, td-RFA1 mre11-H125N (Mre11 nuclease defective) and td-RFA1 sae2Δ strains showed reduced accumulation of the r2 intermediate due to the resection defect, and the r2b fold-back was undetectable; however, the 1.3 kb r1b intermediate was present at higher levels than expected based on the severe resection defect of the strains, consistent with stabilization of fold-back structures in the absence of Mre11 nuclease and Sae2 (Figure 5D, Figure S3). MRX-Sae2 cleavage of ssDNA within the fold-back structure prior to or after ligation could contribute to loss of the 3’ ends seen in the absence of RPA. The 3’ end degradation and fold-back structures were unaffected by exo1Δ or dna2Δ (Figure S3).
Several short (5 to 9-bp) inverted repeats with the potential to form fold-back structures are present within the 3’ tail formed by resection from the HO cut site (Figure 5D, Figure S5). As the repeats are internal to the terminus, after annealing of the inverted repeats the 3’ end would need to be trimmed to create a substrate for DNA synthesis and ligation to form a hairpin that would be denatured in the alkaline gel. To test the requirement for DNA synthesis we used a strain lacking the non-essential subunit of the DNA polymerase δ and ζ complexes, Pol32 (Johansson et al., 2004; Johnson et al., 2012; Makarova et al., 2012). A previous study had shown that micro-homology mediated end joining is defective in the pol32Δ mutant suggesting Pol32-dependent DNA synthesis stabilizes annealed micro-homologies (Lee and Lee, 2007). The r2b intermediate was not detected in the td-RFA1 pol32Δ mutant and the bands corresponding to the r1b intermediates were greatly reduced (Figure 5D). Importantly, 3’ degradation was also reduced in the td-RFA1 pol32Δ mutant indicating 3’ strand loss occurs during formation of the fold-back structures. Fold-back structures were still apparent in td-RFA1 rev3Δ cells; therefore, the Pol32 requirement is through its association with DNA Pol δ.
DISCUSSION
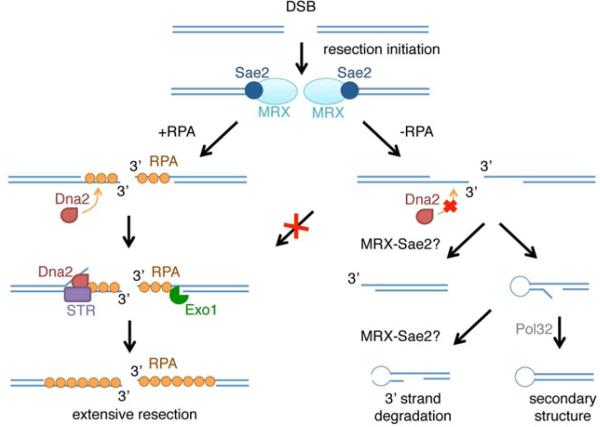
Here we demonstrate the importance of RPA binding to ssDNA to remove secondary structure, to protect ssDNA from nucleases, to recruit Dna2 and to promote extensive resection in vivo (Figure 6). The ssDNA formed by MRX-dependent resection initiation can adopt secondary structure by intramolecular pairing between short repeats present in the 3’ ssDNA overhang, a process that is normally prevented by RPA. We suggest that after annealing of inverted repeats the unpaired 3’ flap is trimmed to create a primer for DNA synthesis, the 3’ end can then be extended and ligated to the resected 5’ end generating a hairpin capped end (Figure 6). The fold-back structures were reduced in the td-RFA1 pol32Δ strain, consistent with the requirement for DNA synthesis (Figure 5D). That DNA synthesis occurs in the absence of RPA is surprising, and the low level of fold-back structures might be due to inefficient DNA synthesis following RPA depletion (Wold, 1997). The inverted repeats identified within the 3’ ssDNA overhang would all require 3’ flap trimming in order to initiate DNA synthesis (Figure S5). This role would normally be carried out by the proofreading function of Pol δ (for short flaps) or by the Rad1-Rad10 nuclease (Paques and Haber, 1997). However, we did not detect a defect in the accumulation of fold-back structures in the td-RFA1 rad1Δ mutant (Figure S3). Rad1-Rad10 is only required for flap removal during single-strand annealing when Rad51 is present (Manthey and Bailis, 2010), and since we found no Rad51 foci following RPA depletion the failure to recruit Rad51 might bypass the need for flap trimming by Rad1-Rad10 (Figure S4).
Figure 6. Model for RPA-mediated coordination of DNA end processing.
Mre11 and Sae2 initiate resection to form short 3’ tails. In the absence of RPA Dna2 is not recruited to the break site, extensive resection is eliminated, and the 3’ overhangs tend to form secondary structures by annealing between short repeats. Following annealing between inverted repeats, the 3’ unpaired flap is trimmed and after DNA synthesis is ligated to the 5’ end to form a hairpin. The 3’ strand loss could occur by Mre11-dependent degradation of 3’ ends or by hairpin cleavage before (shown) or after DNA synthesis and ligation.
The 5’-3’ polarity of end resection was retained in the absence of RPA, but the short 3’ ssDNA overhangs generated were degraded over time (Figure 3). The 3’ degradation appears to result from the formation of stabilized fold-back structures because it was reduced by the pol32 mutation. Mre11 and Sae2 are important to form fold-back structures through their roles in resection, and may also act to destroy fold-back structures by endonucleolytic cleavage of the hairpin ends. The largest of the r1b bands, which would require the least resection and is predicted to have a 9 bp loop, was preserved in the td-RFA1 mre11 and td-RFA1 sae2Δ mutants (Figures 5, S3 and S5). In previous studies showing stabilization of hairpin-capped ends in the absence of the MRX complex, the Mre11 nuclease or Sae2, the inverted repeats were located less than 12 nt apart (Lobachev et al., 2002; Rattray et al., 2005). It is possible that fold-back structures with longer loops, such as r2b, are susceptible to cleavage by nucleases other than Mre11. We favor the hypothesis that Mre11 promotes 3’ end loss by clipping fold-back structures rather than exonucleolytic degradation of 3’ ends because the td-RFA1 mre11-H125N and td-RFA1 sae2Δ mutants exhibit a similar phenotype. Opening of the hairpin-capped end by MRX-Sae2 would create a new substrate for initiation of end resection and might account for the MRX-Sae2 dependent resection of >700 nt observed in the td-RFA1 strain.
The formation of hairpin capped DNA ends via short inverted repeats is expected to be extremely rare because RPA is normally present to prevent their formation. Nevertheless, palindromic gene amplification at uncapped telomeres and HO-induced DSBs has been reported, and is proposed to occur by pairing between closely spaced short (4-11 bp) inverted repeats within the 3’ tail formed by end resection to form a hairpin structure after DNA synthesis and ligation of the 3’ and 5’ ends (Maringele and Lydall, 2004; Rattray et al., 2005). Palindromes result from replication through the hairpin-capped end in the next cell cycle. The palindromic events at an HO-induced DSB were only detected in the absence of the MRX complex, the Mre11 nuclease or Sae2, consistent with hairpin opening by MRX-Sae2 (Rattray et al., 2005). Fold-back structures were also detected by 2D gel electrophoresis during meiotic DSB metabolism in rad52 cells, where DSB ends become hyper-resected due to the failure of second-end capture (Lao et al., 2008). It is possible that RPA is depleted in meiosis when multiple DSBs are formed and become hyper-resected in mutants unable to complete repair, facilitating annealing between short repeats within the 3’ ssDNA overhangs. Depletion of RPA may also account for loss of 3’ ends at later time points after induction of an unrepairable DSB in wild-type mitotic cells (Zierhut and Diffley, 2008). Furthermore, a role for RPA in preventing annealing between micro-homologies might underlie the large increase in chromosome translocations and cancer predisposition observed for rfa1 hypomorphic alleles in yeast and mouse systems (Chen and Kolodner, 1999; Wang et al., 2005).
We were unable to assess the role of RPA in resection initiation because degradation of RPA coincides with HO expression and RPA is still detected by Western blot at the time HO cleaves the MAT locus (Figure S1). It seems unlikely that RPA would be required for resection initiation by MRX-Sae2 because in vitro studies indicate Mre11 and Sae2 degrade DNA in the absence of RPA, and MRX is recruited to DSBs prior to RPA (Lengsfeld et al., 2007; Lisby et al., 2004; Nicolette et al., 2010; Paull and Gellert, 1998). Resection in the td-RFA1 strain is largely Mre11 and Sae2 dependent, consistent with direct initiation by MRX-Sae2.
The formation of hairpin-capped ends could contribute to the block to extensive resection observed when RPA is depleted, but cannot be the only reason because the r2 intermediate is more abundant than the r2b structure (Figure 5B). Furthermore, hairpin formation is reduced in the td-RFA1 pol32Δ mutant yet extensive resection is still defective. Dna2 localization to DSBs was significantly reduced in the absence of RPA suggesting RPA bound to the short 3’ ssDNA tails recruits Dna2. A previous study showed reduced Dna2 localization to DSBs in the mre11 mutant suggesting the MRX complex contributes to Dna2 recruitment or generates a structure bound by RPA (Shim et al., 2010). RPA is essential for Sgs1-catalyzed unwinding (Cejka et al., 2010; Niu et al., 2010). The activity of RPA was only partially substituted by E. coli SSB indicating that protein-protein interactions are important and the effect is not solely due to stabilization of the unwound strands. Sgs1 also interacts with Mre11 and could be recruited to breaks by Mre11 and RPA (Chiolo et al., 2005). Our data confirm the importance of RPA for Sgs1-Dna2 mediated end resection in vivo. Exo1 has no reported interaction with RPA and its activity is not fully dependent on RPA in vitro (Nicolette et al., 2010; Nimonkar et al., 2011); however, we found the Exo1 extensive-resection pathway is also dysfunctional without RPA. The number of cells with Exo1 foci increased after RPA depletion indicating that the role of RPA to promote Exo1-catalyzed resection is not for recruitment and must be at a later step. A previous study showed that the MRX complex is required for localization of Exo1 to DSBs; therefore, the increased Exo1 foci seen in the absence of RPA could be due to persistence of MRX at DSBs, inferred from dependence on MRX-Sae2 for resection initiation and hairpin cleavage. The secondary DNA structure formed after RPA depletion may prevent Exo1 from accessing the correct substrate for end resection.
The formation of ssDNA by resection of DSB ends is essential for Rad51-mediated strand invasion (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008). In vitro studies have shown that RPA can act as a barrier to Rad51 binding, but is also needed to remove secondary structure from ssDNA to allow formation of contiguous Rad51 filaments that are productive for strand exchange (Sung, 1997; Sung and Robberson, 1995). Rad52 acts as a mediator to nucleate Rad51 on RPA-coated ssDNA, and RPA is displaced by the cooperative binding of Rad51 to ssDNA (Kurokawa et al., 2008; Song and Sung, 2000; Sugiyama and Kowalczykowski, 2002). Depletion of RPA results in attenuated recruitment of Rad52 (Lisby et al., 2004), which could be due to the direct interaction between these proteins or due to reduced accumulation of ssDNA in the absence of RPA. Since RPA binding to ssDNA can occlude Rad51, one might expect Rad51 to bind in the absence of RPA, but to form non-contiguous filaments. However, depletion of RPA results in greatly reduced Rad51 foci in yeast and mammalian cells (Figure S4) (Sleeth et al., 2007); thus RPA is required for localization of both Rad51 and Rad52 to damage sites.
The generation of long tracts of ssDNA is important for Ddc2-Mec1 recruitment and cell cycle arrest in response to DSBs (Zhu et al., 2008). A previous study showed RPA stimulates the binding of ATRIP to ssDNA in vitro and ATRIP/Ddc2 localization to damage sites is reduced when RPA is depleted from cells (Zou and Elledge, 2003). Although the resection defect of the td-RFA1 strain following RPA depletion is similar to the sgs1Δ exo1Δ double mutant and consequently one might have expected to reduce Rad53 activation, we found that Rad53 was activated in response to an HO-induced DSB (Figure S2). In budding yeast the Mec1 kinase is primarily responsible for Rad53 activation and cell cycle arrest in response to an unrepaired DSB (Harrison and Haber, 2006). We found that Rad53 activation in the td-RFA1 strain is Mec1 dependent suggesting the residual RPA is sufficient to activate Mec1.
In summary, our data highlight the central role of RPA in DSB repair. RPA is required to generate ssDNA, and to shield ssDNA from degradation and formation of secondary structure that is inhibitory to Rad51 binding. In addition, we suggest the role of RPA in preventing spontaneous annealing between micro-homologies is essential to avoid formation of hairpin-capped ends that can lead to palindromic gene amplification and gross chromosome rearrangements.
EXPERIMENTAL PROCEDURES
Media, growth conditions and yeast strains
Rich medium (yeast extract–peptone–dextrose, YPD), synthetic complete (SC) medium and genetic methods were as described previously (Amberg, 2005). YP medium containing 2% lactate (YPL) was used for the galactose induction of HO. For G2/M arrest, nocodazole was added to the media to 15 g/ml. The yeast strains used were derived from W303 and are listed in Table S1. The Ub-DHFRts-HA-Rfa1-AAAAAAAAG-YFP/CFP fusion (td-Rfa1-YFP/CFP), Dna2-AAAA-YFP and Exo1-4ala-YFP-AAVEL-YFP-RP-YFP was constructed using PCR-based allele replacement essentially as described (Lisby et al., 2004).
Physical monitoring of end resection
Physical analysis of HO-induced DSB end resection was as described previously (Mimitou and Symington, 2010). Cells were grown in YPL and then galactose was added to a final concentration of 2% to induce HO expression. For RPA depletion, cells were switched to 37°C 2 h before HO and UBR1 were induced. For the detection of the HO-cut fragment next to the break site, a probe was generated by PCR amplification of MAT sequences distal to the HO-cut site (coordinates 201176–201580 on chromosome III sequence). For the detection of the fragment 2.6 kb distal to the HO-cut site, the MAT-2.6 kb probe was used (coordinates 204184–204893 on chromosome III). A POX1 probe (coordinates 108631-109001 on chromosome XII) was used for normalization of band intensities using Image J (NIH). DSB end resection for each time point was estimated as a ratio of the signal intensity corresponding to that before induction and represents the mean of 3 independent experiments.
Quantitative analysis of HO-induced DSB end resection by real-time PCR
A real-time PCR assay (Zierhut and Diffley, 2008) using primers flanking the StyI site 0.7 kb away from the HO cut site and the XbaI site 3 kb away was used to quantify end resection. A control primer pair was used to amplify a region on chromosome XV not containing StyI sites. 150 ng of genomic DNA was used for each digestion or mock digestion in a volume of 15 l. DNA was diluted by adding 55 l of ice-cold dH2O to each reaction and 8.8 μl was used for each real-time PCR reaction of 20 μl. PCRs were performed using FastStart universal SYBR Green master mix (Roche Applied Science) with the Mastercycler® ep realplex and corresponding software (Eppendorf). The following program was used for all reactions: 95°C 10 min, 40 cycles of (95°C 15 s and 58°C 60 s), melting curve 10 min. Triplicate reactions were performed for all primer pairs and an average threshold cycle value was then used for each sample. The fraction of DNA resected to 0.7 kb or 3 kb among HO cut DNA was given by x = 2/[(1+2 ΔCt)*f], where ΔCt = Ct,digestion - Ct,mock, and f is the fraction cut by HO determined by southern blot analysis. All Ct values were corrected for DNA concentrations.
Alkaline gel electrophoresis
ssDNA intermediates were analyzed by alkaline gel electrophoresis as described (Mimitou and Symington, 2008). Three additional dsDNA probes (coordinates 201864-202362, 204184–204893, 206829-207310 respectively on chromosome III) were used to determine the range of fragment r1b. MBN was used at 20U for 1 h at 30°C t o treat StyI digested genomic DNA.
Fluorescence microscopy
Cells expressing YFP or CFP tagged proteins in RFA1 and td-RFA1 backgrounds were grown to OD600 = 0.1 in SC medium containing 2% raffinose at 25°C and arrested in G2/M phase by addition of nocodazole for 2 hours. The cultures were shifted to 37°C for 2 hours prior to addition of 2% galactose to induce HO and Ubr1 in the td-RFA1 strains to facilitate degradation of td-Rfa1. Cells from liquid cultures were processed for fluorescence microscopy as described previously (Lisby et al., 2004; Mine-Hattab and Rothstein, 2012). In each field of cells, 15 images were obtained at 0.3 μm intervals along the Z-axis to allow inspection of all focal planes of cells. Image acquisition time for DIC, YFP and CFP were 20 ms, 900 ms and 900 ms, respectively.
Western blot analysis
td-RFA1 strains were grown in YPL media at 23°C with CuSO4 added to a final concentration of 100 μM or at 37°C without adding CuSO4. Galactose was added to a final concentration of 2% to induce UBR1 expression. Samples from different time points were collected and whole cell extracts were prepared by TCA precipitation and analyzed by SDS-PAGE and western blot with anti-S.c.Rfa1 (Agrisera), anti-Rad53 (gift from M. Foiani) and anti-Adh1 for loading control.
Supplementary Material
HIGHLIGHTS.
RPA is required for DNA end resection by Sgs1-Dna2 and Exo1
Recruitment of Dna2 to double-strand breaks requires RPA
RPA stabilizes the ssDNA formed by end resection
Annealing between short homologies is prevented by RPA
ACKNOWLEDGEMENTS
We thank A. Lustig and R. Rothstein for yeast strains, M. Foiani for anti-Rad53 antibodies, I. Gallina for making the Dna2-CFP construct, and W.K. Holloman and members of the Symington lab for comments on the manuscript. We thank J. Miné-Hattab for assistance with microscopy. This study was supported by a grant from the National Institutes of Health (GM041784) to L.S.S. and from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement no 242905 to M.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes five figures and one table.
REFERENCES
- Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold spring Harbor Laboratory Press; 2005. [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae KH, Kim HS, Bae SH, Kang HY, Brill S, Seo YS. Bimodal interaction between replication-protein A and Dna2 is critical for Dna2 function both in vivo and in vitro. Nucleic Acids Res. 2003;31:3006–3015. doi: 10.1093/nar/gkg422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SH, Seo YS. Characterization of the enzymatic properties of the yeast dna2 Helicase/endonuclease suggests a new model for Okazaki fragment processing. J Biol. Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill SJ, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol. Cell. Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nature Genetics. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell. Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nature Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- Eggler AL, Inman RB, Cox MM. The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J Biol. Chem. 2002;277:39280–39288. doi: 10.1074/jbc.M204328200. [DOI] [PubMed] [Google Scholar]
- Firmenich AA, Elias-Arnanz M, Berg P. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol. Cell. Biol. 1995;15:1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc. Natl. Acad. Sci. USA. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Kurokawa Y, Murayama Y, Haruta-Takahashi N, Urabe I, Iwasaki H. Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biol. 2008;6:e88. doi: 10.1371/journal.pbio.0060088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Longhese MP, Plevani P, Lucchini G. Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol. Cell. Biol. 1994;14:7884–7890. doi: 10.1128/mcb.14.12.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey GM, Bailis AM. Rad51 inhibits translocation formation by non-conservative homologous recombination in Saccharomyces cerevisiae. PloS one. 2010;5:e11889. doi: 10.1371/journal.pone.0011889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 2004;18:2663–2675. doi: 10.1101/gad.316504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J Biol. Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nature Cell Biol. 2012;14:510–517. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3' to 5' exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray AJ, Shafer BK, Neelam B, Strathern JN. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nature Rev. Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Diaz A, Kanemaki M, Marchesi V, Labib K. Rapid depletion of budding yeast proteins by fusion to a heat-inducible degron. Sci STKE. 2004;2004:PL8. doi: 10.1126/stke.2232004pl8. [DOI] [PubMed] [Google Scholar]
- Seong C, Sehorn MG, Plate I, Shi I, Song B, Chi P, Mortensen U, Sung P, Krejci L. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J Biol. Chem. 2008;283:12166–12174. doi: 10.1074/jbc.M800763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29:3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Sleeth KM, Sorensen CS, Issaeva N, Dziegielewski J, Bartek J, Helleday T. RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells. J Mol Biol. 2007;373:38–47. doi: 10.1016/j.jmb.2007.07.068. [DOI] [PubMed] [Google Scholar]
- Smith J, Rothstein R. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics. 1999;151:447–458. doi: 10.1093/genetics/151.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Sung P. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J Biol. Chem. 2000;275:15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- Soustelle C, Vedel M, Kolodner R, Nicolas A. Replication protein A is required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 2002;161:535–547. doi: 10.1093/genetics/161.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol. Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Umezu K, Sugawara N, Chen C, Haber JE, Kolodner RD. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen S, Petukhova G, Sigurdsson S, Sung P. Functional cross-talk among Rad51, Rad54, and replication protein A in heteroduplex DNA joint formation. J Biol. Chem. 2002;277:43578–43587. doi: 10.1074/jbc.M205864200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R, Carrion DV, Chin L, Kucherlapati R, Kolodner RD, et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nature Genetics. 2005;37:750–755. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Yan H, Toczylowski T, McCane J, Chen C, Liao S. Replication protein A promotes 5'-->3' end processing during homology-dependent DNA double-strand break repair. J Cell Biol. 2011;192:251–261. doi: 10.1083/jcb.201005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.